Abstract
Fluorescent in situ hybridization (FISH) analysis with a panel of DNA probes for 13q13.1-q14.3 was performed on 20 cases of myeloid malignancies, of which 17 showed a del(13)(q) and three had translocations affecting 13q. By chromosome morphology, deletions consistently involved bands q14 and q21. In addition to confirming the chromosome data, FISH allowed us to delineate a commonly deleted region that was flanked by YAC 833A2 and YAC 854D4. Three cases with 13q translocations unexpectedly showed accompanying cryptic microdeletions of 13q, and in one case the commonly deleted region could be narrowed to a genomic segment, which includes YAC 937C7, RB1, and YAC 745E3. Homozygous deletions were not detected. This region overlaps with the smallest deleted region of 13q14 in chronic lymphocytic leukemia.
CHROMOSOMAL deletions are among the most common genetic events observed in solid tumors and hematologic malignancies. Loss of genetic material in neoplastic cells is suggestive of a recessive mechanism in the pathogenesis of these disorders and is regarded as a hallmark of putative tumor suppressor gene(s) localization. In myeloid disorders, the most frequently reported deletions affect chromosomes 5, 7, and 20.1Deletions of chromosomes 11, 12, and 13 are less frequent, but recurrent.1 Loss of chromosome 13q material is mainly observed in myeloproliferative disorders (MPD), but it also occurs in myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML).2 In most cases the deletions have been described as interstitial and only in a few cases as terminal.2 Attempts to correlate the extent of the deletions with the type of myeloid malignancy have been made and loss of 13q12-q32 bands appeared to be prevalent in MPD, 13q12-q22 in MDS, and the 13q21 band in AML.1 It is noteworthy that one of the well-known tumor suppressor genes, the retinoblastoma gene (RB1), has been assigned to 13q14.3
Recently, some molecular studies on the status of this gene in myeloid malignancies have been published. Ahuja et al4 detected RB1 involvement in rearrangements or intragenic deletions in 5 of 54 cases of AML and in 2 of 18 cases of MDS. In another series of 69 AML cases, RB1 gene rearrangements were not found by Southern blot analysis.5 Similarly, gene rearrangements could not be detected in studies on AML cases in which a significant lack of RB1 protein expression was found.6,7 Also, in a series of 90 cases of MDS, the RB1 gene configuration proved to be germline.8 Finally, in 1 case of AML and one case of agnogenic myeloid metaplasia (AMM), both with a del(13)(q) determined at cytogenetic level, Morris et al9 found heterozygous deletions of the RB1 gene.
Interestingly, rearrangements of chromosome 13, at bands q13-q21, account for 12% of structural changes in chronic lymphocytic leukemia (CLL) with abnormal karyotypes10 and were found in larger numbers of cases when molecular investigations were performed.11,12 The RB1 gene was shown to be deleted in 20% of CLL analyzed by fluorescent in situ hybridization (FISH)11 and in 30% of cases investigated by quantitative Southern blot analysis.12 Homozygous deletions of RB1, however, have been sporadically detected.11 Recently, Brown et al13 reported CLL cases in which RB1 was not lost and drew attention to the D13S25 locus telomeric to RB1 that was lost in 9 of 10 analyzed cases. Additional studies using the LOH (loss of heterozygosity) assay,14,15 quantitative Southern blot,16 and FISH17 strongly suggested that a putative tumor suppressor gene, implicated in the pathogenesis of CLL, might be located between the RB1 gene and the D13S25 marker. Bullrich et al18 restricted the smallest critical region to a segment of 550 kb between 206XF12 and D13S25 marker at band 13q14.
In contrast with the rapid accumulation of molecular findings on 13q deletions in lymphoid disorders, molecular data on 13q chromosome deletions in myeloid malignancies are still scarce, to the extent that no results from molecular cytogenetics are available so far. We report here on 20 cases of various myeloid disorders in which we characterized deletions of 13q by FISH with the aim of determining a commonly deleted region at the molecular level.
MATERIALS AND METHODS
Patients.
Patients with myeloid diseases and deletions or translocations of chromosome 13 were retrieved from the files of the Center for Human Genetics in Leuven, Belgium, Hospital Sant Pau in Barcelona, Spain, and from the Hematology and Bone Marrow Transplantation Unit in Perugia, Italy. All clinical files of the patients were reviewed. A total of 20 cases were included with the following diagnoses: 6 MDS, 5 AMM, 4 AML, 2 essential thrombocythemia (ET), 1 polycytemia vera (PV), 1 Ph-positive CML, and 1 atypical CML. There were 10 female and 10 male patients. Some data about these patients are shown in Table1.
Clinical, Hematologic, and Cytogenetic Data of Cases With Deletions or Translocations Involving 13q
| Patients . | S/A . | Diagnosis . | Karyotype . | Follow-up† . |
|---|---|---|---|---|
| Deletions | ||||
| 1 | F/57 | AMM | 46,XX[3/5] | 47‡ |
| 46,XX,del(13)(q13q21)[2/5] | ||||
| 2 | F/66 | AMM | 46,XX[3/10] | 59 |
| 46,XX,del(13)(q13q21)[7/10] | ||||
| 3 | M/40 | AMM | 46,XY,[4/10] | 46 |
| 46,XY,del(13)(q13q21)[6/10] | ||||
| 4 | M/65 | AMM | 46,XY,t(3;18)(p14;p11),del(5)(q14q34),del(13)(q13q21)[10/10] | 24 |
| 5 | M/55 | AMM | 46,XY,del(13)(q13q21)[15/15] | 19 |
| 6* | M/53 | MDS | 46,XY[6/17] | 12‡ |
| 46,XY,del(13)(q12q21)[11/17] | ||||
| 7 | M/55 | MDS | 46,XY,del(13)(q13q21)[10/10] | Lost |
| 8 | M/63 | MDS | 46,XY,del(13)(q13q21)[8/8] | 22‡ |
| 9 | F/66 | MDS | 46,XX[5/10] | 6 |
| 46,XX,del(5)(q21q32)[1/10] | ||||
| 46,XX,del(5)(q21q32),del(13)(q13q21)[4/10] | ||||
| 10 | F/72 | AML-M6 | 47,XX,del(8)(q?),del(13)(q14q22),+mar[10/10] | 55‡ |
| 11 | F/67 | AML | 47,XX,del(5)(q13q33),+8,del(13)(q12q21)[15/15] | 1‡ |
| 12 | F/36 | AML-M4 | 46,XX[3/10] | Lost |
| 47,XX,+8[3/10] | ||||
| 47,XX,+8,del(13)(q13q22)[4/10] | ||||
| 13* | M/66 | ET | 47,XY, der(5)t(1;5)(p22;q21),+der(6)t(2;6)(q22;p12),del(13)(q14q22),der(17)t(11;17) (q13;p13)[7/7] | Lost |
| 14 | F/69 | ET | 46,XX,del(13)(q13q21)[10/10] | Lost |
| 15* | M/64 | PV | 47,XY,del(1)(p36),+del(1)(p36),del(11)(q22q24),del(13)(q13q21)[7/9] | 177 |
| 46,XY,-5,+9[2/9] | ||||
| 16* | F/67 | CML | 46,XX,t(9;22)(q34;q11)[2/10] | Lost |
| 46,XX,t(9;22)(q34;q11),del(13)(q13q21)[8/10] | ||||
| 17 | F/77 | Atypical CML | 46,XX,del(13)(q13q21)[10/10] | 6‡ |
| Translocations | ||||
| 18 | M/73 | MDS | 43,X,-Y,del(5)(q13q21),der(7)t(7;20)(p11;p11),t(12;13)(p13;q14),17,der(19)t(17;19)(q21;q13),-20[9/16]/ 44,idem,+der(13)t(12;13)(p13;q14)[5/16]/44,idem,+13[2/16] | 2‡ |
| 19 | M/87 | MDS | 45,X,-Y[1/13] | Lost |
| 45,X,-Y,del(5)(q12q34)[1/13] | ||||
| 45,X,-Y,t(2;12;13)(p14;q24;q13),del(5)(q12q34)[11/13] | ||||
| 20 | F/58 | AML-M6 | 52,XX,del(3)(p13p25),del(5)(q23q32),7,der(7)tas(7;13)(7pter→qter→13qter→q14),der(13)t(8;13)(q21;q13),+19, +mar1x2,+mar2,+mar3,+mar4[10/10] | 9‡ |
| Patients . | S/A . | Diagnosis . | Karyotype . | Follow-up† . |
|---|---|---|---|---|
| Deletions | ||||
| 1 | F/57 | AMM | 46,XX[3/5] | 47‡ |
| 46,XX,del(13)(q13q21)[2/5] | ||||
| 2 | F/66 | AMM | 46,XX[3/10] | 59 |
| 46,XX,del(13)(q13q21)[7/10] | ||||
| 3 | M/40 | AMM | 46,XY,[4/10] | 46 |
| 46,XY,del(13)(q13q21)[6/10] | ||||
| 4 | M/65 | AMM | 46,XY,t(3;18)(p14;p11),del(5)(q14q34),del(13)(q13q21)[10/10] | 24 |
| 5 | M/55 | AMM | 46,XY,del(13)(q13q21)[15/15] | 19 |
| 6* | M/53 | MDS | 46,XY[6/17] | 12‡ |
| 46,XY,del(13)(q12q21)[11/17] | ||||
| 7 | M/55 | MDS | 46,XY,del(13)(q13q21)[10/10] | Lost |
| 8 | M/63 | MDS | 46,XY,del(13)(q13q21)[8/8] | 22‡ |
| 9 | F/66 | MDS | 46,XX[5/10] | 6 |
| 46,XX,del(5)(q21q32)[1/10] | ||||
| 46,XX,del(5)(q21q32),del(13)(q13q21)[4/10] | ||||
| 10 | F/72 | AML-M6 | 47,XX,del(8)(q?),del(13)(q14q22),+mar[10/10] | 55‡ |
| 11 | F/67 | AML | 47,XX,del(5)(q13q33),+8,del(13)(q12q21)[15/15] | 1‡ |
| 12 | F/36 | AML-M4 | 46,XX[3/10] | Lost |
| 47,XX,+8[3/10] | ||||
| 47,XX,+8,del(13)(q13q22)[4/10] | ||||
| 13* | M/66 | ET | 47,XY, der(5)t(1;5)(p22;q21),+der(6)t(2;6)(q22;p12),del(13)(q14q22),der(17)t(11;17) (q13;p13)[7/7] | Lost |
| 14 | F/69 | ET | 46,XX,del(13)(q13q21)[10/10] | Lost |
| 15* | M/64 | PV | 47,XY,del(1)(p36),+del(1)(p36),del(11)(q22q24),del(13)(q13q21)[7/9] | 177 |
| 46,XY,-5,+9[2/9] | ||||
| 16* | F/67 | CML | 46,XX,t(9;22)(q34;q11)[2/10] | Lost |
| 46,XX,t(9;22)(q34;q11),del(13)(q13q21)[8/10] | ||||
| 17 | F/77 | Atypical CML | 46,XX,del(13)(q13q21)[10/10] | 6‡ |
| Translocations | ||||
| 18 | M/73 | MDS | 43,X,-Y,del(5)(q13q21),der(7)t(7;20)(p11;p11),t(12;13)(p13;q14),17,der(19)t(17;19)(q21;q13),-20[9/16]/ 44,idem,+der(13)t(12;13)(p13;q14)[5/16]/44,idem,+13[2/16] | 2‡ |
| 19 | M/87 | MDS | 45,X,-Y[1/13] | Lost |
| 45,X,-Y,del(5)(q12q34)[1/13] | ||||
| 45,X,-Y,t(2;12;13)(p14;q24;q13),del(5)(q12q34)[11/13] | ||||
| 20 | F/58 | AML-M6 | 52,XX,del(3)(p13p25),del(5)(q23q32),7,der(7)tas(7;13)(7pter→qter→13qter→q14),der(13)t(8;13)(q21;q13),+19, +mar1x2,+mar2,+mar3,+mar4[10/10] | 9‡ |
Abbreviations: S, sex; A, age; AMM, agnogenic myeloid metaplasia; ET, essential thrombocytemia; PV, polycytemia vera, AML, acute myeloid leukemia; MDS, myelodysplastic syndromes; CML, chronic myeloid leukemia.
Patients in which 13q anomaly was found during the course of disease.
Follow-up in months from the date of diagnosis.
Died.
Cytogenetics.
Bone marrow and/or peripheral blood cells were cultured for 24 or 48 hours without stimulation. Metaphases were R-banded with acridine orange or G-banded with Wright stain. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature (1995).
FISH studies.
FISH was performed as previously described19 using nine biotin-16–dUTP labeled YACs selected from reported STS-based maps.20 21 YACs assigned to the 13q13.1-q14.3 region are ordered as follows: centromere-951A3-765A10-833A2-875A8-911F6-937C7-745E3-935G2-854D4-telomere (Table 2). The RB1 gene was investigated with probe LSI 13/RB1 spectrum orange DNA (Vysis Inc, Downers Grove, IL). A chromosome 13/21 (pUC 1.76) centromeric probe directly labeled with fluorescein-5–dUTP or Texas Red-5–dCTP was used for identifying the der(13) in each case. In some experiments, on case 18, a centromeric probe for chromosome 12 (pRB-12) was also applied. An additional two-color painting with a digoxigenin-11–dUTP labeled library 8, and a biotin-16–dUTP labeled library 13 was performed in case 20. A median of seven abnormal metaphases (range, 3 to 9) were studied by FISH and G-banding for each case. The FISH data were collected on a Leitz fluorescence microscope equipped with a cooled CCD camera (Photometrics) run by SmartCapture software (Vysis, Stuttgart, Germany).
YAC Probes
| YAC* . | STS-Content† . | Genetic Position . | Cytogenetic Position‡ . |
|---|---|---|---|
| 951A3 | MIT: D13S171(U) | 28 cM | |
| WI-5559(U) | — | 13q13.1 | |
| D13S267(O) | 30 cM | ||
| 765A10 | MIT: D13S219(O) | — | |
| WI-9842(D) | 32 cM | 13q13.2 | |
| 833A2 | MIT: WI-8890(U) | — | |
| D13S1233(U) | 42 cM | 13q13.3 | |
| D13S263(U) | 42 cM | ||
| WI-7880(O) | — | ||
| GEN: AFM144xq3 | — | ||
| 875A8 | MIT: GTC17F01(U) | — | |
| D13S1247(O) | 44 cM | 13q13.3 | |
| CHLC.GATA6B07(D) | — | ||
| D13S1297(U) | 44 cM | ||
| D13S1270(U) | 44 cM | ||
| WI-5808(U) | — | ||
| D13S1227(U) | 45 cM | ||
| WI-464(D) | — | ||
| 911F6 | MIT: D13S291(U) | 47 cM | |
| D13S757(U) | — | 13q14.11 | |
| D13S1272(D) | 47 cM | ||
| WI-6833(U) | — | ||
| CHLC.GATA53A06(D) | — | ||
| 937C7 | MIT: WI-2744(D) | — | |
| WI-7773(D) | — | 13q14.12 | |
| D13S328(O) | 49 cM | ||
| WI-3641(D) | — | ||
| 745E3 | MIT: WI-6333(U) | — | |
| WI-9598(U) | — | 13q14.12 | |
| GCT16C05(D) | — | ||
| D13S1269(D) | 51 cM | ||
| WI-5710(D) | — | ||
| D13S262(O) | 51 cM | ||
| GEN: D13S25 | — | ||
| 935G2 | MIT: D13S284(U) | 53 cM | |
| CHLC.GATA29A09(U) | — | 13q14.12 | |
| D13S1325(O) | 51 cM | ||
| GEN: CA2A | — | ||
| D13S227 | — | ||
| D13S31 | — | ||
| 854D4 | GEN: D13S176(D) | 56 cM | 13q14.3 |
| YAC* . | STS-Content† . | Genetic Position . | Cytogenetic Position‡ . |
|---|---|---|---|
| 951A3 | MIT: D13S171(U) | 28 cM | |
| WI-5559(U) | — | 13q13.1 | |
| D13S267(O) | 30 cM | ||
| 765A10 | MIT: D13S219(O) | — | |
| WI-9842(D) | 32 cM | 13q13.2 | |
| 833A2 | MIT: WI-8890(U) | — | |
| D13S1233(U) | 42 cM | 13q13.3 | |
| D13S263(U) | 42 cM | ||
| WI-7880(O) | — | ||
| GEN: AFM144xq3 | — | ||
| 875A8 | MIT: GTC17F01(U) | — | |
| D13S1247(O) | 44 cM | 13q13.3 | |
| CHLC.GATA6B07(D) | — | ||
| D13S1297(U) | 44 cM | ||
| D13S1270(U) | 44 cM | ||
| WI-5808(U) | — | ||
| D13S1227(U) | 45 cM | ||
| WI-464(D) | — | ||
| 911F6 | MIT: D13S291(U) | 47 cM | |
| D13S757(U) | — | 13q14.11 | |
| D13S1272(D) | 47 cM | ||
| WI-6833(U) | — | ||
| CHLC.GATA53A06(D) | — | ||
| 937C7 | MIT: WI-2744(D) | — | |
| WI-7773(D) | — | 13q14.12 | |
| D13S328(O) | 49 cM | ||
| WI-3641(D) | — | ||
| 745E3 | MIT: WI-6333(U) | — | |
| WI-9598(U) | — | 13q14.12 | |
| GCT16C05(D) | — | ||
| D13S1269(D) | 51 cM | ||
| WI-5710(D) | — | ||
| D13S262(O) | 51 cM | ||
| GEN: D13S25 | — | ||
| 935G2 | MIT: D13S284(U) | 53 cM | |
| CHLC.GATA29A09(U) | — | 13q14.12 | |
| D13S1325(O) | 51 cM | ||
| GEN: CA2A | — | ||
| D13S227 | — | ||
| D13S31 | — | ||
| 854D4 | GEN: D13S176(D) | 56 cM | 13q14.3 |
*YAC clones are derived from the Centre d'Etude du Polymorphisme Humain (CEPH) human mark 3 (MEGA) YAC library.
This table only contains STS data that are consistent with contig and cytogenetic data; see e-mail MIT, Whitehead Institute/MIT Center for Genome Research; GEN, Genethon/CEPH; U, unambiguous hit; D, disambiguated hit; O, hits reported by other laboratories (primarily CEPH).
Cytogenetic position according to Chumakov et al.20Except for D13S176 (854D4), these STSes are part of singly-linked contig WC13.1.
RESULTS
Cytogenetics.
Cytogenetic results are shown in Table 1. In all but 4 cases the 13q anomaly was found at diagnosis. In 2 cases del(13)(q) appeared during the course of disease (cases 6 and 16), and the remaining 2 cases (13 and 15) were only studied 2 and 11 years after diagnosis, respectively.
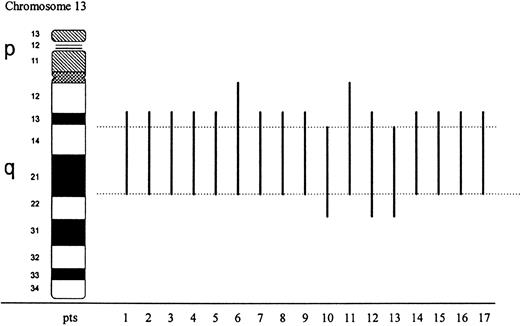
Abnormal metaphases carrying the 13q anomaly ranged from 40% to 100% of analyzed cells. Seventeen cases were classified as interstitial deletions with the following breakpoints: q13q21 in 12 cases, q13q22 in 1 case, q14q22 in 2 cases, q12q21 in 2 cases (Fig1). Based on chromosome morphology, loss of material at bands 13q14-21 was common to all cases with a del(13)(q). Deletion of 13q was present either as the sole abnormality (9 cases) or as part of a complex karyotype (8 cases). In two cases it was associated with trisomy 8 (cases 11 and 12), in 3 cases with a del(5)(q) (cases 4, 9, and 11).
Schematic representation of 13q chromosome interstitial deletions determined by conventional cytogenetics in 17 cases with myeloid malignancies. Black bars show the extent of deletion in each case; dotted lines delimitate the commonly lost region.
Schematic representation of 13q chromosome interstitial deletions determined by conventional cytogenetics in 17 cases with myeloid malignancies. Black bars show the extent of deletion in each case; dotted lines delimitate the commonly lost region.
Translocations involving chromosome 13 were found in 3 cases in which additional karyotypic abnormalities, including a del(5)(q), were observed. Patient 18 investigated at diagnosis, as well as 2 months later, showed a t(12;13)(p13;q14) translocation in 100% of cells in both analyses. Subclones carrying two copies of a der(13)t(12;13) in the presence of a normal 13, or only one der(13)t(12;13) with two normal chromosomes 13, were also identified in this case. A three-way translocation, ie, t(2;12;13)(p14;q24;q13) was found in patient 19. The karyotype of case 20 was characterized by complex chromosomal rearrangements including a der(7)tas(7;13)(7pterqter13qterq14), a der(13)t(8;13)(21;q14), and multiple unidentified markers.
FISH studies.
Results of FISH studies showing the presence or absence of hybridization signals of the applied probes for 13q13.1-q14.3 are summarized in Table 3. Occurrence of del(13)(q) was confirmed by FISH in all 17 cases. Analysis of the deleted regions in these cases allowed for the identification of a commonly deleted region that was proximally flanked by the YAC 833A2 (cases 1 and 8) and distally by the YAC 854D4 (cases 1, 4, 6, 11, 16, and 17). This region corresponds to bands q13.3-q14.3 (Table 2) and is covered by 875A8, 911F6, 937C7, LSI/RB1, 745E3, 935G2. FISH performed in three cases with translocations involving 13q yielded some unexpected results. The hybridization pattern of 13q probes in the two cases with balanced translocations involving 13q is shown in Table 3. In case 18 with a t(12;13)(p13;q14), breakpoints were flanked by 911F6 proximally and 935G2 distally (Fig 2), but the 937C7, RB1, and 745E3 probes hybridized only to the normal chromosome 13. Moreover, both copies of a der(13)t(12;13), present in one of the subclones in this case showed the same hybridization pattern with the applied probes. The chromosome 13 breakpoint in the t(2;12;13)(p14;q24;q13) found in case 19 could not be determined (see Table 3), but loss of one hybridization signal from all applied YACs was observed.
Results of FISH Experiments on 20 Myeloid Malignancies With Deletions or Translocations of Chromosome 13q
| DNA Probes . | Cases . | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deletions . | Translocations . | |||||||||||||||||||
| 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | 19 . | 20 . | |
| 951A3 | ND | − | ND | + | − | ND | − | ND | + | + | ND | ND | − | ND | − | − | ND | ND | ND | + |
| 765A10 | ND | − | − | + | − | ND | − | ND | + | + | ND | ND | − | ND | − | − | ND | ND | ND | ND |
| 833A2 | + | − | ND | − | − | − | − | + | − | − | − | ND | − | ND | − | − | − | + | − | + |
| 875A8 | ND | − | ND | − | − | ND | − | − | ND | − | ND | ND | − | ND | ND | − | − | + | − | + |
| 911F6 | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | − | ND | ND | ND | ND | + | ND | ND |
| 937C7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LSI13/RB1 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 745E3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 935G2 | − | ND | ND | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | − | − | +* | − | ND |
| 854D4 | + | ND | ND | + | − | + | − | − | ND | − | + | ND | − | ND | − | + | + | ND | − | +* |
| DNA Probes . | Cases . | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deletions . | Translocations . | |||||||||||||||||||
| 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 16 . | 17 . | 18 . | 19 . | 20 . | |
| 951A3 | ND | − | ND | + | − | ND | − | ND | + | + | ND | ND | − | ND | − | − | ND | ND | ND | + |
| 765A10 | ND | − | − | + | − | ND | − | ND | + | + | ND | ND | − | ND | − | − | ND | ND | ND | ND |
| 833A2 | + | − | ND | − | − | − | − | + | − | − | − | ND | − | ND | − | − | − | + | − | + |
| 875A8 | ND | − | ND | − | − | ND | − | − | ND | − | ND | ND | − | ND | ND | − | − | + | − | + |
| 911F6 | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | − | ND | ND | ND | ND | + | ND | ND |
| 937C7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LSI13/RB1 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 745E3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 935G2 | − | ND | ND | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | − | − | +* | − | ND |
| 854D4 | + | ND | ND | + | − | + | − | − | ND | − | + | ND | − | ND | − | + | + | ND | − | +* |
Abbreviations: ND, not done; −, deletion; +, two signals.
Two signals of which one is relocated in the translocation partner chromosome.
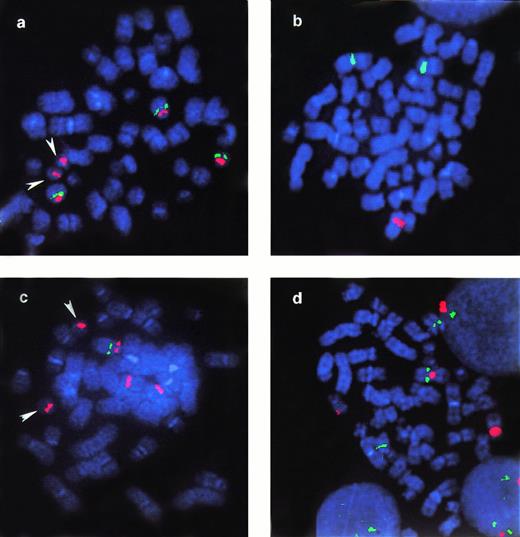
Examples of FISH analysis performed in case 18. Arrowheads in a and c indicate normal chromosome 21 that cohybridize with pUC 1.76 probe for the centromere of chromosome 13. Probes applied: (a) pUC 1.76 (cen 13/21) (red) and 911F6 (green). The picture shows a metaphase with two copies of normal chromosome 13 and one der(13): YAC 911F6 hybridizes on both normal 13 and on the der(13); (b) pBR-12 (cen-12) (green) and RB1 (red). Only one red signal, corresponding to RB1 gene, is kept on normal 13; (c) pUC 1.76 (red) and 937C7 (green). This metaphase represents the subclone with one normal 13 and two copies of der(13): YAC 937C7 is only mantained on normal 13; (d) pBR-12 (red) and 935G2 (green) in a metaphase with two copies of normal 13 and one der(13): YAC935G2 is present in three copies, on the two normal 13 and on the der(12).
Examples of FISH analysis performed in case 18. Arrowheads in a and c indicate normal chromosome 21 that cohybridize with pUC 1.76 probe for the centromere of chromosome 13. Probes applied: (a) pUC 1.76 (cen 13/21) (red) and 911F6 (green). The picture shows a metaphase with two copies of normal chromosome 13 and one der(13): YAC 911F6 hybridizes on both normal 13 and on the der(13); (b) pBR-12 (cen-12) (green) and RB1 (red). Only one red signal, corresponding to RB1 gene, is kept on normal 13; (c) pUC 1.76 (red) and 937C7 (green). This metaphase represents the subclone with one normal 13 and two copies of der(13): YAC 937C7 is only mantained on normal 13; (d) pBR-12 (red) and 935G2 (green) in a metaphase with two copies of normal 13 and one der(13): YAC935G2 is present in three copies, on the two normal 13 and on the der(12).
In case 20, YACs 951A3, 833A2, and 875A8 hybridized to the normal chromosome 13 and to the two copies of the marker defined as mar1 (Fig3). The 854D4 probe gave a signal on the normal chromosome 13 and on the telomeric region of der(7), while 937C7, RB1, and 745E3 probes hybridized only to the normal chromosome 13. These results showed that chromosome 13 was involved in the origin of mar1 and indicated the occurrence of cryptic deletion of 13q in this case. Further FISH studies with a chromosome 13/21 centromeric probe and two-color chromosome painting confirmed the presence of chromosome 13 material on the distal fragment of the der(7), as well as on the proximal region of the two copies of mar1. Chromosome 8 material was detected not only in the der(13)t(8;13)(q21;q14), but also in two copies of the mar1 definitely identified as der(13)t(8;13)(q11;q14) (Fig 3).
Graphical representation of structural rearrangements involving chromosome 13 in case 20 and summary of FISH analysis. Chromosome 7 material is shown in red, chromosome 13 in yellow, and chromosome 8 in green. Order of the applied YACs is indicated next to the normal chromosome 13.
Graphical representation of structural rearrangements involving chromosome 13 in case 20 and summary of FISH analysis. Chromosome 7 material is shown in red, chromosome 13 in yellow, and chromosome 8 in green. Order of the applied YACs is indicated next to the normal chromosome 13.
DISCUSSION
We report results of FISH studies performed on 20 cases of myeloid disorders characterized by deletions (17 cases) or translocations (3 cases) involving chromosome 13. Hematologic diagnoses in the 17 cases with deletions were heterogeneous, including AML, MDS, as well as Ph-positive CML and Ph-negative MPD. Cytogenetic findings were strikingly overlapping (Fig 1), and previous associations between specific breakpoints and distinct myeloid disorders were not confirmed.1 These data suggest that the molecular event(s) undergoing 13q deletions are common to acute and chronic myeloid disorders. Whether a del(13)(q) is related to initiation or progression of myeloid malignancies is unknown. However, the finding of this abnormality in a subclone characterized by a del(5)(q) (case 9), as well as during the course of disease (cases 6 and 16), suggests that loss of 13q material was a secondary event. Cytogenetic results were confirmed by FISH analysis in the 17 cases with cytogenetically discernible 13q deletions. The large region extending from band q13.3 to q14.3 and flanked by YAC 833A2 and YAC 854D4 was lost in all cases.
Microdeletions accompanying translocations affecting the long arm of chromosome 13 found by FISH in three cases (18, 19, and 20) could not be identified by chromosome analysis alone. Similar cryptic deletions of chromosome 13 involved in translocations have been reported in CLL.12,13,17,22 Brown et al13 showed the absence of the D13S25 marker either in the der(13) or in the der(11) contained in hybrids derived from CLL cells with a t(11;13)(p13;q14). Hawthorn et al22 also identified a small region of loss around the D13S25 marker in hybrids derived from a CLL with t(12;13). Interestingly, another chromosomal region undergoing deletions in myeloid malignancies, namely 12p, showed a number of cryptic microdeletions associated with translocations.23 FISH studies of these microdeletions may be highly informative in the molecular mapping of putative tumor suppressor loci. Ours is the first report on cryptic deletions in myeloid cases with only evidence of translocations involving 13q by conventional cytogenetics. Moreover, it appears from present data that loss of 13q material, in addition to translocation, is a consistent event in myeloid malignancies, as it was found in all three cases analyzed in this molecular cytogenetic study.
Two cases of this study deserve a comment because of peculiar features. First, the case of MDS with a t(12;13)(p13;q14) was the most informative because YAC 937C7, the RB1, and YAC 745E3 were exclusively deleted. These results allowed us to define the smallest deleted region common to all cases of this myeloid series to the genomic area corresponding to YAC 937C7, the RB1 gene, and the YAC 745E3.
Second, case 20 provided us with an interesting observation about chromosomal mechanisms underlying involvement of putative suppressor gene(s) in 13q deletions of myeloid malignancies. Despite a partial polysomy of 13q, due to the presence of unbalanced translocations in the karyotype (Fig 3), the three probes, which identified the critical region of loss in all of our myeloid disorders (937C7, LSI/RB1, and 745E3), were maintained only on the normal chromosome 13. Conversely, all of the other analyzed YACs were present in two (854D4) or three (951A3, 833A2, and 875A8) copies, being retained in at least one of the 13q markers derived from the unbalanced translocations. Loss of the region covered by YAC 937C7, LSI/RB1, and YAC 745E3 thus appears to be a critical event in malignant myeloid cells. This large region includes the smallest 13q segment lost in CLL, which is limited by RB1 and D13S25 marker.18
It is a subject of recent controversy if in CLL a proximal region, namely 13q12.3, may be lost in addition to the more terminal one.24 25 Whether the same region is also involved in myeloid disorders remains to be determined. Neverthless, the large extent of lost material shown in this study suggests that more than one suppressor gene may be involved in 13q deletions of myeloid malignancies.
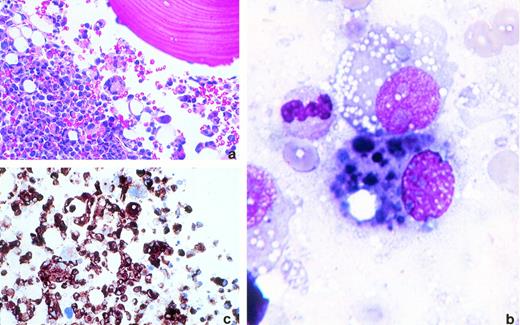
Melanomatous bone marrow invasion. Bone marrow biopsy specimen and aspirate from a patient with disseminated malignant melanoma. (a) Bone marrow biopsy specimens. The bone marrow is almost completely replaced by sheets of neoplastic cells with pleomorphic nuclei, prominent nucleoli, and abundant pink cytoplasm. Some malignant cells display prominent vacuolization (hematoxylin and eosin, original magnification × 200). (b) Bone marrow aspirate. Myeloid precursors containing multiple, irregular, darkly pigmented, melanin-containing vacuoles (Wright's stain, original magnification × 1,000). (c) Bone marrow biopsy specimen. The neoplastic cells are srongly reactive to HMB45 (immunoperoxidase, original magnification × 400). (Courtesy of Niyati Bhagwati, MD, Rommel Seno, MD, and Leslie Oleksowicz, MD, Montefiore Hospital and The Albert Einstein Cancer Center, 111 E 210th St, Bronx, NY 10467.)
Melanomatous bone marrow invasion. Bone marrow biopsy specimen and aspirate from a patient with disseminated malignant melanoma. (a) Bone marrow biopsy specimens. The bone marrow is almost completely replaced by sheets of neoplastic cells with pleomorphic nuclei, prominent nucleoli, and abundant pink cytoplasm. Some malignant cells display prominent vacuolization (hematoxylin and eosin, original magnification × 200). (b) Bone marrow aspirate. Myeloid precursors containing multiple, irregular, darkly pigmented, melanin-containing vacuoles (Wright's stain, original magnification × 1,000). (c) Bone marrow biopsy specimen. The neoplastic cells are srongly reactive to HMB45 (immunoperoxidase, original magnification × 400). (Courtesy of Niyati Bhagwati, MD, Rommel Seno, MD, and Leslie Oleksowicz, MD, Montefiore Hospital and The Albert Einstein Cancer Center, 111 E 210th St, Bronx, NY 10467.)
ACKNOWLEDGMENT
We are thankful to Magda Dehaen and the technicians from the Center for Human Genetics for their expert technical assistance.
Supported in part by “Comitato per la vita Daniele Chianelli,” Pezugio, Italy; and by BIOMED, BMH1-CT94-1703 from the European Community.
Address reprint requests to C. Mecucci, MD, PhD, Hematology, Policlinico Monteluce, via Brunamonti, 06123 Perugia, Italy.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal