Abstract
We have investigated the utility of the green fluorescent protein (GFP) to serve as a marker to assess retroviral gene transfer into hematopoietic cells and as a tool to identify and enrich for cells expressing high levels of the vector-encoded transcript. GFP, by virtue of a naturally occurring chromophore encoded in its primary sequence, displays autonomous fluorescence, thus eliminating the need for antibody or cytochemical staining to detect its expression. A bicistronic murine stem cell virus (MSCV)-based retroviral vector was constructed containing the GFP cDNA and a mutant, human dihydrofolate reductase gene. High-titer, ecotropic retroviral producer cells free of replication competent virus were generated and used to transduce murine bone marrow cells by cocultivation. Within 24 hours after completion of the transduction procedure, a high proportion (40% to 70%) of the marrow cells were intensely fluorescent compared to mock-transduced cells or cells transduced with a control retrovirus. Erythroid and myeloid hematopoietic colonies derived from GFP-transduced marrow were easily scored for retroviral gene transfer by direct in situ fluorescence microscopy. Clonogenic progenitors expressing increased levels of antifolate drug resistance could be enriched from the GFP-transduced marrow population by fluorescence activated cell sorting of cells expressing high levels of GFP. In vivo, splenic hematopoietic colonies and peripheral blood cells from animals transplanted with GFP-transduced marrow displayed intense fluorescence. These results show that GFP is an excellent marker for scoring and tracking gene-modified hematopoietic cells and for allowing rapid selection and enrichment of transduced cells expressing high levels of the transgene.
THE POTENTIAL use of recombinant retroviral vectors as gene transfer vehicles for delivery of therapeutic genes to primitive hematopoietic cells continues to hold promise for the treatment of many inherited, single-gene disorders such as sickle cell anemia, the thalassemias, and the immunodeficiency syndromes.1-5 In addition, various genetic therapy strategies that rely on gene transfer into stem cells are under development for the treatment of acquired immunodeficiency syndrome and cancer.6,7 Although high levels of retroviral gene transfer into murine primitive hematopoietic progenitors are now routinely achieved,8,9 efficient gene transfer into long-term repopulating hematopoietic cells of larger animals, including humans, has been problematic.10-12 One strategy that may help overcome these problems is the inclusion of a selectable marker gene in retroviral vectors to allow identification, enumeration, and enrichment of transduced cells.
Genes encoding resistance to toxic drugs such as neomycin,13 puromycin,14 and hygromycin15 have been widely used in retroviral vectors to aid in the identification of transduced cells in vitro. However, this approach is limited by the potential for nonspecific drug toxicity and the inability to easily measure transgene expression levels. In addition, the use of drug-resistance genes to facilitate enrichment of transduced hematopoietic cells before transplantation has not been generally used for a variety of reasons, including the necessity for prolonged culture periods to obtain a differential selection of transduced versus nontransduced cells. Alternatively, cytochemical markers such as β-galactosidase16 and alkaline phosphatase17 have been investigated as marker genes but are also somewhat problematic because of their endogenous expression in some cell types,18 the requirement to transport flourogenic substrates across the cell membrane for detection,19 and the potential for passive transfer to nontransduced cells.
Recently, several groups have begun to use a variety of cell-surface markers, including the multidrug resistance protein,20 the low-affinity nerve growth factor receptor,21 CD24 antigen,22 and the murine heat-stable antigen,23 to identify transduced cells by immunofluorescent antibody staining techniques. With this approach, transduced cells and their subpopulations can be enumerated, characterized, and enriched without drug toxicity using multiparameter flow cytometry. Using CD24 as a marker of gene transfer, Pawliuk et al22 were able to identify and enrich for transduced murine hematopoietic cells. Both spleen colony-forming cells and peripheral blood (PB) cells in mice transplanted with virally transduced, CD24-enriched hematopoietic precursors had a higher level of CD24 positivity compared to animals transplanted with unsorted cells.
One of the most promising, new markers currently being used to investigate gene transfer is the green fluorescent protein (GFP). Cloned from the jellyfish Aequorea victoria,24 the GFP gene encodes a 238-amino acid polypeptide containing a naturally occurring chromophore consisting of an imidazolone ring formed by the autocyclization of amino acid residues serine-65, tyrosine-66, and glycine-67.25 Various codon usage changes and point mutations in the wild-type GFP gene have led to several versions of GFP with improved fluorescence intensity in mammalian cells.26,27 Because fluorescence from GFP requires no substrates, cofactors, or additional gene products, cells expressing GFP can be easily scored in real-time by flow cytometry or fluorescence microscopy without cell manipulation.28 GFP has been readily detected in the living tissues of mice with a GFP-encoding transgene.29 Recently, both adenoviral26 and retroviral vectors30 31 containing a GFP cassette have been used to evaluate gene transfer into human endothelial cells, human tumor cell lines, and murine fibroblasts. This led us to evaluate the ability of GFP to act as a marker for retroviral gene transfer into hematopoietic progenitor cells.
In this study, we report the ability to achieve high-level gene transfer and expression of GFP in murine bone marrow (BM) as assessed by flow cytometry, in clonal hematopoietic colonies as evaluated by fluorescence microscopy, in hematopoietic progenitors as reflected by enrichment of transduced cells after cell sorting, and in progenitor-derived spleen colonies and PB cells of all lineages in vivo. GFP appears to be an excellent marker for evaluating gene transfer and for isolating, enriching, and tracking genetically marked cells.
MATERIALS AND METHODS
Cell lines and vector construction.The ecotropic packaging cell line GP + E8632 was used for the generation of helper-free recombinant retroviruses. JZenCD24tkNEO viral producer cells22 (CD24 producer cells) were a generous gift of R.K. Humphries (Terry Fox Laboratory, Vancouver, British Columbia, Canada). GP + E86, CD24 producer cells, 293T,33 and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mmol/L glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in a 5% CO2, humidified atmosphere.
The neomycin coding cassette in MSCVNEO2.134 (kindly provided by R. Hawley, Sunnybrook Research Institute, Toronto, Ontario, Canada) was replaced with a DNA fragment containing an internal ribosome entry site (ir) from the encephalomyocarditis virus35 linked to a gene encoding a human dihydrofolate reductase variant containing a leucine to tyrosine substitution in codon 22 (termed L22Y). When expressed in murine myeloid progenitors, the L22Y gene confers to cells a 100-fold increase in trimetrexate (TMTX) resistance.36 The GFP gene from the EGFP-1 plasmid (Clontech Laboratories, Palo Alto, CA) was cloned into the resulting plasmid MirL22Y to generate the bicistronic vector MGirL22Y (Fig 1B) using standard methodology.
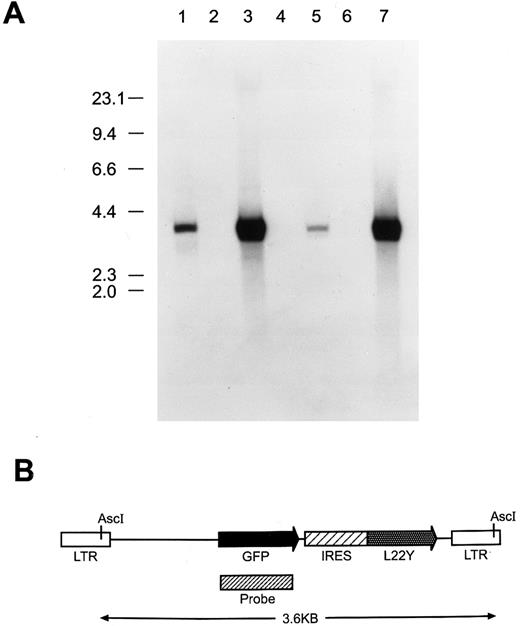
(A) Southern blot analysis to assess proviral integrity and copy number in cells genetically modified with the MGirL22Y retroviral vector. Genomic DNA from cells was restricted with Asc I which cuts once within each viral LTR, liberating the full-length, 3.6-kb retroviral genome. DNAs analyzed (15 μg) in each lane are: 1, GP + E86 DNA containing 50 pg of the MGirL22Y vector plasmid DNA; 2, GP + E86; 3, GP + E86-derived MGirL22Y viral producer; 4, NIH 3T3; 5, GFP vector-transduced NIH 3T3; 6, CD24 vector-transduced BM; 7, GFP vector-transduced BM. The migration patterns of the molecular-weight markers are shown at left. (B) Schematic of the MGirL22Y retroviral vector which consists of the MSCV retroviral backbone containing the coding sequence of GFP followed by an internal ribosomal entry site (IRES or ir) from the encephalomyocarditis virus linked to a mutant dihydrofolate reductase gene (L22Y). The designated probe region depicts the fragment used in the Southern analysis.
(A) Southern blot analysis to assess proviral integrity and copy number in cells genetically modified with the MGirL22Y retroviral vector. Genomic DNA from cells was restricted with Asc I which cuts once within each viral LTR, liberating the full-length, 3.6-kb retroviral genome. DNAs analyzed (15 μg) in each lane are: 1, GP + E86 DNA containing 50 pg of the MGirL22Y vector plasmid DNA; 2, GP + E86; 3, GP + E86-derived MGirL22Y viral producer; 4, NIH 3T3; 5, GFP vector-transduced NIH 3T3; 6, CD24 vector-transduced BM; 7, GFP vector-transduced BM. The migration patterns of the molecular-weight markers are shown at left. (B) Schematic of the MGirL22Y retroviral vector which consists of the MSCV retroviral backbone containing the coding sequence of GFP followed by an internal ribosomal entry site (IRES or ir) from the encephalomyocarditis virus linked to a mutant dihydrofolate reductase gene (L22Y). The designated probe region depicts the fragment used in the Southern analysis.
Generation of high-titer, ecotropic MGirL22Y virus producer cells.Conditioned media containing high-titer, amphotropic MGirL22Y vector particles was derived by cotransfection of 293T cells with the plasmid containing the MGirL22Y sequences and a helper plasmid containing the required gag, pol, and env retroviral genes driven by a Moloney leukemia virus LTR (D. Persons and E. Vanin, manuscript in preparation). This media was used to transduce GP + E86 cells a total of six times over 3 days. The cells were then allowed to recover for 24 hours and subsequently the media was changed to contain 10% thymidine phosphorylase-treated, heat-inactivated FCS and 100 nmol/L trimetrexate. After 7 days in culture, all cells on control plates were killed and the polyclonal trimetrexate-resistant population of vector-transduced cells was analyzed for GFP expression by flow cytometry. The highest expressing 10% of cells were sterilely sorted and subsequently expanded. The resulting viral titer of conditioned media from this producer population was 5 × 106 trimetrexate-resistant colony-forming units per milliter as assayed using NIH 3T3 cells. These virus-producing cells were free of replication competent retrovirus as demonstrated by the failure of trimetrexate resistance to be transferred from virally transduced NIH 3T3 cells to naive NIH 3T3 cells. Specifically, 10 mL of conditioned media from the GFP producer cells was used to transduce 105 NIH 3T3 cells which were then passaged in culture for 3 weeks without selection. Ten milliliters of conditioned media from these cells was then used to transduce 105 naive NIH 3T3 cells, and 48 hours later selection was applied with 100 nmol/L trimetrexate in DMEM supplemented with 10% thymidine phosphorylase-treated, heat-inactivated FCS. No trimetrexate resistant colonies were observed.
DNA analysis.Genomic DNA from the MGirL22Y producer cells, transduced NIH 3T3 cells, and transduced murine BM was purified using the Pure Gene DNA Isolation Kit (Gentra Systems, Minneapolis, MN) according to the manufacturer's specifications. Southern blot analysis of restricted DNA was performed by standard methods.37 Densitometric quantitation was performed using a PhosphoImager 425F (Molecular Dynamics, Sunnyvale, CA) and ImageQuant software.
Retroviral transduction of BM cells.Retroviral transduction of murine BM cells was performed as previously described.38 Briefly, BM was harvested from 8- to 12-week-old female C57/B16 mice 2 days after treatment with 150 mg/kg 5-fluoruracil (5-FU; Pharmacia, Kalamazoo, MI). Marrow cells were prestimulated with 20 ng/mL murine interleukin-3 (IL-3), 50 ng/mL human IL-6, and 50 ng/mL rat stem cell factor (all generously provided by Amgen, Thousand Oaks, CA) in DMEM supplemented with 15% heat-inactivated FCS (Hyclone, Logan, UT). BM cells were subsequently cocultured with irradiated (1,200 cGy) viral producer cells using the above culture media supplemented with 6 μg/mL polybrene. Forty-eight hours later, nonadherent BM cells were gently rinsed off the producer cell monolayers, pelleted, and resuspended in fresh culture medium with the above cytokines. Cells were cultured for an additional 24 to 48 hours before analysis for expression of GFP or CD24 as described below, plated into methylcellulose media, or used for transplantation.
In vitro clonogenic progenitor assays.Unsorted and sorted BM cells, in a volume not exceeding 150 μL, were suspended in 3 mL methylcellulose culture media (M3434; Stem Cell Technologies, Vancouver, British Columbia, Canada), which had been pretreated with 3 U thymidine phosphorylase at 37°C for 1 to 2 hours. Where specified, a stock solution of trimetrexate was added to the medium to obtain the desired drug concentration. After thorough mixing, cells were plated into 35-mm dishes. Cultures were incubated at 37°C in a 5% CO2 , humidified atmosphere and colonies enumerated after 7 to 10 days.
Transplantation of retroviral vector-transduced BM cells.Transduced BM cells were washed twice and resuspended in phosphate-buffered saline (PBS) containing 2% FCS. Three to 5 × 106 cells were transplanted by tail-vein injection into lethally irradiated (900 cGy), congenic WB/B6 F1 +/+ recipient mice which differed in hemoglobin phenotype from the donor mice. Four weeks posttransplantation, PB obtained by retroorbital sinus puncture was analyzed for expression of GFP or CD24 as described below. Complete hematology parameters including hematocrit, platelet count, and total leukocyte counts were obtained by standard methods. Leukocyte differentials were obtained by scoring Wright-Giemsa–stained blood films. Hemoglobin analysis, to assess hematopoietic reconstitution by donor marrow, was performed by standard methods.39
Evaluation of GFP expression in cells and tissues.In vitro cultured cells (fibroblasts and BM) were resuspended as single cells and directly subjected to flow cytometry with a FACS Vantage (Becton Dickinson, San Jose, CA) using excitation at 488 nm and fluorescence detection at 530 ± 30 nm (for GFP) or 575 ± 26 nm (for phycoerythrin). Propidium iodide (PI; Sigma, St Louis, MO) was added to all samples before analysis to allow identification and elimination of dead cells from the analysis. CD24 antibody staining of transduced marrow was performed using a murine IgG2a, phycoerythrin-conjugated, anti-CD24 monoclonal antibody (30785X; PharMingen, San Diego, CA) and was compared with staining using an isotype-matched control antibody. Before antibody staining of leukocytes, Fc receptors were blocked with a rat anti-CD16 antibody (Fc Block, 02141A; PharMingen). Plates containing hematopoietic colonies growing in methylcellulose were directly evaluated for GFP expression by visualization using a standard fluorescence microscope.
Expression of GFP in spleen colony-forming cells was evaluated by fluorescence microscopy using a wide-band green filter on fresh frozen, noncoverslipped 10-μm sections of spleens obtained from WB/B6 F1 +/+ animals that had been transplanted, after irradiation with 900 cGy, with 104 to 105 GFP- or CD24-transduced BM cells 13 days earlier. Photomicrographs were obtained using a Hamamatsu Low Light Level Camera (Hamamatsu Photonics K.K., Hamamatsu City, Japan) attached to a PC-based Leica Imaging System (Vashaw Scientific, Norcross, GA). Adjacent sections to those used for evaluation of GFP were stained with hematoxylin and eosin, dehydrated, coverslipped, and photographed using a Kontron digital camera attached to a Roche Image Analysis System (Roche Imaging Systems, Inc, Elon College, NC). Once all of the digital images were captured, they were imported and compiled into final figures using Adobe Photoshop software.
PB cells were analyzed for GFP expression as follows: whole blood was diluted 1:100 in PBS and used to assess GFP expression in red blood cells (RBCs) and platelets. Flow cytometry routinely revealed two easily distinguishable and characteristic populations, representing RBCs and platelets, based on forward and side light scatter parameters. These regions were used to gate on the respective populations for evaluation of GFP expression. As the contaminating leukocytes make up 1% or less of the total cell population, their presence did not significantly influence the analysis. To analyze GFP expression in the total leukocyte population, 300 to 500 μL of blood was depleted of RBCs by standard lysis procedures. After multiple washings with PBS/2% FCS, each followed by low-speed centrifugation, PI was added to the samples and the PI-dull, total leukocyte population was analyzed by flow cytometry.
RESULTS
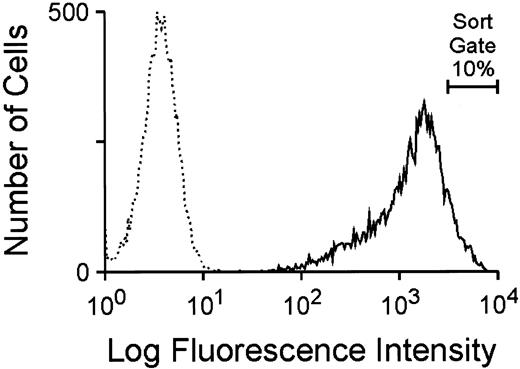
Utilization of the GFP marker to generate high-titer, ecotropic GFP retroviral vector producer cells.To investigate the potential utility of GFP as a selectable marker and reporter for gene transfer into hematopoietic cells, a murine stem cell virus (MSCV)-based retroviral vector, depicted in Fig 1B, was constructed as described in Materials and Methods. This bicistronic vector, MGirL22Y, contains the complete 714-bp GFP cDNA sequence linked by the encephalomyocarditis internal ribosome entry site to a human dihydrofolate reductase variant cDNA (termed L22Y), which confers high-level resistance to antifolate drugs such as trimetrexate. Conditioned media from transiently transfected 293T cells containing a high titer of amphotropic vector particles was used to multiply transduce the ecotropic packaging cell line GP+E86 and cells were subsequently selected for trimetrexate resistance. Flow cytometric analysis of the resulting trimetrexate-resistant polyclonal population, shown in Fig 2, showed intense fluorescence of all cells, demonstrating a concordance of expression of the two genes in the bicistronic vector. The brightest 10% of cells expressing GFP (Fig 2) was FACS sorted and expanded in culture. The viral titer of conditioned media obtained from this cell population, as assessed by transfer of trimetrexate resistance to NIH 3T3 cells, was approximately 5 × 106 colony-forming units (CFU)/mL. No toxic effects of GFP expression were observed despite extensive passaging of the cells in culture, and conditioned media from the producer cells exhibited a stable viral titer with passage of cells.
Flow cytometric analysis of GFP expression in GP + E86 cells transduced with amphotropic MGirL22Y vector particles after 7 days of selection with 100 nmol/L trimetrexate. The broken line profile designates the fluorescence of parental GP + E86 control cells while the solid line profile indicates the fluorescence of the GFP-transduced population. The proportion of cells sorted for high level GFP expression is indicated.
Flow cytometric analysis of GFP expression in GP + E86 cells transduced with amphotropic MGirL22Y vector particles after 7 days of selection with 100 nmol/L trimetrexate. The broken line profile designates the fluorescence of parental GP + E86 control cells while the solid line profile indicates the fluorescence of the GFP-transduced population. The proportion of cells sorted for high level GFP expression is indicated.
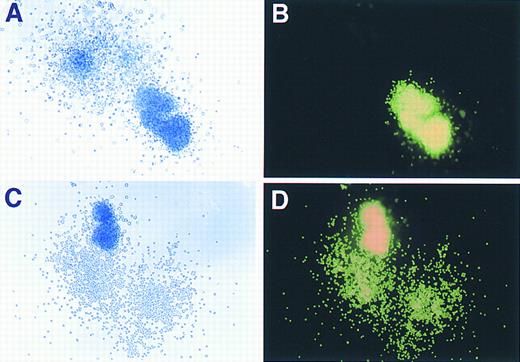
Phase contrast and fluorescence microscopy evaluation of GFP expression in murine hematopoietic colonies. (A) and (C) show colonies, derived from marrow transduced with the GFP vector, as visualized under phase contrast (original magnification × 80), while (B) and (D) demonstrate the same colonies under fluorescence microscopy performed on the identical fields shown in (A) and (C).
Phase contrast and fluorescence microscopy evaluation of GFP expression in murine hematopoietic colonies. (A) and (C) show colonies, derived from marrow transduced with the GFP vector, as visualized under phase contrast (original magnification × 80), while (B) and (D) demonstrate the same colonies under fluorescence microscopy performed on the identical fields shown in (A) and (C).
Southern blot analysis performed on MGirL22Y producer cell DNA showed the presence of an unrearranged proviral genomic band of expected size compared with the plasmid standard (Fig 1, lanes 3 and 1, respectively). Densitometry revealed the presence of an average of 15 proviral copies per cell in the MGirL22Y producer cells. Analysis of DNA from NIH 3T3 cells transduced with a low viral multiplicity of infection and subsequently selected with trimetrexate (to generate a cell line with approximately one proviral copy per cell) demonstrated faithful transmission of an intact, nonrearranged proviral genome (Fig 1, lane 5). Interestingly, on shorter exposures of the blot, a weakly intense, minor band was observed that was slightly smaller than the full-length provirus (data not shown). This probably represented a spliced, but transmitted, viral genome.
Assessment of retroviral gene transfer into murine myeloid and erythroid clonogenic progenitors using the GFP marker.As shown in Fig 3, both myeloid and erythroid colonies derived from transduced progenitors expressed high levels of GFP as judged by direct visualization with phase contrast and fluorescence microscopy. Positive and negative colonies were easily discriminated from each other (Fig 3A and B) as there was no background autofluorescence observed in nontransduced cells. Colonies derived from BM cells transduced with a CD24 retroviral control vector also showed no detectable fluorescence (data not shown), indicating the specificity of GFP expression as a marker for retroviral gene transfer. Scoring of plates from three separate experiments showed a greater than 90% (618 of 656 hematopoietic colonies) overall transduction efficiency into clonogenic progenitors. A spectrum of intensity of GFP expression was readily observed ranging from dim, slightly positive colonies to colonies with very intense fluorescence.
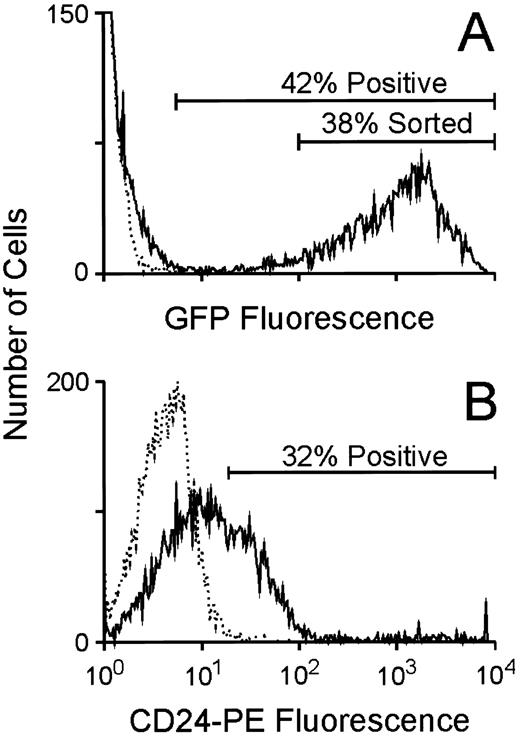
Utilization of the GFP marker to enrich for clonogenic progenitors expressing increased levels of a linked, antifolate drug-resistance gene.To evaluate the potential of using GFP as a selectable marker to enrich for hematopoietic cells expressing high levels of a second gene in a bicistronic transcript, murine BM cells were cocultivated with the GFP producer cells as above and subsequently analyzed for GFP expression by flow cytometry. As shown in a representative experiment, approximately 40% of the marrow cells (60% and 70%, respectively, in two other experiments) were very strongly positive for GFP expression (Fig 4A), while unstained, CD24-transduced BM cells showed no fluorescence. When stained with an anti-CD24 phycoerytherin-conjugated antibody, CD24-transduced cells consistently displayed fluorescence intensity at least two logs lower in magnitude than that measured for GFP-expressing cells (Fig 4B). Cells expressing high levels of GFP (Fig 4A) were FACS purified and plated in clonogenic progenitor cultures in the presence or absence of trimetrexate. As shown in a representative experiment, these cells displayed increased resistance to high concentrations of trimetrexate compared to the unsorted population (Fig 5), despite the fact that greater than 90% of unsorted progenitors were transduced as scored by GFP fluorescence in the absence of drug. Therefore, this strategy enables the efficient elimination of nontransduced and transduced, but low-level–expressing cells from cells expressing high levels of the bicistronic vector genome. As expected, all drug-resistant colonies showed GFP fluorescence, again confirming a high degree of concordance of expression of the two genes.
Flow cytometric analysis of GFP and CD24 expression in BM cells transduced with either the MGirL22Y vector or the CD24 vector. (A) Evaluation of GFP expression in BM cells 24 hours after completion of the transduction period. The broken line indicates the fluorescence profile of unstained, CD24-transduced control marrow while the solid line indicates the fluorescence profile of the GFP-transduced marrow. The extremely high level of fluorescence of the GFP-expressing cells necessitated the use of lower fluorescence amplification when analyzing for GFP expression. The proportion of positive cells in the population as well as the population expressing high levels of GFP that was FACS purified is shown. (B) Evaluation of CD24 expression in BM cells 24 hours after completion of the transduction period. The broken line indicates the fluorescence profile of GFP-transduced control marrow stained with an anti-CD24 antibody while the solid line indicates the fluorescence profile of the CD24-transduced marrow. The proportion of positive cells in the CD24-transduced population is shown.
Flow cytometric analysis of GFP and CD24 expression in BM cells transduced with either the MGirL22Y vector or the CD24 vector. (A) Evaluation of GFP expression in BM cells 24 hours after completion of the transduction period. The broken line indicates the fluorescence profile of unstained, CD24-transduced control marrow while the solid line indicates the fluorescence profile of the GFP-transduced marrow. The extremely high level of fluorescence of the GFP-expressing cells necessitated the use of lower fluorescence amplification when analyzing for GFP expression. The proportion of positive cells in the population as well as the population expressing high levels of GFP that was FACS purified is shown. (B) Evaluation of CD24 expression in BM cells 24 hours after completion of the transduction period. The broken line indicates the fluorescence profile of GFP-transduced control marrow stained with an anti-CD24 antibody while the solid line indicates the fluorescence profile of the CD24-transduced marrow. The proportion of positive cells in the CD24-transduced population is shown.
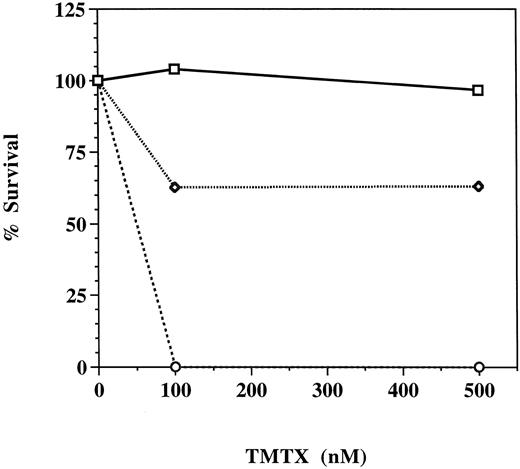
Trimetrexate resistance of hematopoietic progenitors transduced with the MGirL22Y retroviral vector. Unsorted GFP-transduced marrow cells (dotted line), GFP-transduced marrow cells FACS sorted for high level expression of GFP (solid line), and mock-transduced marrow cells (broken line) were plated into methylcellulose cultures containing 0, 100, or 500 nmol/L trimetrexate as described in Materials and Methods. Five to eight days later, colonies were counted and percent survival, derived from the number of trimetrexate-resistant colonies divided by the number of colonies observed in the absence of drug, was determined.
Trimetrexate resistance of hematopoietic progenitors transduced with the MGirL22Y retroviral vector. Unsorted GFP-transduced marrow cells (dotted line), GFP-transduced marrow cells FACS sorted for high level expression of GFP (solid line), and mock-transduced marrow cells (broken line) were plated into methylcellulose cultures containing 0, 100, or 500 nmol/L trimetrexate as described in Materials and Methods. Five to eight days later, colonies were counted and percent survival, derived from the number of trimetrexate-resistant colonies divided by the number of colonies observed in the absence of drug, was determined.
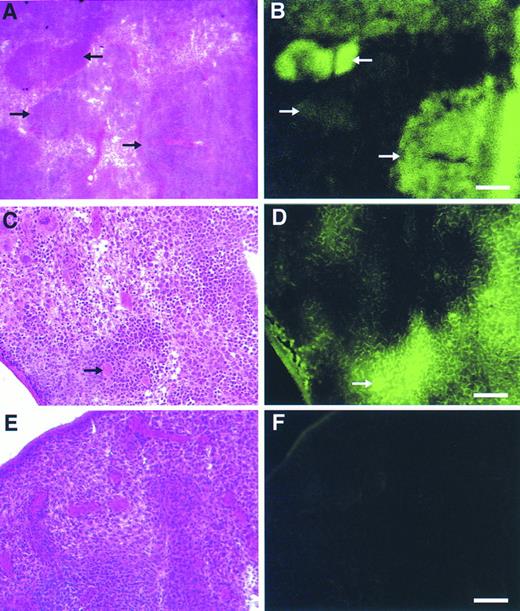
Detection of retroviral gene transfer into day 13 spleen colony-forming cells (CFU-S) using the GFP marker.BM cells transduced with the GFP vector or the CD24 vector as a control were injected into irradiated mice and 13 days later animals were sacrificed and spleens procured. Fresh frozen sections of spleens from GFP-transplanted and control-transplanted animals were then evaluated by fluorescence microscopy. Spleen colonies, identified morphologically by staining alternate sections with hematoxylin-eosin, derived from GFP-transduced progenitors displayed a range of fluorescence (Fig 6A and B), from very intense to dim. Higher magnification views (Fig 6C and D) showed the specificity of GFP marking in individual hematopoietic cells compared with the background splenic tissue. Not only were marked myeloid and erythroid cells discernible, but megakaryocytes were also intensely fluorescent (Fig 6C and D, arrow). No significant background fluorescence was detectable in spleens from control animals (Fig 6E and F). In an independent analysis, macroscopic, well-isolated spleen colonies from GFP-transplanted and control-transplanted animals were dissected and subsequently analyzed by flow cytometry. A significant proportion of cells (33% to 84%) in all 10 colonies evaluated which were derived from GFP-transduced progenitors displayed intense fluorescence, whereas control colonies contained no cells with a detectable fluorescent signal (data not shown).
Detection of gene transfer into day 12 spleen colony-forming cells using the GFP marker. Alternate, serial fresh frozen sections of spleens from animals transplanted with the GFP vector (A through D) or a control, CD24 vector (E and F) were stained with hematoxylin-eosin (A, C, and E) or directly visualized by fluorescence microscopy (B, D, and F) as described in Materials and Methods. In (A) and (B) the different arrows mark spleen hematopoietic colonies demonstrating a range of fluorescence intensities. In (C) and (D), the bold arrow denotes a GFP-marked megakaryocyte. Scale bars in (A) and (B) are equal to 325 μm, in (C) through (F) they are equal to 165 μm.
Detection of gene transfer into day 12 spleen colony-forming cells using the GFP marker. Alternate, serial fresh frozen sections of spleens from animals transplanted with the GFP vector (A through D) or a control, CD24 vector (E and F) were stained with hematoxylin-eosin (A, C, and E) or directly visualized by fluorescence microscopy (B, D, and F) as described in Materials and Methods. In (A) and (B) the different arrows mark spleen hematopoietic colonies demonstrating a range of fluorescence intensities. In (C) and (D), the bold arrow denotes a GFP-marked megakaryocyte. Scale bars in (A) and (B) are equal to 325 μm, in (C) through (F) they are equal to 165 μm.
Transplantation of GFP-transduced BM cells into lethally irradiated mice leads to reconstitution of hematopoiesis with GFP-marked PB cell progeny.To evaluate the potential of using GFP to track hematopoietic progeny in vivo after BM transplantation, a cohort of lethally irradiated mice was transplanted with congenic MGirL22Y-transduced or control CD24-transduced BM cells. In addition, several mice were transplanted with a 1:1 mixture of GFP- and control CD24-transduced marrow. Southern blot analysis performed on DNA from GFP-transduced BM showed high-level transfer of the unrearranged proviral genome (Fig 1, lane 7).
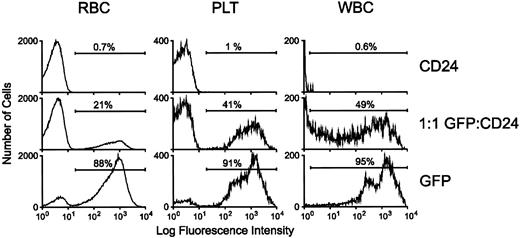
PB samples from reconstituted animals were analyzed by flow cytometry for GFP expression 4 weeks posttransplantation. RBCs, platelets, and leukocytes from animals reconstituted with GFP-transduced marrow displayed high-level fluorescence (Fig 7). Mice reconstituted with a 1:1 mixture of GFP- and CD24-transduced marrow also had brightly fluorescent cells in all lineages. In contrast, blood elements from mice reconstituted with CD24-transduced marrow had no significant level of background fluorescence.
Reconstitution of hematopoiesis with GFP-marked PB cells in lethally irradiated animals transplanted with GFP-transduced BM. Flow cytometric analysis of GFP expression in RBCs, platelets, and leukocytes was performed as described in Materials and Methods. For the white blood cell (WBC) panel, very high-level GFP fluorescence in expressing cells necessitated analysis using lower fluorescence amplification for all the WBC samples. The profiles of representative animals transplanted with marrow transduced with the control CD24 vector, the MGirL22Y vector, or an equal number of MGirL22Y- and CD24-transduced marrow cells 4 weeks posttransplantation are shown as indicated. The proportion of GFP-positive cells within each respective population is indicated.
Reconstitution of hematopoiesis with GFP-marked PB cells in lethally irradiated animals transplanted with GFP-transduced BM. Flow cytometric analysis of GFP expression in RBCs, platelets, and leukocytes was performed as described in Materials and Methods. For the white blood cell (WBC) panel, very high-level GFP fluorescence in expressing cells necessitated analysis using lower fluorescence amplification for all the WBC samples. The profiles of representative animals transplanted with marrow transduced with the control CD24 vector, the MGirL22Y vector, or an equal number of MGirL22Y- and CD24-transduced marrow cells 4 weeks posttransplantation are shown as indicated. The proportion of GFP-positive cells within each respective population is indicated.
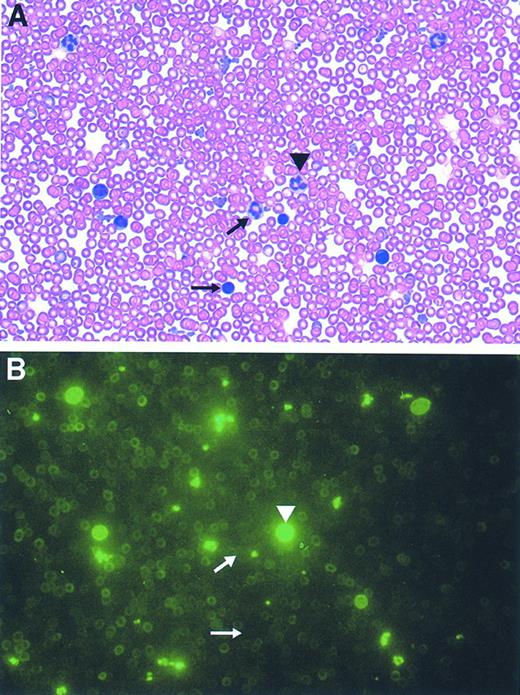
Most of the mice reconstituted with GFP-transduced marrow (n = 8) typically displayed a 70% to 90% range of GFP-marking of cells within each lineage, consistent with the degree of engraftment of donor cells as assessed in the erythroid lineage by hemoglobin electrophoresis (data not shown). Mice reconstituted with a 1:1 mixture of GFP- and control-transduced marrow (n = 4) displayed a range of 30% to 50% marking in leukocytes and platelets and 20% to 25% in RBCs. PB films from these animals showed concordance of the flow cytometry data with the degree of cell positivity estimated by direct visualization using fluorescence microscopy (Fig 8, see page 1780). Intense fluorescence was easily observed in individual leukocytes and platelets, while RBCs appeared dimmer. In contrast, evaluation of blood films obtained from animals transplanted with marrow transduced with the control, CD24 vector demonstrated the absence of detectable cell fluorescence (data not shown). No abnormalities in hematopoiesis have been observed in mice reconstituted with GFP-transduced marrow as judged by engraftment, complete blood counts, and blood cell morphology (data not shown).
Direct microscopic visualization of genetically modified PB cell progeny using the GFP marker. Photomicrographs (original magnification × 2,000) of the identical field from a PB film obtained from an animal transplanted with GFP-transduced marrow both before (B), and after, staining with Wright-Giemsa (A) are shown. The arrowhead denotes a granulocyte exhibiting bright fluorescence while the arrows mark a very weakly fluorescent granulocyte (top arrow) and a mononuclear cell lacking fluorescence (bottom arrow).
Direct microscopic visualization of genetically modified PB cell progeny using the GFP marker. Photomicrographs (original magnification × 2,000) of the identical field from a PB film obtained from an animal transplanted with GFP-transduced marrow both before (B), and after, staining with Wright-Giemsa (A) are shown. The arrowhead denotes a granulocyte exhibiting bright fluorescence while the arrows mark a very weakly fluorescent granulocyte (top arrow) and a mononuclear cell lacking fluorescence (bottom arrow).
DISCUSSION
Our work has shown that the GFP gene can serve as a very useful marker of genetic modification of hematopoietic cells. Selection of primitive hematopoietic cells ex vivo, based on the presence and level of GFP expression, provides a population of transduced cells with the high level of expression of a linked, drug-resistance marker gene encoded with GFP in a bicistronic transcript. By virtue of its intrinsic fluorescence, GFP is readily detected in tissues and PB cells, providing a convenient strategy to track the behavior of genetically modified cells.
Although the human CD24 molecule and its murine homologue, the heat-stable antigen,40 have been shown by others to be useful as selectable markers for identifying and enriching transduced hematopoietic cells,22,23 each has associated limitations. The use of the CD24 marker in human cells is restricted because of its endogenous expression on hematopoietic and immune tissues.41 Both CD24 and heat-stable antigen, which has been proposed as an alternate marker for use in human cells,23 suffer shortcomings because of their putative roles as immune regulatory molecules.41-44 Interestingly, human CD24 has also recently been reported to be a major ligand for P-selectin,45 which is involved in platelet adherence to leukocytes and endothelial cells. Unlike GFP, these markers cannot be used to directly determine gene transfer into in vitro clonogenic progenitors grown in methylcellulose because of the requirement for antibody staining. Other advantages of using the GFP marker are the elimination of potential undesired effects of antibody staining (crosslinking of cell-surface membrane proteins), as well as its associated costs. Furthermore, the lack of necessity for extensive cell manipulation, as occurs during antibody staining protocols, when preparing GFP-transduced cells for FACS purification is not only a theoretical benefit, but a practical one when large numbers of purified cells are desired.
Another issue with regard to the use of proteins such as CD24, which are attached to the cell membrane via a glycophosphoinositol (GPI) anchor, is their propensity to be transferred from expressing to nonexpressing cells.46 47 Indeed, we have observed that the proportion of leukocytes that display the CD24 marker consistently exceeds the proportion of marked RBCs and platelets in animals transplanted with CD24-transduced BM cells (J. Allay and B. Sorrentino, unpublished observations, November 1996). Preliminary analysis has led to the detection of leukocytes expressing both the CD24 and GFP markers in our animals which were transplanted with the mixture of CD24- and MGirL22Y-transduced cells (unpublished observations, May 1997). Our prediction is that such double-marked cells will contain the MGirL22Y genome but lack the genome for the CD24 virus.
As suggested by others,21-23 the use of retroviral vectors containing markers not normally present on target cells to facilitate enrichment of transduced cells can enable BM transplantation or adoptive immunotherapy to be performed with pure populations of genetically modified cells. For this to be practical on even a modest preclinical scale, transduced cells must be easily discriminated from nontransduced cells, and if desired, low-level–expressing, transduced cells. In this regard, the GFP marker, which demonstrates markedly intense fluorescence in transduced marrow cells in our studies, appears to be an outstanding selectable marker. In contrast, we have found that the CD24 marker (Fig 4), as have others with both the CD24 and HSA markers,22 23 as detected by immunofluorescence techniques, display at least two logs lower fluorescence intensity than that of GFP in transduced marrow cells. Although this could be due in part to vector design and viral producer cell titer, it would be highly unusual for an antibody staining technique to yield fluorescence intensity comparable to that observed with GFP in these studies. In this regard, the fluorescence intensity of the GFP marker in transduced BM cells facilitates discrimination of genetically modified cells from unmodified cells. For FACS purification strategies, this enables sort gates to be less stringently designated, even for high-level–expressing, positive cells, leading to an increased yield of highly purified cells. Preliminary experiments have shown the ability to obtain large numbers of purified, transduced BM cells expressing high levels of GFP. The GFP marker could potentially prove to be a powerful tool to highly enrich large numbers of cells expressing a linked therapeutic gene.
In addition to its utilities as a selectable marker and in detecting the progeny of genetically modified hematopoietic cells of all lineages in the spleen and PB, the GFP marker gene holds significant potential in facilitating the in vivo tracking of migration of lymphoid and monocytic cells derived from transplanted marrow. In this regard, gene-modified B and T lymphocytes display high-level GFP expression in mice 6 weeks posttransplantation with GFP-transduced marrow cells (J. Riberdy, J. Allay, D. Persons, and P. Doherty, unpublished observations, May 1997). Experiments designed to follow the movements of various phentoypically defined lymphocyte subpopulations in immune tissues in response to a wide array of antigenic challenges are underway. Likewise, tracking of gene-corrected phagocytic cells into the central nervous system in the setting of murine models of lipid storage diseases48 may yield information pertinent to the design of improved gene therapy strategies for these diseases.
Recent reports have emphasized the importance of a potential host immune response to the transgene product in genetically modified cells.4 49-52 Such immune responses have not generally been documented in irradiated mice that are recipients of genetically modified cells. Therefore, the relative immunogenicity of GFP compared with other markers such as murine heat-stable antigen or the low-affinity nerve growth factor receptor remains to be determined. Experiments designed to directly assess the immunogenecity of GFP in nonhuman primates transplanted with MGirL22Y-transduced BM may be informative.
Our studies show that GFP can serve as an excellent marker of gene transfer in hematopoietic tissues. Its utilization in human gene-marking trials will allow purification of genetically modified cells ex vivo and the tracking of such cells following transplantation. Evaluation of various methodologies for gene transfer into immunophenotypically defined, rare cell populations, such as the hematopoietic stem cell, should be facilitated by the use of the GFP marker.
ACKNOWLEDGMENT
The authors thank Elio Vanin and John Cunningham for helpful advice and stimulating discussions. We thank John Zachar for his help in fluorescence microscopy and Jean Johnson for help in preparing this manuscript.
Supported in part by National Heart, Lung and Blood Institute Program Project P01 HL 53749-01, Cancer Center Support CORE Grant P30 CA 21765, National Institutes of Health Grant No. CA 70089, and American Lebanese Syrian Associated Charities (ALSAC).
Address reprint requests to Arthur W. Nienhuis, MD, Director, St Jude Children's Research Hospital, 332 N Lauderdale Dr, Memphis, TN 38105-2794.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal