Abstract
The involvement of 11q23-balanced translocations in acute leukemia after treatment with drugs that inhibit the function of DNA topoisomerase II (topo II) is being recognized with increasing frequency. We and others have shown that the gene at 11q23 that is involved in all of these treatment-related leukemias is MLL (also called ALL1, Htrx, and HRX). In general, the translocations in these leukemias are the same as those occurring in de novo leukemia [eg, t(9; 11), t(11; 19), and t(4; 11)], with the treatment-related leukemias accounting for no more than 5% to 10% of any particular translocation type. We have cloned the t(11; 16)(q23; p13.3) and have shown that it involves MLL and CBP (CREB binding protein). The CBP gene was recently identified as a partner gene in the t(8; 16) that occurs in acute myelomonocytic leukemia (AML-M4) de novo and rarely in treatment-related acute myeloid leukemia. We have studied eight t(11; 16) patients, all of whom had prior therapy with drugs targetting topo II with fluorescence in situ hybridization (FISH) using a probe for MLL and a cosmid contig covering the CBP gene. Both probes were split in all eight patients and the two derivative (der) chromosomes were each labeled with both probes. Use of an approximately 100-kb PAC located at the breakpoint of chromosome 16 from one patient revealed some variability in the breakpoint because it was on the der(16) in three patients, on the der(11) in another, and split in four others. We assume that the critical fusion gene is 5′MLL/3′CBP. Our series of patients is unusual because three of them presented with a myelodysplastic syndrome (MDS) most similar to chronic myelomonocytic leukemia (CMMoL) and one other had dyserythropoiesis; MDS is rarely seen in 11q23 translocations either de novo or with t-AML. Using FISH and these same probes to analyze the lineage of bone marrow cells from one patient with CMMoL, we showed that all the mature monocytes contained the fusion genes as did some of the granulocytes and erythroblasts; none of the lymphocytes contained the fusion gene. The function of MLL is not well understood, but many domains could target the MLL protein to particular chromatin complexes. CBP is an adapter protein that is involved in regulating transcription. It is also involved in histone acetylation, which is thought to contribute to an increased level of gene expression. The fusion gene could alter the CBP protein such that it is constitutively active; alternatively, it could modify the chromatin-association functions of MLL.
ONE OF THE UNFORTUNATE consequences of the successful treatment of cancer/leukemia is the subsequent development of a treatment-related myelodysplastic syndrome (t-MDS) or acute leukemia, usually myeloblastic or less often lymphoblastic (treatment-related acute myeloid leukemia [t-AML] or treatment-related acute lymphoblastic leukemia [t-ALL]).1,2 Treatment of patients with alkylating agents (with or without radiation) has previously been found to result in t-MDS or t-AML with an associated loss of chromosomes 5 and/or 7 (reviewed in Pedersen-Bjergaard and Rowley3 ). More recently, balanced translocations have been identified after the use of drugs that target DNA topoisomerase II (topo II), notably the epipodophyllotoxins (VP16 and VM26), particularly when used in high doses. These translocations usually involve the MLL gene (also called ALL1, Htrx, or HRX) at chromosome band 11q23 or less often the AML1 (CBFA2) gene at 21q22.4-8 The most common translocations are t(9; 11)(p22; q23) or t(11; 19)(q23; p13) in t-AML and t(4; 11) (q21; q23) in t-ALL; these are also the most common MLL translocations in AML and ALL de novo. We recently described two patients who had a t(11; 16)(q23; p13.3) associated with t-MDS that evolved to t-AML and then to T-cell ALL in one patient9 and t-AML(M4) in the second patient.10 We have used genomic cloning of the breakpoint and reverse transcriptase-polymerase chain reaction (RT-PCR) to identify the gene on chromosome 16 involved in this translocation.11 We have obtained cytogenetic material from six patients6,13 14 in addition to the two we previously described and we report here on our analysis of the breakpoint region on chromosome 16 and show that the same gene, CBP, is involved in all eight patients using fluorescence in situ hybridization (FISH).
MATERIALS AND METHODS
Patient samples were obtained with informed consent at the University of Chicago (patient no. 1),9 Tufts Medical Center (patient no. 2),1 the University of Texas Southwestern Medical Center (patients no. 36 and 4), the University of Kiel (patient no. 5),13 the Medical University of South Carolina Medical Center (patients no. 6 and 714), and the St Jude Children's Research Hospital (patient no. 8). The bone marrow specimens were processed for cytogenetic analysis using standard procedures. In some cases, material was also frozen for subsequent use for DNA and/or RNA.
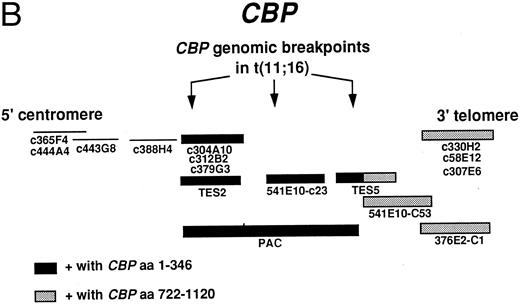
FISH was performed as previously reported.14 Initially, we used several YACs to try to map the breakpoint. YAC 615F4 from NAD labeled 16p13 very weakly, with additional stronger signals on 17q23 and 12 centromere. One other YAC from JB also was not useful; YAC 541E10 from the CEPH library gave a weak signal on chromosome 16, band p13, and signals on the satellites of chromosomes 13 and 22. A genomic probe cloned from the der(16) chromosome breakpoint junction from patient no. 1 mapped near the CBP gene11 (Fig 1). FISH analysis was pursued using a series of cosmid clones from JB that span the CBP locus (Tes2, 541E10-C23, Tes5, 541E10-C53, and 376E2-C1) and cosmids from NAD (C365F4, C444A4, C443G8, C388H4, C304A10, C312B2, C379G3, C330H2, C58E12, and C307E6). Rearrangements of the MLL gene were identified using the MLL probe (Oncor, Inc, Gaithersburg, MD).
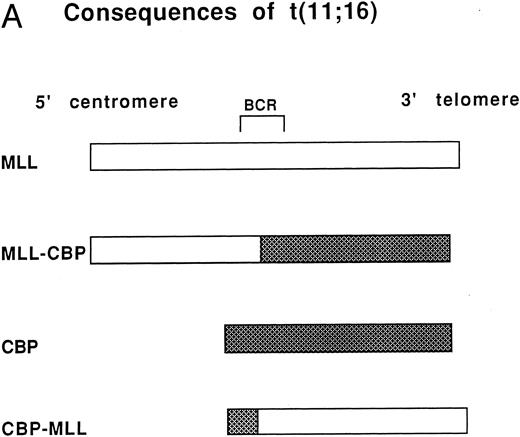
(A) Consequences of the t(11; 16). MLL on chromosome 11 and CBP on chromosome 16 are the genes involved in the t(11; 16) (q23; p13.3). The der(11) product contains 5′ MLL fused to 3′ CBP and the der(16) product contains 5′ CBP fused to 3′ MLL. BCR refers to the MLL breakpoint cluster region. (B) Cosmids and a PAC that span CBP were used for FISH analysis of patients with the t(11; 16). The arrows indicate the putative location of patient genomic breakpoints within CBP based on variability in FISH staining pattern using the CBP PAC.
(A) Consequences of the t(11; 16). MLL on chromosome 11 and CBP on chromosome 16 are the genes involved in the t(11; 16) (q23; p13.3). The der(11) product contains 5′ MLL fused to 3′ CBP and the der(16) product contains 5′ CBP fused to 3′ MLL. BCR refers to the MLL breakpoint cluster region. (B) Cosmids and a PAC that span CBP were used for FISH analysis of patients with the t(11; 16). The arrows indicate the putative location of patient genomic breakpoints within CBP based on variability in FISH staining pattern using the CBP PAC.
Studies correlating FISH results with morphology were undertaken using methods previously published.16 17 Briefly, freshly made bone marrow aspirate smears were stained with Wright's stain and coverslipped with Pro Tex (Lerner Labs, Pittsburgh, PA), and images of the Wright-stained cells were videotaped. After the coverslips were removed, the slides were soaked in Xylene for 5 minutes, air-dried, and processed for FISH. The results of the FISH analysis were correlated with morphology by comparing the stored images of the Wright-stained cells and the FISH results. The cells were scored as showing one of three patterns: (1) der(11)/der(16) (1 red, 1 green, and 2 fusion signals); (2) normal pattern (2 red signals and 2 green signals); and (3) indeterminate.
RESULTS
Clinical features.All eight of the patients had been treated previously for a variety of malignancies, including leukemia, lymphoma, and other solid tumors (summarized in Table 1). Every patient's treatment included a drug that targeted topo II, usually etoposide or teniposide, but also doxorubicin; the doses ranged from 2,200 mg/m2 to 8,100 mg/m2. In addition, six of the patients had received alkylating agents. All of the patients achieved a complete remission as a result of their initial therapy but developed a secondary hematologic disorder within 6 to 60 months. Every patient had a t(11; 16) often accompanied by other abnormalities (6 patients). The secondary hematologic disease was variable with four patients having t-AML and three having t-MDS. One other patient (no. 7) had dyserythropoiesis that resolved without treatment concommittant with normalization of his karyotype.14 MDS appeared to be most compatible with chronic myelomonocytic leukemia (CMMoL). Seven of eight patients received treatment for their secondary disease, including bone marrow transplantation in four children or young adults. Four patients are undergoing treatment or are in remission and three are dead.
Summary of t(11; 16)(q23; p13) in Patients With Treatment-Related Acute Leukemia
| Patient No. . | Age*/Sex . | Primary Malignancy . | Topo II Inhibitors . | Time to Leukemia . | Type of Leukemia . | Karyotype . | Length of Survival/Present Status . | PAC Results . | Reference . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10/F | AML-M2; t(8; 21) | Etoposide and Doxorubicin (600 mg/m2) + aa | 13 mo | t-MDS/t-AML (M4) to t-T-ALL | 46,XX,t(11; 16)(q23;p13)[18]46,idem,+12[14] | 26 mo†, post-BMT | Der(11) | Patient no. 19 |
| 2 | 64/F | Breast cancer | Doxorubicin + aa, G-CSF | 10 mo | t-AML (M4) | 46,XX,t(11; 16)(q23; p13.3)[23]/45,idem,−18[2] | Alive in remission as of 2/97 | Der(16) | 1 |
| 3 | 3/F | PreB-ALL; hyperdiploid | Etoposide (8,100 mg/m2) | 33 mo | t-AML (M2) | 46,XX,t(1; 12)(p36.1; q13),t(11; 16)(q23; p13.3)[13]/46,idem, del(7)(q31)[7] | 10 mo+; post-BMT | Split | Patient no. 76 |
| 4 | 11/M | Pre B-ALL | Etoposide (5,555 mg/m2) | 37 mo | t-AML (M4) | 46,XY,del(1)(p34.3p36.1), t(11; 16)(q23; p13.3)[12]/46,idem, t(2; 3)(p21; q21)[8] | 25 mo; post BMT† | Der(16) | |
| 5 | 74/M | Mantle cell lymphoma | Etoposide (2,200 mg) + aa | 19 mo | t-AML (M5a) | 47,XY,+8, t(11; 16)(q23; p13)[11]/48,idem,+8[3] | CR but early relapse <1 mo† | Der(16) | Patient no. 213 |
| 6 | 16/F | PNET | Etoposide (2,950 mg/m2) doxorubicin (375 mg/m2) + aa | 19 mo | t-MDS (CMML) | 46,XX,t(11; 16)(q23; p13)[17]/46,idem,i(17)(q10)[2]/46,XX[1] | BMT | Split | |
| 7 | 19/M | Ewing's sarcoma | Etoposide (5,000 mg/m2) doxorubicin (600 mg/m2) aa, x-ray, G-CSF | >6 mo | t-MDS | 46,XY,t(11; 16)(q23; p13)[13]/46,XY[7]‡ | Normal, healthy | Split | Patient no. 214 |
| 8 | 5/F | Neuroblastoma | Epipodophyllotoxin, aa | 28 mo | t-MDS | 46,XX,t(11; 16)(q23; p13)[19]/46,XX[1] | 13 mo | Split | |
| 9 | 14/F | T-ALL; 46,XX,t(11; 14) (p13; q11) | Teniposide | 5 yr | t-MDS t-AML (M4) | 46,XX,del(6)(p23),t(11; 16)(q23; p13)[4]/46,XX[6] | 2 yr+ | Patient no. 618 | |
| 10 | M | T-ALL; 46,XY | Teniposide and doxorubicin | 21 mo | t-B-ALL | 46,XY,t(5; 8)(q33; q12),t(11; 16)(q23; p13)[21]/46,XY[3] | Patient 312 | ||
| 11 | 7/M | B-ALL; 46,XY | Pirarubicin (300 mg/m2) | 29 mo | t-MDS (CMML) | 46,XY,t(11; 16)(q23; p13)[?] | 19 |
| Patient No. . | Age*/Sex . | Primary Malignancy . | Topo II Inhibitors . | Time to Leukemia . | Type of Leukemia . | Karyotype . | Length of Survival/Present Status . | PAC Results . | Reference . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10/F | AML-M2; t(8; 21) | Etoposide and Doxorubicin (600 mg/m2) + aa | 13 mo | t-MDS/t-AML (M4) to t-T-ALL | 46,XX,t(11; 16)(q23;p13)[18]46,idem,+12[14] | 26 mo†, post-BMT | Der(11) | Patient no. 19 |
| 2 | 64/F | Breast cancer | Doxorubicin + aa, G-CSF | 10 mo | t-AML (M4) | 46,XX,t(11; 16)(q23; p13.3)[23]/45,idem,−18[2] | Alive in remission as of 2/97 | Der(16) | 1 |
| 3 | 3/F | PreB-ALL; hyperdiploid | Etoposide (8,100 mg/m2) | 33 mo | t-AML (M2) | 46,XX,t(1; 12)(p36.1; q13),t(11; 16)(q23; p13.3)[13]/46,idem, del(7)(q31)[7] | 10 mo+; post-BMT | Split | Patient no. 76 |
| 4 | 11/M | Pre B-ALL | Etoposide (5,555 mg/m2) | 37 mo | t-AML (M4) | 46,XY,del(1)(p34.3p36.1), t(11; 16)(q23; p13.3)[12]/46,idem, t(2; 3)(p21; q21)[8] | 25 mo; post BMT† | Der(16) | |
| 5 | 74/M | Mantle cell lymphoma | Etoposide (2,200 mg) + aa | 19 mo | t-AML (M5a) | 47,XY,+8, t(11; 16)(q23; p13)[11]/48,idem,+8[3] | CR but early relapse <1 mo† | Der(16) | Patient no. 213 |
| 6 | 16/F | PNET | Etoposide (2,950 mg/m2) doxorubicin (375 mg/m2) + aa | 19 mo | t-MDS (CMML) | 46,XX,t(11; 16)(q23; p13)[17]/46,idem,i(17)(q10)[2]/46,XX[1] | BMT | Split | |
| 7 | 19/M | Ewing's sarcoma | Etoposide (5,000 mg/m2) doxorubicin (600 mg/m2) aa, x-ray, G-CSF | >6 mo | t-MDS | 46,XY,t(11; 16)(q23; p13)[13]/46,XY[7]‡ | Normal, healthy | Split | Patient no. 214 |
| 8 | 5/F | Neuroblastoma | Epipodophyllotoxin, aa | 28 mo | t-MDS | 46,XX,t(11; 16)(q23; p13)[19]/46,XX[1] | 13 mo | Split | |
| 9 | 14/F | T-ALL; 46,XX,t(11; 14) (p13; q11) | Teniposide | 5 yr | t-MDS t-AML (M4) | 46,XX,del(6)(p23),t(11; 16)(q23; p13)[4]/46,XX[6] | 2 yr+ | Patient no. 618 | |
| 10 | M | T-ALL; 46,XY | Teniposide and doxorubicin | 21 mo | t-B-ALL | 46,XY,t(5; 8)(q33; q12),t(11; 16)(q23; p13)[21]/46,XY[3] | Patient 312 | ||
| 11 | 7/M | B-ALL; 46,XY | Pirarubicin (300 mg/m2) | 29 mo | t-MDS (CMML) | 46,XY,t(11; 16)(q23; p13)[?] | 19 |
Abbreviations: AML-M2, acute myeloblastic leukemia; ALL, acute lymphoblastic leukemia; BMT, bone marrow transplant; CMML, chronic myelomonocytic leukemia; CR, complete remission; MDS, myelodysplastic syndrome; PNET, peripheral neuroepithelioma; aa, alkylating agents.
Age at diagnosis of first malignancy.
Dead.
Without treatment, percentage of abnormal metaphase cells decreased and disappeared 12 months after original pancytopenia; his peripheral counts also became normal.
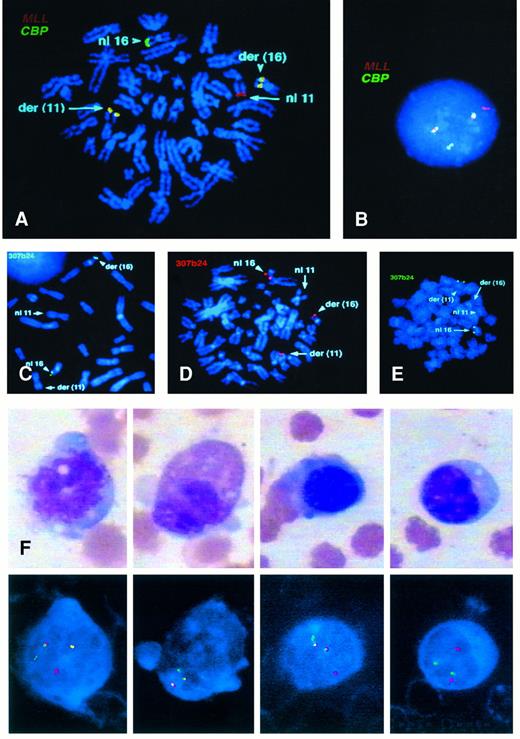
FISH.We performed FISH using the MLL probe on all eight patients. Patients no. 1, 2, and 5 had previously been shown to have an MLL rearrangement.1,9,13 Cells from all patients showed a split MLL probe that labeled both the der(11) and der(16) chromosomes. We used some or all of the cosmid probes for the CBP gene and showed that these probes labeled the normal chromosome 16 as well as both the der(16) and der(11) chromosomes, indicating that the CBP gene was split by the translocation (Figs 1 and 2, upper row, A and B). Our FISH analysis with the PAC genomic probe, which is about 100 kb and which contains CBP coding sequences11 (Fig 1B) detected some variability in the breakpoint in CBP because the genomic probe remained on the der(16) in three patients, was translocated to the der(11) in one patient, and was split in four others (Fig 2, middle row, C, D, and E). In the latter patients, more of the PAC remained on the der(16) than was translocated to the der(11). The fusion gene on the der(11) chromosome, which we believe contains the critical junction consists of 5′ MLL and 3′CBP, and the der(16) would contain 5′CBP and 3′MLL. Using RT-PCR, we cloned the MLL-CBP junction from patients no. 1 and 6; the CBP breakpoint in patient no. 1 is at the same amino acid as in the MOZ-CBP fusion, whereas in patient no. 6 it is more telomeric, occurring just before the bromodomain of CBP.11
(A) Upper Left. Metaphase cell from patient no. 8 labeled with MLL (chromosome 11 in red) and CBP (chromosome 16 in green). The two derivative chromosomes show a fusion signal. (B) an interphase cell from patient no. 6 labeled as in (A). (C) Middle left. Metaphase cell from patient no. 2 showing the PAC probe (307624) labeling the normal 16 and the der(16). (D) Center. Metaphase cell from patient no. 8 showing the PAC probe splitting and labeling both the der(11) and der(16) as well as the normal 16. (E) Right. Metaphase cell from patient no. 1 showing the PAC on the normal 16 and on the der(11). Lineage analysis in patient no. 6. Lower panel. Monocyte, granulocyte, erythroid precursor, and mature lymphocyte (F, G, H, and I, respectively) and the corresponding FISH results below from patient no. 6 with CMMoL-like tMDS. The monocyte, granulocyte, and erythroid precursor show the pattern of one red, one green, and two fusion signals, indicating the presence of MLL/CBP translocation on the der(11) and der(16) chromosomes, whereas the lymphocyte shows the pattern of a normal (nonclonal) cell with two red and two green signals.
(A) Upper Left. Metaphase cell from patient no. 8 labeled with MLL (chromosome 11 in red) and CBP (chromosome 16 in green). The two derivative chromosomes show a fusion signal. (B) an interphase cell from patient no. 6 labeled as in (A). (C) Middle left. Metaphase cell from patient no. 2 showing the PAC probe (307624) labeling the normal 16 and the der(16). (D) Center. Metaphase cell from patient no. 8 showing the PAC probe splitting and labeling both the der(11) and der(16) as well as the normal 16. (E) Right. Metaphase cell from patient no. 1 showing the PAC on the normal 16 and on the der(11). Lineage analysis in patient no. 6. Lower panel. Monocyte, granulocyte, erythroid precursor, and mature lymphocyte (F, G, H, and I, respectively) and the corresponding FISH results below from patient no. 6 with CMMoL-like tMDS. The monocyte, granulocyte, and erythroid precursor show the pattern of one red, one green, and two fusion signals, indicating the presence of MLL/CBP translocation on the der(11) and der(16) chromosomes, whereas the lymphocyte shows the pattern of a normal (nonclonal) cell with two red and two green signals.
The lineage analysis in patient no. 6 using FISH showed that the t(11; 16) (MLL/CBP) was present in the expanded monocytic series and that it was also present in a subpopulation of the mature granulocytic and erythroid elements. Virtually all of the monocytes showed the der(11) der(16) pattern, whereas only a subset of granulocytes (20%) and erythroid precursors (10%) were involved. None of the small mature-appearing lymphocytes evaluated showed the fusion (Table 2 and Fig 2F, G, H, and I, upper and lower panels).
Lineage Analysis in Patient No. 6: FISH Analysis: Number of Cells (%) With Each FISH Pattern
| Cell Type . | RGFF . | RRGG . | Indeterminate . |
|---|---|---|---|
| Monocytes | 67 (100) | 0 (0) | 0 (0) |
| Granulocytes | 6 (20.7) | 15 (51.7) | 8 (27.5) |
| Erythroid precursors | 4 (10.5) | 25 (65.8) | 9 (23.7) |
| Lymphocytes | 0 (0) | 77 (96.3) | 3 (3.7) |
| Cell Type . | RGFF . | RRGG . | Indeterminate . |
|---|---|---|---|
| Monocytes | 67 (100) | 0 (0) | 0 (0) |
| Granulocytes | 6 (20.7) | 15 (51.7) | 8 (27.5) |
| Erythroid precursors | 4 (10.5) | 25 (65.8) | 9 (23.7) |
| Lymphocytes | 0 (0) | 77 (96.3) | 3 (3.7) |
Abbreviations: RGFF, red, green, fusion, fusion; RRGG, red, red, green, green.
DISCUSSION
In addition to the eight patients whom we have studied, we are aware of three other patients with a t(11; 16), patients no. 9 through 11 in Table 1.18 19 The t(11; 16) appears to be unusual, because all of the cases of leukemia or myelodysplasia of which we are aware are treatment related. Thus, material from these t(11; 16) patients provides a special opportunity to study the unique features of the CBP gene that render it unusually sensitive to exposure to drugs that inhibit topo II. For the other translocations, such as the t(9; 11) and t(4; 11), the vast majority (>95%) occur in acute leukemia de novo, with only a very small percentage seen in treatment-related leukemias. As with our patients, these three patients had a primary malignancy (T-ALL [2] or B-ALL [1]) for which they received a topo II inhibitor. Their clinical picture is similar to that of our eight patients. One unusual feature of the group as a whole is the high incidence of a myelodysplastic syndrome at presentation of the treatment-related disorder. In two patients, it was classified as CMMoL. Most patients with secondary hematologic disease and MLL translocations are diagnosed with t-AML or less commonly t-ALL with no preceding MDS. Thus, the MLL-CBP fusion is unique for its association with t-MDS.
The lineage study showing that the MLL/CBP translocation was present in mature/maturing hematopoietic elements in patient no. 6 is in keeping with the patient's diagnosis of myelodysplasia (CMMoL). In myelodysplasia, clonal cells are capable of differentiation. It is interesting that the MLL /CBP translocation can be found in mature cells (suggesting that the gene product does not completely interfere with cell differentiation), but that it can also be seen in purely blastic proliferations such as B-ALL or T-ALL. In this regard, patient no. 1 is of particular importance as an MDS process transformed to an AML (M4) and then to T-ALL. The mechanisms through which MLL/CBP results in clones with differentiating capacity, with blocked differentiation (blastic proliferations), and with the potential to switch lineage from myeloid to lymphoid will be of interest for further studies. The absence of MLL/CBP in the mature lymphocytes in patient no. 6 was surprising, because we suspect that the transformed progenitor is a pluripotent stem cell capable of myeloid and lymphoid differentiation. It may be that the MLL/CBP gene product conferred a selective disadvantage to mature lymphoid cells in this case.
The presence of the MLL/CBP fusion in maturing cells is in keeping with the data showing the detection of expression of the MLL/AF9 fusion gene in mature cells of multiple origins in transgenic mice.20 These observations suggest that the presence of other factors (lineage-specific cofactor) may be involved in the proliferative expansion seen in the monocytic series in our patient studies. Study of differentiation capacity (as shown here) could have important implications regarding the mechanism through which translocations of 11q23 result in acute/blastic proliferations.
The CBP gene has recently been shown to be involved in the t(8; 16)(p11; p13.3) in which the MOZ (monocytic leukemia zinc finger) gene is fused to CBP at almost exactly the same nucleotide in the cDNA as the MLL-CBP junction in patient no. 1.11,21 The cDNA junction in the MOZ-CBP fusion involves the addition of two nontemplated nucleotides (two Gs) to maintain the fusion cDNA in frame. MOZ is a large multidomain protein with C4HC3 zinc fingers similar to those of MLL; it is a putative histone acetyltransferase. In the MLL-CBP fusion, the break in MLL occurs after a codon triplet and therefore the fusion with CBP is in frame with no nucleotides added to maintain the open reading frame.11 While we were cloning the translocation junctions and had evidence that CBP was the involved gene on chromosome 16, we became aware of the successful cloning of this junction by Satake et al19 from patient no. 11 (Table 1). The breakpoint in CBP is identical to our patient no. 1. In addition, CBP appears to be the gene responsible for Rubinstein-Taybi syndrome.22 In patients with this disease, the CBP gene shows evidence of genomic instability, in that affected patients have translocations, inversions, and deletions.
As a result of the t(11; 16), the carboxy portion of CBP is fused to the amino portion of MLL. The chimeric protein produced would contain the AT-hooks and repression domain of MLL fused to most of CBP, including the CREB binding domain, bromodomain, histone acetyltransferase domain, and E1A-binding domain in patients no. 1 and 11; the fusion protein in patient no. 6 contains less of CBP because the breakpoint is just before the bromodomain. CBP can serve as a multifunctional transcriptional coactivator that coordinates signals from many sequence-specific activators to modulate transcription. Furthermore, because it possesses histone acetyltransferase activity, it may contribute to transcriptional regulation via targeted acetylation of chromatin. When these functions of CBP are brought to the amino terminal portion of MLL that presumably retains its ability to bind AT-rich DNA, the coadaptor and chromatin modification functions of CBP could be redirected to inappropriate genomic regions.
All 26 patients with t(8; 16) reviewed by Hanslip et al23 had AML, usually M4 or M5 with erythrophagocytosis; the incidence is estimated as 4 per 1,000 patients with AML. Three of these cases with t-AML are included in a review by Quesnel et al,24 which reports on a total of six cases with t-AML and t(8; 16). Four of the six had received doxorubicin often in combination with alkylating agents. The interval for onset of treatment to leukemia was 7 to 23 months (7, 8, 9, 23, and 23 months). The subtype of leukemia was mainly M4, with one M5b; only one patient had a preceding MDS phase. Thus, for the t(8; 16) as for most of the translocations with MLL, those with t-AML are a small fraction of the total number of patients.
In our comparison of the location of the genomic breakpoint in MLL in various translocations, we noted that 75% of patients with acute leukemia de novo had breaks in the centromeric half of the 8.3-kb breakpoint cluster region, whereas 75% of the treatment-related acute leukemia patients had breaks in the telomeric half of the MLL breakpoint cluster region.25 This 3′ telomeric portion contains a scaffold attachment region (SAR) and six of seven topo II consensus cleavage sites. We postulated that this chromatin structure made this region more vulnerable to translocations in the presence of drugs that target topo II. In contrast, we know nothing about the location of SARs or putative topo II cleavage sites in CBP, because the genomic region of the CBP breakpoints has not been sequenced. However, as noted by others,21 22CBP appears to be genomically very unstable.
A critical practical concern is whether patients develop treatment-related leukemia because of chance (bad luck) or because of an unusual sensitivity to the topo II inhibitors. One way to resolve the question is to save material on all such patients so that the translocation breakpoints can be cloned and sequenced. This information would provide data regarding the presence of unusual polymorphisms that could lead to genomic instability in a manner analogous to the triplet repeats in the fragile X syndrome. It has been noted by Winick et al6 that 7 of 15 of their patients with t-AML were of Mexican origin. Whether this is a coincidence or evidence for a polymorphism that would cause no phenotype in the absence of unusual stress such as exposure to epipodophyllotoxins will only be clarified by further studies.
CD34+ cells purified from normal bone marrow using an antibody column. Cytocentrifuge preparation. (Courtesy of Alberto A. Marmont, MD, and Giovanna Piaggio, MD, Ospedale San Martino, Centro Trapianti di Midollo Osseo, Divisione Ematologia 2, 16132 Genova, Italy.)
CD34+ cells purified from normal bone marrow using an antibody column. Cytocentrifuge preparation. (Courtesy of Alberto A. Marmont, MD, and Giovanna Piaggio, MD, Ospedale San Martino, Centro Trapianti di Midollo Osseo, Divisione Ematologia 2, 16132 Genova, Italy.)
ACKNOWLEDGMENT
The authors acknowledge the critical assistance of many colleagues for the information on the karyotype and the clinical status of the patients, including Drs Michelle Le Beau, Diane Roulston (University of Chicago, Chicago, IL), Carol Rosenberg, Herman Wyandt (Boston University, Boston, MA), Naomi Winick (UT Southwestern, Dallas, TX), and Andrew Carroll (University of Alabama, Birmingham, AL).
Supported by grants from the National Institutes of Health (CA42557 to J.D.R. and CA72734 to J.C.B.), the Department of Energy (DE-FG0286ER60408 to J.D.R.), and the Wilhelm Sander Stiftung (to B.S.). J.B. is supported by the Leukemia Society of America.
Address reprint requests to Janet D. Rowley, MD, DSc, Blum-Riese Distinguished Service Professor of Medicine and of Molecular Genetics and Cell Biology, University of Chicago, Medical Center, Department of Medicine, Section of Hematology/Oncology, 5841 S Maryland Ave, MC2115, Chicago, IL 60637-1470.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal