Abstract
Systemic release of tumor necrosis factor (TNF) and lymphotoxin-α (LTα) has been found to contribute to the severity of non-Hodgkin's lymphoma (NHL). We investigated whether genetic polymorphisms in the TNF locus, previously shown to influence TNF and LTα genes expression, might contribute to these cytokines production and to the clinical course of NHL. Genomic DNA from 273 lymphoma patients was typed for TNF (−308) polymorphism using an allele-specific polymerase chain reaction (PCR) and for LTα (+252) polymorphism with a PCR-based restriction fragment length polymorphism. The presence of the TNF allele involved in increased TNF gene transcription was associated with higher plasma levels of this cytokine at the time of lymphoma diagnosis (χ2 test, P = .013). An extended haplotype analysis showed that the presence of at least two TNF or LTα high-producer alleles constituted a risk factor for first-line treatment failure (χ2 test, P = .021), shorter progression-free survival (log-rank test, P = .0007), and overall survival (log-rank test, P = .012). In the subgroup of 126 patients with diffuse large-cell lymphoma, the presence of two or more TNF/LTα high producing alleles contributed significantly to a higher rate of relapse and progression (log-rank test, P = .045 and P = .027). In multivariate Cox regression models including the variables of the International Prognostic Index, the TNF/LTα haplotype status was found to be an independent risk factor for progression-free survival (relative risk 2.33, 95% confidence interval [1.17 to 4.64], P = .0053) and overall survival (relative risk 1.92, 95% confidence interval [0.63 to 5.80],P = .081) of large-cell lymphoma patients. These results indicate that genetic polymorphism leading to increased TNF production influences the outcome of NHL and suggest a pathophysiological role for the genetic control of the immune response in lymphoid malignancies.
TUMOR NECROSIS FACTOR (TNF) and lymphotoxin-α (LTα) (formerly known as TNF-β) have been identified to play an important role in the development and in the function of normal lymphoid tissue.1-3 TNF is also known to be one of the earliest cytokine produced in inflammatory processes, generating a cytokine cascade that includes the production of interleukin-1, interleukin-6, and other mediators, as well as TNF itself.4,5 It has also been shown that patients with malignant lymphomas have high circulating levels of both cytokines and that higher plasma levels of TNF are associated with poor disease outcome.6,7 Several studies showed that excessive TNF production may influence the host status, including weight loss, cachexia, modification of the immune response, and anemia, therefore hampering the patient ability to tolerate his tumor and his treatment; other experimental data indicate that TNF may promote the growth of certain lymphoid cells.6-10
The TNF and LTα genes lie within a 7-kb DNA locus in the class III region of the major histocompatibility complex, telomeric to the class II and centromeric to the class I region. Until now, four polymorphic sites in the promoter of TNF gene and one polymorphic site within the first intron of LTα gene have been described.11-14 A polymorphism that directly affects TNF expression in vitro was located at nucleotide position −308. This genetic variation results in two allelic forms, in which the presence of guanine (G) defines the common variant TNF1 and the presence of adenine (A) defines the less common variant TNF2.11 Interestingly, the presence of TNF1 allele defines a 10-bp sequence homologous to the consensus binding site of activator protein-2 (AP-2), which is disrupted in the TNF2 variant.15 Functional assays demonstrated that AP-2 can repress the activity of the TNF promoter in Jurkat T-cell line, suggesting that the −308 polymorphism influences the TNF gene expression.16 Transfection studies in human B-cell lines showed that the presence of TNF2 allele results in higher constitutive and inducible levels of TNF expression compared with TNF1 allele, confirming the importance of this site in the transcriptional regulation of the TNF gene.17
A polymorphism that affects LTα expression was found in the first intron of the gene at nucleotide position +252.14 Because this genetic variation results in the disappearance of a Nco I restriction site by replacing A by G, both allelic forms are referred to, respectively, as LTα (5.5 kB) and LTα (10.5 kB). This LTα +252 polymorphism, conserved in both human and mice, is located within a phorbol ester-responsive DNA element (TRE) with high affinity for the AP-1, jun, and c-fos heterodimer transcription factor family. The presence of LTα (5.5 kB) allele was shown to result in significantly higher LTα production by phytohemagglutinin-stimulated peripheral blood mononuclear cells due to increased LTα gene transcription.14
Several studies have demonstrated the linkage disequilibrium between both TNF (−308) and LTα (+252) polymorphic sites and with other allelic markers within the cluster of HLA gene.11,12,18Both TNF2 and LTα (5.5 kB) high-producer alleles are linked with the extended haplotype HLA A1-B8-DR3-DQ2, which was found to be associated with autoimmunity and rapid progression of HIV infection.11,18,19 In patients with infectious and autoimmune disorders, different studies reported a higher frequency of the TNF2 allele in cases with severe disease, suggesting that genetic variations within the TNF locus may be functionally relevant in vivo.20-23
In the present study, we report that the genetic background inducing increased TNF production influences treatment outcome, progression-free survival, and overall survival of non-Hodgkin's lymphoma (NHL) patients. These findings emphasize the contribution of innate immunity in the pathophysiology of malignant disorders.
MATERIALS AND METHODS
Subjects.
The study comprised 273 patients with NHL and 96 unrelated healthy controls who provided samples available for genetic analysis after informed consent. Patients positive for human immunodeficiency virus (HIV) testing were excluded from this study. The plasma of 85 of these patients, without active infections and without any history of autoimmune disease or steroid treatment, was tested at the time of lymphoma diagnosis for TNF and LTα levels (Medgenix Diagnostics [Fleurus, Belgium] and R & D Systems [Minneapolis, MN], respectively). All the specimens used in the study were coded, and the patients' confidentiality was preserved according to the guidelines for studies of human subjects.
Pathological confirmation of the diagnosis and clinical history were available in every case. Histological diagnoses were classified according to the International Lymphoma Study Group (Revised European American Lymphoma classification).24 The initial medical evaluation consisted of a complete history and physical examination; chest radiographic examination; computed tomographic scan of the chest, abdomen, and pelvis; and blood chemistry. The extent of disease and presence of B symptoms were categorized according to the Ann Arbor classification, and performance status was assessed using criteria of the Eastern Cooperative Oncology Group (ECOG). Clinical characteristics of the patients are shown in Table 1.
Characteristics of 273 Patients With NHL
| Characteristic . | No. (%) . |
|---|---|
| Sex | |
| Male | 133 (49) |
| Female | 140 (51) |
| Age | |
| ≤60 yrs | 161 (59) |
| >60 yrs | 112 (41) |
| Performance status (ECOG score) | |
| <2 | 216 (79) |
| ≥2 | 57 (21) |
| Ann Arbor stage | |
| I, II | 73 (27) |
| III, IV | 200 (73) |
| No. of extranodal sites | |
| <2 | 190 (70) |
| ≥2 | 83 (30) |
| Serum LDH level | |
| ≤1 × normal | 146 (55) |
| >1 × normal | 119 (45) |
| Unknown | 8 |
| Serum β2-microglobulin level | |
| ≤3.0 mg/mL | 168 (66) |
| >3.0 mg/mL | 86 (34) |
| Unknown | 19 |
| Serum albumin level | |
| ≥35.0 g/L | 215 (85) |
| <35.0 g/L | 38 (15) |
| Unknown | 20 |
| B symptoms | |
| Absent | 205 (75) |
| Present | 68 (25) |
| Histology | |
| Lymphocytic/lymphoplasmocytoid | 22 |
| Follicular | 96 |
| Mucosal-associated (MALT) | 2 |
| Marginal zone | 7 |
| Mantle cell | 18 |
| Diffuse large cell | 126 |
| Burkitt cell | 2 |
| Characteristic . | No. (%) . |
|---|---|
| Sex | |
| Male | 133 (49) |
| Female | 140 (51) |
| Age | |
| ≤60 yrs | 161 (59) |
| >60 yrs | 112 (41) |
| Performance status (ECOG score) | |
| <2 | 216 (79) |
| ≥2 | 57 (21) |
| Ann Arbor stage | |
| I, II | 73 (27) |
| III, IV | 200 (73) |
| No. of extranodal sites | |
| <2 | 190 (70) |
| ≥2 | 83 (30) |
| Serum LDH level | |
| ≤1 × normal | 146 (55) |
| >1 × normal | 119 (45) |
| Unknown | 8 |
| Serum β2-microglobulin level | |
| ≤3.0 mg/mL | 168 (66) |
| >3.0 mg/mL | 86 (34) |
| Unknown | 19 |
| Serum albumin level | |
| ≥35.0 g/L | 215 (85) |
| <35.0 g/L | 38 (15) |
| Unknown | 20 |
| B symptoms | |
| Absent | 205 (75) |
| Present | 68 (25) |
| Histology | |
| Lymphocytic/lymphoplasmocytoid | 22 |
| Follicular | 96 |
| Mucosal-associated (MALT) | 2 |
| Marginal zone | 7 |
| Mantle cell | 18 |
| Diffuse large cell | 126 |
| Burkitt cell | 2 |
Treatment.
Therapeutic attitudes were defined according to the initial histological subtypes and prognostic factors. Among the 126 patients with diffuse large-cell lymphoma, 53 received CHOP (cyclophosphamide, doxorubicin, vincristin, prednisone) or CHOP-like regimen, 66 received ACVBP (doxorubicin, cyclophosphamide, vindesine, bleomycin, prednisone, intrathecal metrotexate) or ACVBP-like high-dose regimen, and 7 older patients received multiple drug combination chemotherapy without anthracyclin. All patients were treated according to prospective trials, except for 3 that were referred to the department at the time of their first progression. A complete treatment response was defined as the disappearance of all disease manifestations and normalization of all laboratory values. Progression-free survival was calculated from onset of treatment until relapse, disease progression, or last follow-up evaluation. Freedom from relapse survival was equal to progression-free survival for patients that achieved a complete remission after their first-line treatment. Overall survival was measured as the time between the beginning of treatment and death or the date of the last follow-up evaluation.
Genotyping analysis.
Genomic DNA from peripheral blood mononuclear cells or lymph nodes was extracted with the QIAmp kit (QIAGEN Inc, Chatsworth, CA) or by the standard phenol-chloroform method. The primer pairs F1(5′TCTCGGTTTCTTCTCCATCG) and R1 (5′ ATAGGTTTTGAGGGGCATGG) and F1-R2(5′ATAGGTTTTGAGGGGCATGA) were used to amplify a 184-bp fragment of the TNF gene, which includes the polymorphic site at the nucleotide position −308. Each sample was tested with both primers pairs (F1-R1 and F1-R2). Primer pair F1-R3(5′GAGTCTCCGGGTCAGAATGA) was used to amplify a TNF gene's fragment of 531 bp as an internal control in the allele-specific polymerase chain reaction (ASPCR) and as a genomic DNA template for sequencing. Primer R3 was also used as a competitor for the primer pairs F1-R1 and F1-R2 to improve the specificity of the ASPCR assay25 (Fig 1A). After heating at 95°C for 10 minutes, PCR reactions were performed for 31 cycles consisting of heat denaturation (95°C for 90 seconds), annealing for 150 seconds (62°C for primer pair F1-R1and 60°C for primer pair F1-R2 ), and extension (72°C for 60 seconds).
Identification of the TNF (−308) gene polymorphism with ASPCR (A) and LTα (+252) gene polymorphism with PCR-RFLP (B). Genotypes were identified as homozygous, TNF1/1 or TNF2/2 if the single 184-bp fragment appeared exclusively in the F1-R1 or F1-R2 primer sets' reactions (A, lanes 1 and 2 and 3 and 4, respectively). Genotypes were identified as heterozygous, TNF1/2, if the 184-bp fragment appeared in both F1-R1 and F1-R2 primer sets' reactions (A, lanes 5 and 6). The 531-bp fragment, amplified with F1-R3primers, served as an internal control and competitor for the ASPCR. Lanes 1 through 3 on the (B) show the LTα (+252) polymorphic genotypes. The 368-bp fragment cleaved with Nco I (133- and 235-bp fragments) represented the LTα (5.5) allele, and that not cleaved represented the LTα (10.5) allele. Genotypes were identified as homozygous for LTα (10.5/10.5) if the single 368-bp fragment appeared (B, lane 1) and homozygous for LTα (5.5/5.5) if two 133- and 235-bp fragments were present (B, lane 2). The presence of noncleaved (368 bp) and cleaved fragments (133 and 235 bp) in the same sample identified the heterozygous genotype, LTα (10.5/5.5) (B, lane 3).
Identification of the TNF (−308) gene polymorphism with ASPCR (A) and LTα (+252) gene polymorphism with PCR-RFLP (B). Genotypes were identified as homozygous, TNF1/1 or TNF2/2 if the single 184-bp fragment appeared exclusively in the F1-R1 or F1-R2 primer sets' reactions (A, lanes 1 and 2 and 3 and 4, respectively). Genotypes were identified as heterozygous, TNF1/2, if the 184-bp fragment appeared in both F1-R1 and F1-R2 primer sets' reactions (A, lanes 5 and 6). The 531-bp fragment, amplified with F1-R3primers, served as an internal control and competitor for the ASPCR. Lanes 1 through 3 on the (B) show the LTα (+252) polymorphic genotypes. The 368-bp fragment cleaved with Nco I (133- and 235-bp fragments) represented the LTα (5.5) allele, and that not cleaved represented the LTα (10.5) allele. Genotypes were identified as homozygous for LTα (10.5/10.5) if the single 368-bp fragment appeared (B, lane 1) and homozygous for LTα (5.5/5.5) if two 133- and 235-bp fragments were present (B, lane 2). The presence of noncleaved (368 bp) and cleaved fragments (133 and 235 bp) in the same sample identified the heterozygous genotype, LTα (10.5/5.5) (B, lane 3).
A substitution of A to G at the nucleotide position +252 of LTα gene creates a Nco I restriction fragment length polymorphism (RFLP). With a PCR-based RFLP assay, DNA samples were analyzed forNco I restriction site that is present in the LTα (5.5 kB) allele but not in the LTα (10.5 kB) allele. PCR-amplified products of 368 bp were obtained with the use of forward (5′CTCCTGCACCTGCTGCCTGGATC) and reverse (5′GAAGAGACGTTCAGGTGGTGTCAT) primers. After heating at 95°C for 10 minutes, PCR reactions were performed for 31 cycles consisting of heat denaturation (96°C for 60 seconds), annealing (65°C for 60 seconds), and extension (72°C for 120 seconds). Then, PCR-amplified products were directly digested with 3 U of restriction enzyme. The band cleaved with Nco I (133- and 235-bp fragments) represented the LTα (5.5 kB) allele, and the noncleaved (368 bp) band represented the LTα (10.5 kB) allele (Fig 1B).
To confirm the accuracy of the ASPCR and PCR-RFLP assays, amplification products from 3 individuals homozygous for each given allele (n = 12 in total) were purified from the gel, ligated into pGEM-T vector (Promega, Madison, WI) and subcloned. Recombinant plasmid DNAs were sequenced using Sequenase (Amersham, Braunchweig, Germany) with Cy5-labeled primers and analyzed on an automated laser fluorescent sequencer (ALF-Express; Pharmacia Biotech, Uppsala, Sweden). Reproducibility of genotyping was obtained in every case.
Statistical analysis.
Distribution and allele frequency and their associations with other variables were compared using the Yates corrected χ2test. The progression-free survival and overall survival of the patients were estimated by the Kaplan-Meier method, and statistical differences were assessed using the log-rank test. Multivariate regression analysis using the Cox model was performed to assess the influence of TNF/LTα polymorphic haplotype status on lymphoma outcome along with prognostic variables validated by the International Prognostic Index.26 Statistical tests with a Pvalue smaller than 5% were considered significant in the whole population. Statistical analysis were performed using the BMDP package (Statistical Software, Los Angeles, CA).
RESULTS
Distribution and allele frequency of TNF(−308) and LTα (+252) polymorphisms.
Distribution and allele frequency of TNF (−308) and LTα (+252) polymorphisms among the 273 lymphoma patients were similar to those observed in healthy controls (Table 2). In both populations, an expected association was found between the presence of allele TNF1 with LTα (10.5) and allele TNF2 with LTα (5.5) (χ2 test, P < .0001). This translated into a strong genotype association between the TNF1/1 and LTα (10.5/10.5), TNF1/2 and LTα (10.5/5.5), and TNF2/2 with LTα (5.5/5.5) haplotypes (not shown).
Distribution and Allele Frequency of TNF (−308) and LTα (+252) Polymorphisms Among 96 Healthy Controls and 273 Patients With NHL, Including 85 Patients With Available Cytokines' Plasma Levels at the Time of Diagnosis
| . | Allele Frequency . | Genotype Distribution . | |||
|---|---|---|---|---|---|
| TNF1 . | TNF2 . | TNF1/1 . | TNF1/2 . | TNF2/2 . | |
| Controls | 0.84 | 0.16 | 69 (72%) | 24 (25%) | 3 (3%) |
| Patients | 0.86 | 0.14 | 203 (74%) | 65 (24%) | 5 (2%) |
| TNF ≤33.0 pg/mL | 0.94 | 0.06 | 38 (90%) | 3 (7%) | 1 (2%) |
| TNF >33.0 pg/mL | 0.83 | 0.17 | 28 (65%) | 15 (35%) | 0 |
| LTα (10.5) | LTα (5.5) | LTα (10.5/10.5) | (10.5/5.5) | (5.5/5.5) | |
| Controls | 0.70 | 0.30 | 48 (50%) | 38 (40%) | 10 (10%) |
| Patients | 0.71 | 0.29 | 140 (51%) | 110 (40%) | 23 (9%) |
| LTα ≤10.0 pg/mL | 0.73 | 0.27 | 22 (53%) | 17 (40%) | 3 (7%) |
| LTα >10.0 pg/mL | 0.66 | 0.34 | 19 (44%) | 19 (44%) | 5 (12%) |
| . | Allele Frequency . | Genotype Distribution . | |||
|---|---|---|---|---|---|
| TNF1 . | TNF2 . | TNF1/1 . | TNF1/2 . | TNF2/2 . | |
| Controls | 0.84 | 0.16 | 69 (72%) | 24 (25%) | 3 (3%) |
| Patients | 0.86 | 0.14 | 203 (74%) | 65 (24%) | 5 (2%) |
| TNF ≤33.0 pg/mL | 0.94 | 0.06 | 38 (90%) | 3 (7%) | 1 (2%) |
| TNF >33.0 pg/mL | 0.83 | 0.17 | 28 (65%) | 15 (35%) | 0 |
| LTα (10.5) | LTα (5.5) | LTα (10.5/10.5) | (10.5/5.5) | (5.5/5.5) | |
| Controls | 0.70 | 0.30 | 48 (50%) | 38 (40%) | 10 (10%) |
| Patients | 0.71 | 0.29 | 140 (51%) | 110 (40%) | 23 (9%) |
| LTα ≤10.0 pg/mL | 0.73 | 0.27 | 22 (53%) | 17 (40%) | 3 (7%) |
| LTα >10.0 pg/mL | 0.66 | 0.34 | 19 (44%) | 19 (44%) | 5 (12%) |
Cytokines' plasma levels equal to or lower versus higher than median values observed in NHL patients.
When analysis was restricted to the 85 lymphoma patients available for both enzyme-linked immunosorbent assay (ELISA) and genotyping assays at the time of diagnosis, the frequency of allele TNF2 was found to be higher among the patients with TNF higher plasma levels than the median value compared with those with TNF plasma levels below this limit (0.17v 0.06, χ2 test, P = .013). Frequency of allele LTα (5.5) was also found to be higher in patients with TNF higher levels (not shown, P = .037). No association was found between the allelic frequency within LTα (+252) polymorphism or TNF (−308) and LTα initial plasma levels in lymphoma patients (Table 2).
TNF/LTα polymorphic haplotype status in lymphoma patients.
The strong association between TNF (−308) and LTα (+252) polymorphic alleles and the possible contribution of both polymorphic sites in increased production of TNF led us to analyze these genetic markers as haplotypes rather than as individual alleles in a predictive model for the patients' outcome. The low-risk haplotype denoted therefore the presence of less than two alleles associated with increased TNF or LTα production, including the haplotypes typed as TNF1/1 and LTα (10.5/10.5), TNF1/1 and LTα (5.5/10.5), or TNF1/2 and LTα (10.5/10.5). The high-risk haplotype denoted the presence of two or more alleles associated with increased cytokines' production, including haplotypes typed as TNF1/2 and LTα (5.5/10.5), TNF1/1 and LTα (5.5/5.5), TNF2/2 and LTα (10.5/10.5), TNF1/2 and LTα (5.5/5.5), TNF2/2 and LTα (5.5/10.5), or TNF2/2 and LTα (5.5/5.5).
The differences in the frequency of the TNF/LTα polymorphic extended haplotypes between the case and the control group were not statistically significant (not shown). No statistically significant association was found between the presence of a given TNF/LTα haplotype status and prognostic variables such as age, performance status, disease stage, B symptoms, the number of extranodal sites of disease, serum levels of lactate dehydrogenase (LDH), albumin, and β2-microglobulin or with the categories of the international prognostic index (not shown).
Although there were no statistically significant differences in TNF haplotype distribution when all lymphoma histological subtypes were considered, there were less patients with follicular lymphoma carrying the high-risk haplotype (19% high-risk v 81% low-risk) when compared with all other histology (31% high-risk v 69% low-risk; χ2 test, P = .02) or to the large-cell lymphoma subgroup (33% high-risk v 67% low-risk; P = .023). Haplotype distribution in the large-cell lymphoma subgroup was not different from the remaining patients with other subtypes than follicular lymphoma (P = .9) or with the group of healthy controls (69% v 31%, P = .75).
Outcome of lymphoma patients according to the risk groups defined by TNF/LTα polymorphic haplotype status.
Among 273 NHL patients, 161 (61%) achieved a complete response to treatment, whereas 103 (39%) did not. Nine patients were not yet available for therapy response. Of 273 patients, 153 (56%) patients have experienced disease progression, whereas 120 (44%) have not, and 87 (32%) patients died. The median follow-up for the patients remaining alive was 28 months.
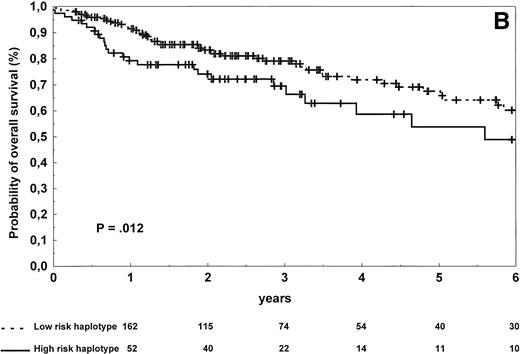
The patients carrying low-risk haplotype achieved a complete response to first-line therapy more frequently than the patients with high-risk haplotype (66% v 49%; χ2 test, P = .021). The estimated 2-year and 4-year progression-free survival rates in the high-risk and low-risk groups were, respectively, 46% and 23% versus 62% and 43% (log-rank test, P = .0007; Table 3 and Fig2A). The estimated 2-year and 4-year overall survival rates in the groups of patients carrying high-risk and low-risk haplotypes were, respectively, 74% and 59% versus 83% and 72% (log-rank test,P = .012; Table 3 and Fig 2B). A higher rate of relapse from complete remission was also observed in patients with the high-risk haplotype (log-rank test, P = .027).
Outcome of NHL Patients According to Risk Groups Defined by TNF/LT Polymorphic Haplotype Status
| Risk Group . | No. of High-Producer Alleles . | Distribution of Patients (%) . | Complete Response Rate (%) . | Progression-Free Survival (%) . | Overall Survival (%) . | ||
|---|---|---|---|---|---|---|---|
| 2-yr Rate (SE) . | 4-yr Rate (SE) . | 2-yr Rate (SE) . | 4-yr Rate (SE) . | ||||
| All patients enrolled in the study (n = 273) | 61 | 57.2 ± 3.2 | 37.0 ± 3.8 | 80.8 ± 2.5 | 68.4 ± 3.6 | ||
| Low | 0 or 1 | 197 (72%) | 66 | 61.6 ± 3.7 | 42.7 ± 4.6 | 83.3 ± 2.8 | 71.9 ± 4.1 |
| High | 2-4 | 76 (28%) | 49 | 45.8 ± 6.1 | 23.3 ± 6.3 | 74.1 ± 5.3 | 58.7 ± 7.7 |
| Patients with large cell lymphoma (n = 126) | 69 | 57.4 ± 4.8 | 44.0 ± 6.5 | 69.2 ± 4.6 | 59.6 ± 6.3 | ||
| Low | 0 or 1 | 84 (67%) | 74 | 62.4 ± 5.7 | 55.3 ± 6.9 | 70.4 ± 5.8 | 63.8 ± 6.8 |
| High | 2-4 | 42 (33%) | 61 | 47.7 ± 8.5 | 14.3 ± 11.7 | 67.1 ± 8.0 | 46.7 ± 15.0 |
| Risk Group . | No. of High-Producer Alleles . | Distribution of Patients (%) . | Complete Response Rate (%) . | Progression-Free Survival (%) . | Overall Survival (%) . | ||
|---|---|---|---|---|---|---|---|
| 2-yr Rate (SE) . | 4-yr Rate (SE) . | 2-yr Rate (SE) . | 4-yr Rate (SE) . | ||||
| All patients enrolled in the study (n = 273) | 61 | 57.2 ± 3.2 | 37.0 ± 3.8 | 80.8 ± 2.5 | 68.4 ± 3.6 | ||
| Low | 0 or 1 | 197 (72%) | 66 | 61.6 ± 3.7 | 42.7 ± 4.6 | 83.3 ± 2.8 | 71.9 ± 4.1 |
| High | 2-4 | 76 (28%) | 49 | 45.8 ± 6.1 | 23.3 ± 6.3 | 74.1 ± 5.3 | 58.7 ± 7.7 |
| Patients with large cell lymphoma (n = 126) | 69 | 57.4 ± 4.8 | 44.0 ± 6.5 | 69.2 ± 4.6 | 59.6 ± 6.3 | ||
| Low | 0 or 1 | 84 (67%) | 74 | 62.4 ± 5.7 | 55.3 ± 6.9 | 70.4 ± 5.8 | 63.8 ± 6.8 |
| High | 2-4 | 42 (33%) | 61 | 47.7 ± 8.5 | 14.3 ± 11.7 | 67.1 ± 8.0 | 46.7 ± 15.0 |
Progression-free survival (A) and overall survival (B) of 273 NHL patients according to the risk groups defined by TNF (−308) and LTα (+252) polymorphic haplotype status. The initial number of patients at risk for disease progression or death was 197 for the low-risk haplotype and 76 for the high-risk haplotype. The number of patients remaining at risk is shown below each time point. Pdenotes the log-rank test value.
Progression-free survival (A) and overall survival (B) of 273 NHL patients according to the risk groups defined by TNF (−308) and LTα (+252) polymorphic haplotype status. The initial number of patients at risk for disease progression or death was 197 for the low-risk haplotype and 76 for the high-risk haplotype. The number of patients remaining at risk is shown below each time point. Pdenotes the log-rank test value.
Prognostic significance of TNF/LTα polymorphism according to histological subtypes.
Because large-cell lymphoma and follicular lymphoma patients represented the largest homogeneous categories of patients, we determined the effects of TNF/LTα haplotype status on lymphoma outcome in these two patients' subgroups.
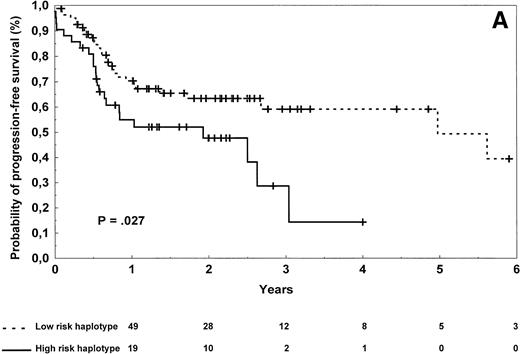
Diffuse large-cell lymphoma patients carrying the high-risk haplotype achieved complete remission after first-line treatment in 61% of the cases versus 74% for those carrying the low-risk haplotype, a difference that was not statistically significant (P = .22). Progression-free survival was significantly shorter in diffuse large-cell lymphoma patients (Table 3 and Fig 3A) carrying the high-risk versus low-risk haplotype (log-rank test, P = .027), but no statistical difference in overall survival was observed (Table 3,P = .27). Of note, patients with the high-risk haplotype also relapsed more frequently from complete remission (log-rank test,P = .045). To evaluate the importance of TNF/LTα haplotype status on lymphoma outcome, a multivariate regression analysis was performed in this subgroup of 126 patients. The Cox model was tested by introducing the TNF/LTα haplotype status along with prognostic variables of the International Prognostic Index, ie, age (≤60 years of age v older), disease stage (I/II v III/IV), performance status (0 or 1 v 2 or more), serum LDH level (normal v abnormal), and number of extranodal sites of disease (0 or 1 v 2 or more). The TNF/LTα polymorphic haplotype status was found to be an independent risk factor for both progression-free survival (relative risk 2.33, 95% confidence interval [1.17 to 4.64], P = .0053; global χ2 = 23.33,P < .0001) and overall survival (relative risk 1.92, 95% confidence interval [0.63 to 5.80], P = .081; global χ2 = 32.52, P < .0001).
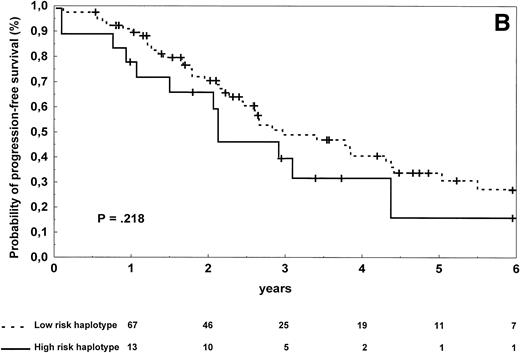
Progression-free survival of 126 patients with diffuse large-cell (A) and 96 patients with follicular (B) lymphoma according to the risk groups defined by TNF (−308) and LTα (+252) polymorphic haplotype status. The initial number of patients at risk for disease progression with diffuse large-cell lymphoma and carrying low-risk haplotype was 84, whereas the number of those with high-risk haplotype was 42. The initial number of patients at risk for disease progression with follicular lymphoma and carrying low-risk haplotype was 78, whereas the number of those with high-risk haplotype was 18. The number of patients remaining at risk is shown below each time point. P denotes the log-rank test value.
Progression-free survival of 126 patients with diffuse large-cell (A) and 96 patients with follicular (B) lymphoma according to the risk groups defined by TNF (−308) and LTα (+252) polymorphic haplotype status. The initial number of patients at risk for disease progression with diffuse large-cell lymphoma and carrying low-risk haplotype was 84, whereas the number of those with high-risk haplotype was 42. The initial number of patients at risk for disease progression with follicular lymphoma and carrying low-risk haplotype was 78, whereas the number of those with high-risk haplotype was 18. The number of patients remaining at risk is shown below each time point. P denotes the log-rank test value.
In follicular lymphoma patients, there were no significant differences in terms of complete response rate (P = .13), progression-free survival (P = .218; Fig 3B), and overall survival (P = .40) according to the TNF/LT haplotype status.
DISCUSSION
We found that in lymphoma patients the presence of the rare TNF2 allele involved in increased TNF gene transcription was associated with higher TNF plasma levels at the time of diagnosis and that the presence of at least two TNF or LTα high-producer alleles constituted an independent risk factor for first-line treatment failure, shorter progression-free survival, and overall survival of the patients. These findings were more significant in the larger subgroup of patients presenting diffuse large-cell lymphoma. In patients with other histological subgroups, this high-risk haplotype was not significantly associated with an adverse outcome, although these observations could be limited by the lower number of patients analyzed.
TNF (−308) and LTα (+252) allelic frequencies and genotype distributions in the present cohort of healthy individuals were similar to those previously reported in different ethnical groups.20 27-29 Because they were also similar in NHL patients, it is unlikely that polymorphic variations in the TNF locus increase the susceptibility for lymphoma occurrence. The presence of two alleles associated with increased TNF production seemed to be less frequently found in follicular lymphoma patients than in other patients or control individuals. Whether follicular lymphoma may develop in a population of patients with this particular genotype in which high-risk alleles are less represented needs to be investigated in larger epidemiological studies.
The results of the present study and those described previously demonstrating increased plasma levels of TNF and LTα in a subset of lymphoma patients with adverse outcome indicate that their excessive production influence the clinical course of the disease.6,7The higher frequency of TNF2 allele in NHL patients with elevated plasma levels of TNF as well as the independent prognostic significance of TNF/LTα polymorphic haplotype status suggest that inherited ability of the host to regulate these cytokines' production contributes to this phenomenon. However, higher plasma levels of TNF and LTα but not their specific polymorphic alleles were also associated in previous studies with numerous variables reflecting increased tumor burden, including high serum values of β2-microglobulin and LDH, advanced disease stage, or large tumor mass.6,7 This suggests that elevated TNF and LTα plasma levels may also originate from their autonomous production by malignant cells. In addition, TNF and LTα activate the transcription factor NF-κB, which, in turn, may further perpetuate their production in an autocrine fashion. Several factors, including proinflammatory cytokines, chemokines, and adhesion molecules, might modulate this amplification loop and thus indirectly influence the overall TNF and LTα expression.5 It seems likely therefore that genetic variability in the TNF locus is important, although not the sole factor that influences TNF and LTα production in lymphoma patients.
These data raise some questions regarding how overproduction of TNF and LTα can influence the clinical course of NHL. It is coherent that individuals who are genetically predisposed to increased TNF and LTα production are at higher risk for chronic immune activation upon tumor challenge, which further results in the presence of systemic symptoms, anemia, hypoalbuminemia, cachexia, and poor performance status.6-9 All of these adverse conditions may affect the ability of the host to tolerate treatment and consequently preclude disease's poor outcome. Increased TNF and LTα levels could also impair the efficiency of antitumor cellular immune response.30,31 TNF has been shown in vitro to promote the differentiation of dendritic cells,2 but its excessive production may hamper their ability for effective antigen presentation.32,33 It has also been demonstrated that increased endogenous TNF production by tumor cells could contribute to the chemotherapeutic drug resistance.34 Finally, TNF and LTα were also shown in some models to stimulate the growth of malignant B cells.10,35 All of these biological observations can contribute to our findings that inherited susceptibility to excessive TNF production increases the risk for lymphoma patients to fail achieving a complete remission and to relapse after remission. However, it is also possible that TNF/LTα haplotype, which is linked to other extended haplotypes such as the HLA A1/2-B8-DR3-DQ2 alleles associated with autoimmunity and HIV disease progression,11,19 is in linkage desequilibrium with other genes that can influence lymphoma outcome. Other studies regarding the influence of TNF polymorphism on the clinical course of autoimmune or infectious diseases have also reported controversial results,20-23 36-39 indicating that a single polymorphism in a given gene cannot probably account for the variability of disease's course.
In conclusion, this report indicates that genetic susceptibility to increased TNF production contributes to the clinical course of malignant lymphomas. Both TNF (−308) and LTα (+252) polymorphic markers may be considered as candidates to design new prognostic models for NHL based on biological factors.40 They may also be helpful in selecting the patients for whom new treatment approaches, especially those based on immunomodulation41 or TNF inhibitors,42 43 could be advised.
ACKNOWLEDGMENT
The authors are indebted to Dr Margaret A. Shipp and Dr Gilbert M. Lenoir for their helpful comments.
Supported by the Hospices Civils de Lyon-PHRC (96.044) and by INSERM (Paris, France). K.W. was supported by a grant founded by the Fondation de France (Paris) and P.R. by the European Society for Medical Oncology (ESMO, Lugano).
Address reprint requests to Gilles Salles, MD, PhD, Service d'Hématologie, Centre Hospitalier Lyon-Sud, 69495 Pierre-Benite Cedex, France; e-mail: gs@hematologie.univ-lyon1.fr.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal