Abstract
Extrahepatic sites capable of supporting hepatitis C virus (HCV) replication have been suggested. We analyzed the influence of virological factors such as viral genotype and viral load, and cellular factors such as cell phenotype, on the detection rate of HCV sequences in hematopoietic cells of infected patients. Thirty-eight chronically infected patients were included in the study: 19 infected by genotype 1 isolates (1a and 1b), 13 by nongenotype 1 isolates (including genotypes 2 a/c, 3a, and 4), and 6 coinfected by genotype 1 and 6 isolates. Polymerase chain reaction (PCR) detection efficiency of viral genomic sequences, both the positive and negative strand RNA, was evaluated using RNA transcripts derived from genotype 1, 2, 3, and 4 cloned sequences and found to be equivalent within one log unit. The serum viral load, ranging from less than 2 × 105 Eq/mL to 161 × 105 Eq/mL, did not influence the detection rate of either strand of RNA in patients' peripheral blood mononuclear cells (PBMCs). Positive and negative strand RNA were found in PBMCs of all 3 cohorts of patients with a detection rate ranging from 15% to 100% and from 8% to 83.3% for the positive and negative strand RNA, respectively. Coinfected patients showed a detection rate in all cases greater than 80%. Patients infected with genotype 1 isolates showed a higher detection rate of either strands of RNA when compared with patients infected with other genotypes (P < .001 andP < .04). Both strands were found restricted to polymorphonuclear leukocytes, monocytes/macrophages, and B (but not T) lymphocytes. These data show that HCV genomic sequences, possibly reflecting viral replication, can be detected in PBMCs of chronically infected patients independent of the viral load and that specific associated cell subsets are implicated in the harboring of such sequences.
INFECTION CAUSED BY hepatitis C virus (HCV), a single-stranded RNA virus belonging to the Flaviviridae family,1 evolves in more than 70% of cases towards a chronic carrier state that is responsible for liver cirrhosis in approximately 20% of patients.2 HCV has also been found to be closely associated with the development of hepatocellular carcinomas.3
At least six different genotypes (1 to 6) and 52 subtypes of the virus have been identified based on differences in the nucleotide sequences (for review see Bukh et al4). Many studies suggest that the course and severity of the disease may depend on the infecting genotype, although it does remain a matter of controversy.5,6 These studies point toward the development of more aggressive liver disease when infection is due to subtype 1b. Infection with this subtype has also repeatedly been found associated with more severe graft injury in patients who have undergone transplantation.7 Moreover, different HCV strains may vary in their responsiveness to interferon therapy, subtypes 1b and possibly 1a being the poorest responders.8 9
HCV, like other hepatitis viruses, is predominantly a hepatotropic virus. Nonetheless, previous work by several groups including ours suggests that HCV genomic sequences, both the positive and negative strand RNA (the presumed replicative intermediate),1 can be detected in peripheral as well as medullar hematopoietic cells, mainly peripheral blood mononuclear cells (PBMCs) from infected patients.10-17 However, very few studies have attempted to further discriminate the cell populations harboring these sequences.18-20 Such studies implicate mainly B cells as a potential reservoir for viral replication. A more recent study has, on the contrary, failed to document any evidence of HCV replication in hematopoietic cells or cells from any tissues other than the liver in human and chimpanzee samples.21 All of these reports illustrate the actual controversies and difficulties in providing clear and specific evidence on the subject of the possible existence of extrahepatic reservoirs capable of supporting HCV replication. A disturbing observation is the wide variation in the detection rates of HCV genomic sequences found in PBMCs of patients observed in the different published studies. Such rates can range from 0% to 100%.10-21 Most of the conclusions reached to date have to be interpreted with caution because they involved a very limited number of patients or because detection techniques used may have been subjected to artifacts resulting in erroneous observations.10 21 In particular, most of these studies used very poorly validated methods for detection of the negative strand RNA. In addition, comparison of the different studies reported has been hampered by the fact that the patient populations analyzed were not well characterized, in particular with respect to the infecting viral load and genotype. Analysis of such factors is important, not only to better define a putative difference in the biology of HCV viruses belonging to different genotypes, but also to allow for adequate comparison of data provided by different studies.
Because total PBMCs represent a heterogeneous cellular population, including lymphocytes and monocytes/macrophages (M/M) that show different functions related to host defense against infection, it is particularly important to gain further insights in the cellular as well as the molecular events involved in the apparent infection of these cells by HCV. In this study, we looked for evidence of HCV genomic sequences (both the positive and the negative strand RNA) in PBMCs in relation to the infecting viral load and genotype, as well as with the phenotype of cell subsets harboring these sequences.
MATERIALS AND METHODS
Patients.
A total of 38 patients, 17 females and 21 males, ages 24 to 75 years (mean 46.9 ± 2.1 years) were studied. All were positive for HCV antibodies as detected by ELISA (third generation) and RIBA III assays (both Ortho Diagnostic Systems Inc, Raritan, NJ). Viral infection was confirmed in all patients by polymerase chain reaction (PCR) detection of HCV RNA in patients' sera.10 Except for 1 case, all had chronic hepatitis (n = 7, with cirrhosis). No evidence of other active viral infection (HIV, HBV, HAV) was documented in these patients. The infection mode was related to blood transfusion, parenteral drug abuse, occupational exposure, or was unknown in 14, 9, 1, and 14 cases, respectively. Three patients were under combination immunosuppressive therapy (steroids and cyclosporines) because of a previous hepatic transplantation (from the group of coinfected patients, see Results). All patients underwent a complete clinical assessment including liver biopsy.
Determination of genotypes.
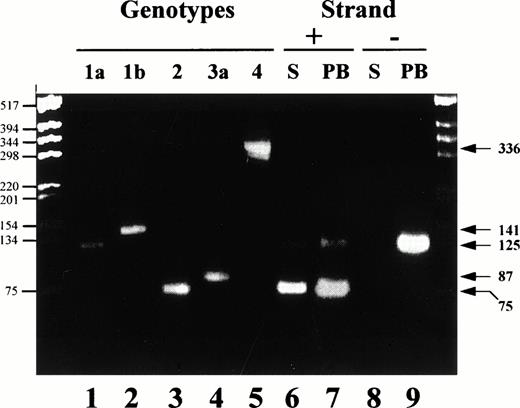
The genotype of infecting strains was analyzed from sera, total PBMCs, or hematopoietic cell subsets of infected patients by using three different assays. Two assays are PCR based, the commercial HCV INNO-LIPA assay (INNOGENETICS, Gent, Belgium) and the CAP-PCR assay. This latter assay uses genotype-specific primers from the nucleocapsid (CAP) region and was adapted from the original Okamoto's technique.22 We designed a number of original primers, including 186 NTER, 132 N, 104 IIa, 134 N, 104 IIIa, 339N, and 104Va. Complementary DNA (cDNA) synthesis was performed using the 186NTER primer (ATAGAGAAAGAGCAACCGGG) and the sense primer 256, as described.22 The amplified product (1/50) was used for the second (nested) PCR using a mix of 10 primers specific for the amplification of 4 different HCV genotypes: 1 (a and b), 2, 3 (a), and 4. Detection of genotypes 5 and 6 is not achievable with this technique. Four sense primers, 104 AGGAAGACTTCCGAGCGGTC, 104 IIa AGGAAGACTTCGGAGCGGTC, 104 IIIa CGTAAAACTTCTGAACGGTC, and 104 IVa CGAAAGACTTCGGAGCGGTC, and six antisense primers, 132 Nbis GCAGCCCTCATTGCCATA, 133 Nbis GCCATCCTGCCCACCCCATG, 134 Nbis1 ACTTGCCAGTGGAGCGCCG, 134 Nbis2 ATTTGCCAGTGGAGCGCCG, 339N GCTGAGCCCAGGACCGGCCR, and 465 TCCCGTCCTCCACAGCCCRG were used. Primer combinations, expected size of products, and corresponding detected genotypes/subtypes were as follows: 104/132N, 125 bp, 1a; 104/133, 141 bp, 1b; 104 IIa/134N, 75 bp, 2; 104IIIa/339N, 87 bp, 3a; and 104 IVa/465, 336 bp, 4a. An illustration of the results obtained with this technique is shown in Fig 1.
Analysis of the genotype distribution in the serum and PBMC of a HCV patient harboring a dual infection. Nested RT-PCR was performed using Cap derived primers for the amplification of genotype specific products either from the positive or the negative strand viral RNA. Control human sera included positive strand RNA amplified from patients infected with genotypes 1a, 1b, 2(a/c), 3 and 4 (lanes 1, 2, 3, 4, and 5, respectively). Serum (S, lanes 6 and 8) and total PBMC (PB, lanes 7 and 9) from a patient infected with a genotype 1a and a genotype 2 were used for amplification of both the positive (+) and the negative (−) strand RNA. PCR products were fractionated on a 3% agarose gel and stained by ethidium bromide. Markers are shown on the left hand side (base pair, bp) while expected sizes of amplified products specific for the different genotypes are shown on the right hand side (bp).
Analysis of the genotype distribution in the serum and PBMC of a HCV patient harboring a dual infection. Nested RT-PCR was performed using Cap derived primers for the amplification of genotype specific products either from the positive or the negative strand viral RNA. Control human sera included positive strand RNA amplified from patients infected with genotypes 1a, 1b, 2(a/c), 3 and 4 (lanes 1, 2, 3, 4, and 5, respectively). Serum (S, lanes 6 and 8) and total PBMC (PB, lanes 7 and 9) from a patient infected with a genotype 1a and a genotype 2 were used for amplification of both the positive (+) and the negative (−) strand RNA. PCR products were fractionated on a 3% agarose gel and stained by ethidium bromide. Markers are shown on the left hand side (base pair, bp) while expected sizes of amplified products specific for the different genotypes are shown on the right hand side (bp).
The third assay, the MUREX 1-6 test (MUREX Inc, Dartford, UK) is based on the detection of genotype-specific antibodies. All genotypes presented in the study were deduced from concordant results obtained with at least two of the three assays run. Distribution of the different genotypes/subtypes was: 1a, n=5; 1b, n=14; 2a/c, n=4; 3a, n=7; 4, n=2; and 1a + 2 a/c, n=5; 1b + 2 a/c, n=1.
Assessment of viral loads.
Positive strand HCV RNA was quantified by the branched (bDNA) assay (Chiron HCV RNA 2.0 Assay; Chiron Corp, Emeryville, CA) using 50 μL of serum. This second-generation assay has been shown to correct for genotype differences in amplification efficiency. Patients' viral loads varied from less than 2 × 105 to greater than 107 genomic equivalents (Eq)/mL of serum.
Extraction of nucleic acids and cDNA synthesis.
They were essentially performed as described.10 RNA was extracted from 250 μL of serum and different amounts of total PBMCs (see Table 1) or hematopoietic cell subsets using two phenol/acid guanidium thiocyanate extraction steps, followed by a chloroform extraction step and precipitation with ethanol. cDNAs were synthesized with specific primers as described below and as reported elsewhere. Distilled water, normal sera, and normal total PBMCs or cell subsets were used as negative controls.
Detection of HCV Positive and Negative Strand RNA in Different Subsets of Peripheral Hematopoietic Cells
| Cells* . | Phenotype† . | RT-PCR Strand Specificity . | Cell Range‡ (×106 cells) . | |
|---|---|---|---|---|
| Positive . | Negative . | |||
| Total PBMC | x | 11/11 (100%) | 3/11 (27%) | 1.30 to 15.00 |
| Granulocytes | CD15+ | 8/9 (89%) | 3/10 (30%) | 0.03 to 12.50 |
| Monocytes/Macrophages | CD14+ | 5/9 (56%) | 1/10 (10%) | 0.03 to 4.00 |
| B lymphocytes | CD19+ | 5/8 (63%) | 2/9 (22%) | 0.06 to 1.80 |
| T lymphocytes | CD3+ | 0/7 (0%) | 0/7 (0%) | 0.34 to 4.40 |
| Thrombocytes | negative fraction1-153 | 0/6 (0%) | 0/6 (0%) | x |
| Cells* . | Phenotype† . | RT-PCR Strand Specificity . | Cell Range‡ (×106 cells) . | |
|---|---|---|---|---|
| Positive . | Negative . | |||
| Total PBMC | x | 11/11 (100%) | 3/11 (27%) | 1.30 to 15.00 |
| Granulocytes | CD15+ | 8/9 (89%) | 3/10 (30%) | 0.03 to 12.50 |
| Monocytes/Macrophages | CD14+ | 5/9 (56%) | 1/10 (10%) | 0.03 to 4.00 |
| B lymphocytes | CD19+ | 5/8 (63%) | 2/9 (22%) | 0.06 to 1.80 |
| T lymphocytes | CD3+ | 0/7 (0%) | 0/7 (0%) | 0.34 to 4.40 |
| Thrombocytes | negative fraction1-153 | 0/6 (0%) | 0/6 (0%) | x |
*Cellular subsets are deduced according to their phenotype.
Monoclonal antibodies directed against those surface molecules were used to purify corresponding cell type as described in Materials and Methods.
Range of cells used in the PCR assays.
Negative fraction collected at the end of the magnetic selection and corresponding to the CD15, CD14, CD19, and CD3− cells. This fraction was depleted in nucleated cells and contained principally thrombocytes.
Cloning, sequencing, and in vitro transcription.
Briefly, total RNA was extracted from 4 patients' sera (250 μL23) harboring subtypes 1b, 2a/c, 3a, and 4 strains as determined by the INNO-LIPA assay. cDNA was synthesized with genotype-specific primers located in the C-terminal end of CAP and designed according to Bukh et al.24 RNA samples were preheated first for 10 minutes at 70°C, with 0.1 μm of primer and 10 U RNasin (Promega, Madison, WA), and cDNA synthesis was then performed as described.10 Samples were then heated to 95°C for 30 minutes. A unique-sense primer (nt 37-56), from the highly conserved 5′UTR was used for PCR amplification of the cDNA. Final products encompassed nt 37-901, 37-900, 37-904 for genotype 1b-, 3a-, and 4-derived sequences, respectively. For the genotype 2a/c sequence, a nested PCR was performed with 1/25 of the first amplification product using internal primers. The final product encompassed nt 45-884. PCR products were cloned in the pGEM-T vector (TA Cloning Kit; Promega) according to the manufacturer's instructions. Plasmid DNA was purified (QIAgen plasmid preparation system, Qiagen) and sequenced, using a DNA Sequencer Stretch (Applied Biosystems, Foster City, CA). Sequences were compared with published databases to confirm the genotype/subtype of infecting isolates (courtesy of L. Jarvis and P. Simmonds). The sequence from the isolate 2a/c was confirmed to be 2c and for isolate 4 to be 4c. A clone encompassing the entire structural region of a genotype 1a strain, strain H, was also used.25GenBank accession numbers for the sequences derived in this study areU94722-U94724.
Three μgs of plasmid DNA were linearized with SphI or NotI and in vitro transcription was performed with SP6 or T7 RNA polymerases, respectively, using a T7-SP6 Riboprobe System (Promega). A 15-minute digestion with Klenow DNA polymerase (GIBCO, Grand Island, NY) of the 3′ protruding ends generated by SphI was performed at 22°C before in vitro transcription, in order to avoid synthesis of RNA molecules that were initiated at the terminus of the template.26 Digestion of template DNA was performed using RQ1 DNase (Promega) at the concentration of 1 U/μg DNA twice at 37°C for 15 minutes. The obtained synthetic RNAs were quantitated using a UV spectrophotometer at 260 and 280 nm. Integrity of the products as well as predicted concentrations were controlled on ethidium bromide-stained agarose gel. These analyses were systematically performed in duplicate. RNAs were serially diluted in DEPC-treated water before reverse transcription-polymerase chain reaction (RT-PCR) amplification. Absence of residual plasmid DNA was controlled by performing PCR amplification without prior cDNA synthesis using primers located in the 5′ NCR. In all cases, residual DNA could be detected at a concentration of 109 to 106RNA molecules per assay and thus did not interfere with the quantitative analysis described thereafter. All analyses were performed in duplicate, independent experiments.
PCR amplification of cDNA.
One-eighth of the generated cDNA was amplified as previously described10 (Inchauspé et al, submitted) using primer pairs and conditions specific for the amplification of positive (using primers from the 5′ noncoding region, 5′ NCR primers) and negative (using primers from the nucleocapsid region, CAP primers) strand HCV RNA.
For detection of the positive strand RNA, one-fourth of the PCR product was analyzed by agarose gel and Southern hybridization with a 32 P-labeled probe internal to the PCR primers.10 The probe was derived from a genotype 1a sequence. All samples were analyzed at least in duplicate experiments. Hybridizations were done as previously described.10
For the detection of the negative strand RNA, typically a RT-PCR nested amplification of CAP viral sequences was performed. Primers for cDNA synthesis and first PCR are those described for the CAP-assay (used in a reverse order). These primers show a high degree of conservation ranging from 85% to 100% between genotypes. For the second PCR, the cocktail of 10 different primers (5 pairs) described for the CAP-assay were used. Primers displayed 100% homology with the corresponding prototype sequences.24 PCR conditions consisted of an initial cycle at 95°C for 5 minutes, 30 cycles at 95°C for 1 minute, 63°C for 1 minute (negative strand); or 55°C for 1 minute (positive strand), 72°C for 1 minute 30 seconds, and a final extension cycle of 72°C for 10 minutes. One-fourth of the PCR product was analyzed by agarose gel. In some cases (dual infection), a RT-PCR using the reverse set of primers was performed for amplification and direct genotyping from the positive strand RNA.
Purification of PBMCs and peripheral hematopoietic cell subsets.
Peripheral venous blood cells were collected in EDTA-treated tubes. Mononuclear cells were obtained as described,10 ie, after Ficoll separation at 300g for 15 minutes (the collected fraction still contained granulocytes and erythrocytes, up to 20%). When specified, peripheral hematopoietic cell subsets were separated and purified using immunomagnetic positive selection with antibodies directed against specific surface molecules. Mononuclear cells were washed and the cell pellet was resuspended in 80 μL phosphate-buffered saline (PBS) buffer containing 0.5% bovine serum albumin (BSA) (wt/vol) and 5 mmol/L EDTA (PBBE) per 107cells. Twenty μL of anti-CD3 (Leu4 clone, Becton Dickinson, San Jose, CA) coupled with magnetic microbeads (Miltenyi Biotech Inc, Sunnyvale, CA) per 107 cells was added and the mixture was gently mixed by rotation for 15 minutes at 4°C. Cells were then washed and resuspended in 100 μL of PBBE per 107 cells. Depletion of T lymphocytes (CD3+ cells) was performed using a VarioMacs column-type BS (Miltenyi Biotech Inc) according to the manufacturer's instructions.
CD3− cells were further separated using MiniMacs columns (Miltenyi Biotech Inc). CD3− cells were first incubated 20 minutes at 4°C with an anti-CD15 antibody (Leu-M1; Becton Dickinson), washed, and incubated 15 minutes at 4°C with goat antimouse Ig-conjugated microbeads (Miltenyi Biotech Inc). CD15+ cells were purified and collected. CD15− cells were then incubated with an anti-CD14 antibody (LeuM3, Becton Dickinson) and further separated as mentioned above. CD14− cells were finally incubated with an anti-CD19 antibody (LeuM12; Becton Dickinson). Those sequential separations allowed us to purify four different cell subsets: CD3, CD15, CD14, and CD19+ cells. The last negative fraction consisted mainly of platelets and dead cells. The viability of cells was tested after each purification by the mean of trypan blue exclusion. Purity of each cell subset was assessed by morphological (May Grunwald Giemsa staining) and phenotypical analysis by immunofluorescence study of membrane antigens with a FACscan flow cytometer (Becton Dickinson, Baltimore, MD). Results of a typical purification are summarized in Table 2 and show that all fractions contained more than 95% of the expected cells. Dry pellets of those cell subsets were stored at -80°C before RNA extraction.
FACscan Analysis of Immunofluorescence Assays Performed on PBMC and Cell Subsets Purified From HCV-Infected Patients
| Antibodies* . | CD Tested . | Cell-151 . | Percent Detection . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Before Selection . | After Selection Using Antibodies Directed Against*: . | ||||||||
| Total Events . | CD45+ -152 . | CD3 . | CD15 . | CD19 . | CD14 . | Neg Fraction-153 . | |||
| M833 | CD45RB | PBMC | 62.3 | 100 | 100 | 100 | 95-155 | 100 | 38¶ |
| Leu4 | CD3 | T lymphocytes | 35.3 | 59.3 | 99 | 3 | <1 | <1 | 37 |
| Leu12 | CD19 | B lymphocytes | 6.9 | 11.6 | <1 | <1 | 94 | <1 | <1 |
| LeuM1 | CD15 | Granulocytes | 20.5 | 24.9 | <1 | 96 | <1 | <1 | <1 |
| LeuM3 | CD14 | Monocytes | 8.2 | 13.8 | <1 | <1 | <1 | 79 | <1 |
| CD42a | Platelets | 9.5 | x | <1 | <1 | <1 | <1 | 52 | |
| Antibodies* . | CD Tested . | Cell-151 . | Percent Detection . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Before Selection . | After Selection Using Antibodies Directed Against*: . | ||||||||
| Total Events . | CD45+ -152 . | CD3 . | CD15 . | CD19 . | CD14 . | Neg Fraction-153 . | |||
| M833 | CD45RB | PBMC | 62.3 | 100 | 100 | 100 | 95-155 | 100 | 38¶ |
| Leu4 | CD3 | T lymphocytes | 35.3 | 59.3 | 99 | 3 | <1 | <1 | 37 |
| Leu12 | CD19 | B lymphocytes | 6.9 | 11.6 | <1 | <1 | 94 | <1 | <1 |
| LeuM1 | CD15 | Granulocytes | 20.5 | 24.9 | <1 | 96 | <1 | <1 | <1 |
| LeuM3 | CD14 | Monocytes | 8.2 | 13.8 | <1 | <1 | <1 | 79 | <1 |
| CD42a | Platelets | 9.5 | x | <1 | <1 | <1 | <1 | 52 | |
*Immunomagnetic selection and antibodies as described in Materials and Methods, 10,000 events were counted and analyzed for each antibody.
Subset of cells expressing the corresponding CD.
Percentage of fluorescent cells when CD45+ cells = 100%.
Negative fraction collected at the end of the last immunomagnetic selection.
The CD45− cells were dead cells (<5%) and erythrocytes (23% of the events).
¶The CD45− cells were dead cells.
Statistical analysis.
Genotype distribution was analyzed with the χ2or test.
RESULTS
Efficiency of PCR-based detection assays for the positive and negative strand RNA.
Two RT-PCR assays were typically used in this study; for the detection of the positive strand RNA, primers were located in the 5′ NCR, whereas for the negative strand RNA, they were located in the CAP region. Although the 5′ NCR is the most conserved domain in the HCV genome (>97%),25 its use as a target for the amplification of HCV negative strand RNA has been limited because it has been shown that artifactual priming of the template could result in erroneous interpretation of the results.10,21,27 Most of the described artifacts can nonetheless be eliminated when primers for the PCR-amplification are derived from the less structured CAP region.10 This region, although less conserved than the 5′ NCR, is the most conserved region in the HCV genome open-reading frame (90% to 95%).25
Primers used for cDNA synthesis and PCR amplification of the positive strand RNA displayed 100% homology with genotype 1 to 4 sequences found in published data bases. The primer used as probe in the Southern blotting analysis displayed 91% to 100% homology with genotype 1 to 4 described sequences from the same databases (Gene Bank24,28). Primers used for the detection of the negative strand RNA had been derived from conserved regions deduced from published consensus sequences (see Materials and Methods). CAP primers used for cDNA synthesis and first PCR showed 85% to 100% homology when compared with genotype 1a, 1b, 2a/c, 3a, and 4 templates, respectively, for the antisense primer, and 0 nt, 1 nt, 1 nt, 3 nt, and 2 nt mismatch, respectively, for genotypes 1a, 1b, 2a/c, 3a, and 4 templates for the sense primer. Primers used for the nested CAP-PCR showed 100% homology when compared with genotype consensus sequences described by Bukh et al.25
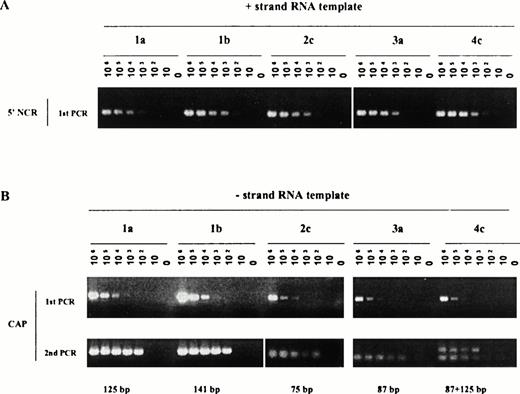
It was particularly important in the frame of our study and with respect to the above remarks to assess the detection efficiency of both strands of RNA specific for each genotype included in our study. Such evaluation was performed using synthetic RNAs. Figure 2 illustrates the sensitivity of detection of the positive strand (Fig 2A) or the negative strand (Fig2B) RNA using transcripts derived from genotype 1 (a or b), 2c, 3a, and 4c cloned sequences. The following observations could be made: sensitivity of detection of the positive strand RNA observed after single PCR amplification ranged from 102 to 103template copies per reaction. It was the lowest (103) for genotypes 1a- and 3a-derived RNA. The level of detection was unchanged after Southern blot hybridization (data not shown). The sensitivity of detection of the negative strand RNA also ranged from 102to 103 template copies per reaction after nested amplification, the lowest sensitivity (103) being detected for the genotype 4c-derived RNA. For genotype 4c-derived sequences, duplicate bands could be observed, after nested PCR, possibly reflecting annealing of primers at two different positions on the synthetic template yielding lower size products than the expected ones. These bands were nonetheless never detected using biological samples (see, for example, Fig 1, lane genotype 4 below) and did not affect sensitivity of the assays. In presence of biological contexts (such as serum or liver extracts), an additional loss of half a log to one log in sensitivity was noticed independent of the genotype tested and as previously reported.10
Efficiency of detection of positive (A) and negative (B) strand RNA from genotype 1 to 4 derived templates. Synthetic RNA, encompassing the near full length 5' NCR and CAP sequences from genotypes 1 to 4 HCV sequences, were derived as described in Materials and Methods and used in serial dilution assays. Assays included amplification of 0 to 10 6 RNA copies per reaction. PCR products obtained after single rounds of amplification (positive strand RNA, 1A) or nested rounds of amplification (negative strand RNA, 1B) were fractionated on 2.5% agarose gel and stained with ethidium bromide.
Efficiency of detection of positive (A) and negative (B) strand RNA from genotype 1 to 4 derived templates. Synthetic RNA, encompassing the near full length 5' NCR and CAP sequences from genotypes 1 to 4 HCV sequences, were derived as described in Materials and Methods and used in serial dilution assays. Assays included amplification of 0 to 10 6 RNA copies per reaction. PCR products obtained after single rounds of amplification (positive strand RNA, 1A) or nested rounds of amplification (negative strand RNA, 1B) were fractionated on 2.5% agarose gel and stained with ethidium bromide.
Thus, under experimental conditions used in our study, a maximal difference of 1 log unit in sensitivity of RNA template detection could be observed for either the positive or the negative strand RNA and for all the genotypes tested.
Detection of HCV genomic RNA in total PBMCs of chronic carriers: Association with serum viral load and genotype.
The presence of positive and negative strand HCV RNA in total PBMCs was analyzed from 38 chronically infected patients and results interpreted with respect to the viral load and viral genotype of the patients. All genotypes indicated were confirmed by at least 2:3 independently run genotyping assays (see Materials and Methods). Overall results are shown in Fig 3A and 3B.
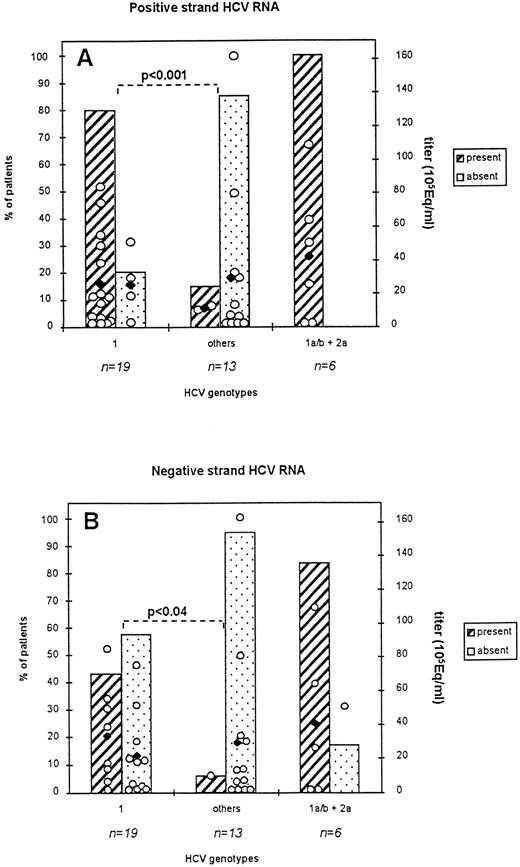
Percent of patients harboring HCV RNA sequences in total PBMC: correlation with viral load and viral genotype. Serum (250 μL) and PBMC (2 × 106 cells) from 38 patients were processed as described in Materials and Methods. Titration of HCV RNA in serum was performed by means of the bDNA assay (HCV RNA 2.0 assay). All indicated genotypes were deduced fom concordant results obtained from three genotyping assays. Titers < 2 × 105 genome equivalents/ml (Eq/mL) were considered equal to 2 × 105 Eq/ml for representation in the figure. (2A): detection of the positive strand RNA; (2B): detection of the negative strand RNA. Subtype distribution in co-infected patients was: subtype 1a, n = 5, subtype 1b, n = 1. Open circles indicate individual viral titers for each patient while black symbols represent the mean titers for each group of patients represented.
Percent of patients harboring HCV RNA sequences in total PBMC: correlation with viral load and viral genotype. Serum (250 μL) and PBMC (2 × 106 cells) from 38 patients were processed as described in Materials and Methods. Titration of HCV RNA in serum was performed by means of the bDNA assay (HCV RNA 2.0 assay). All indicated genotypes were deduced fom concordant results obtained from three genotyping assays. Titers < 2 × 105 genome equivalents/ml (Eq/mL) were considered equal to 2 × 105 Eq/ml for representation in the figure. (2A): detection of the positive strand RNA; (2B): detection of the negative strand RNA. Subtype distribution in co-infected patients was: subtype 1a, n = 5, subtype 1b, n = 1. Open circles indicate individual viral titers for each patient while black symbols represent the mean titers for each group of patients represented.
Detection of the positive strand RNA (2A).
Positive strand RNA was detected in all three groups of patients: the genotype 1 infected group, the nongenotype 1 infected group (others), and the groups of coinfected patients. Detection rates were 79% (15:19) for genotype 1 infected patients, 15% (2:13) for nongenotype 1 infected patients, and 100% (6:6) for patients from the coinfected group. These latter patients harbored a dual infection due to genotype 1a/b and 2a/c viruses. More precisely, positive signals were detected in 2:5, 13:14, 1:4, 1:7 genotype 1a, 1b, 2a/c, and 3a infected patients. The difference in detection rates between genotype 1 and other genotype infected patients was statistically significant (P < .001). As shown in the figure, viral loads between the groups of patients harboring HCV sequences in their PBMCs and the ones who did not were comparable. They ranged from less than 2.105 Eq/mL to 161 × 105 Eq/mL, whereas mean titers varied from 11 ± 15 to 26 ± 7 105 Eq/mL for the groups displaying positive strand RNA in their PBMCs and from 25 ± 10 to 30 ± 15 105 Eq/mL for the groups who did not display positive strand RNA in these cells. Patients with titers below the level of detection (ie, <2.105 Eq/mL) represented a very limited number (n=6) and were mostly found in the group 2a/c infected patients (n=3). The mean titers of coinfected patients were comparable to those from other groups of infected patients (42 ± 17 105 Eq/mL).
Detection of negative strand RNA (2B).
Negative strand RNA could also be detected in all three groups of patients although to a lower frequency than observed for detection of the positive strand RNA. Detection rates were 42%, 8%, and 83.3% in the genotype 1, the nongenotype 1, and the coinfected group of patients, respectively. More precisely, positive signals were found in 8:14 and 1:7 genotype 1b- and 3a-infected patients. The difference in detection rates between the two first groups of patients was statistically significant (P < .04). Viral loads for all groups of patients were comparable. Excluding patients with titers below the level of detection, mean titers varied from 33 105 Eq/mL for patients harboring an HCV negative strand in their PBMCs and from 21 ± 7 to 29 ± 14 105 Eq/mL for patients who did not harbor the HCV negative RNA.
Overall results indicate that, independent of the viral load and viral genotype, HCV genomic sequences can be found in PBMCs of chronically infected patients. Under the experimental conditions used in our study, genotype 1-derived sequences were predominantly detected.
Analysis of the genotype distribution in PBMCs of coinfected patients.
There were 6 patients in our study who were infected by two genotypes, genotype 1a (n = 5) or 1b (n = 1) and 2a/c viruses, as confirmed with two of the three genotyping assays used. To identify whether sequences from one genotype or both were present in the PBMCs, we determined genotypes of the PBMC-associated RNA using two genotyping assays, the INNO-LIPA and CAP-PCR assays. Both assays were performed using both the positive as well as the negative strand viral RNA as templates. Genotypes were also similarly and concomitantly analyzed from patients' sera. Results obtained with all samples were concordant between the two assays used.
RT-PCR results obtained with the CAP-derived assay for one representative patient (infected with a genotype 1a and 2a/c) are shown in Fig 1. Lanes 1 through 4 illustrate the different size products obtained when HCV positive strand RNA is amplified using this assay from genotype 1a, 1b, 2a/c, 3a, and 4, respectively, containing sera (chosen randomly). The expected size product for genotype 1a is 125 bp; for 1b, 141 bp; for 2, 75 bp; for 3a, 87 bp; and for 4, 336 bp (see Materials and Methods). In the serum as well as in the PBMCs of this patient, products specific for genotype 1a and 2 were amplified from the positive strand RNA (lanes 6 and 7, respectively). No product was obtained after amplification of the negative strand RNA from the serum of the patient (lane 8). On the other hand, a product specific for genotype 1a (125 bp) could be amplified using the negative strand RNA from PBMCs of the patient (lane 9) but no bands corresponding to genotype 2 amplified products could be detected when amplification was performed from the negative strand RNA. Overall, identical results were obtained for all 6 coinfected patients; the negative strand RNA for genotype 2 viruses was never documented in PBMCs, whereas the one for genotype 1 was systematically found. In contrast, products specific for both genotypes were amplified from PBMCs of the patients when PCR was performed from the positive strand RNA.
In conclusion, the data indicate that different genotypes/subtypes can be identified in a biological sample depending on the (genomic) RNA template used in the assay.
Detection of HCV RNA sequences in different subsets of hematopoietic cells.
In hematopoietic cells, three cellular subsets theoretically have the capacity to phagocytose viral particles: monocytes, macrophages, and granulocytes. Because the presence of HCV genomic RNA in PBMCs was clearly documented, we investigated which hematopoietic cell type may actually harbor HCV sequences and whether viral sequences could be documented in cells without phagocytic skills. We purified three different peripheral hematopoietic cell subsets from total PBMCs of 11 different patients: monocytes (and macrophages), B lymphocytes, and T lymphocytes. The purification protocol that we followed resulted in the purification of a granulocyte-rich fraction in addition to the CD15 fraction (a fraction that could still contain some activated B cells and monocytes). In our study, natural killer cell populations (CD2+, CD3−) were not selected and would likely be found in the CD3 population as well as the negative fraction. Results of the detection of HCV sequences are shown in Table 1.
Positive strand RNA was detected in three different cell subsets: granulocytes, monocytes, and B lymphocytes from respectively 89%, 56%, and 63% of patients. Even when a low number of cells were used in the PCR (3 × 104 to 6 × 104) positive signals could be detected. Positive strand RNA was not detected in T lymphocytes (0/7), even when more than 4 × 106 cells were tested, or in thrombocytes (0/6). Similarly, the negative strand RNA could be amplified from granulocytes, monocytes, and B lymphocytes from respectively 3:10, 1:10, and 2:9 patients. The two patients harboring negative strand RNA in B lymphocytes displayed well-characterized severe cryoglobulinemia. Thus, different PBMC-associated cell subsets are capable of harboring HCV genomic sequences, either the positive or the negative strand RNA.
DISCUSSION
Two main conclusions can be drawn from our study: first, that PBMCs from HCV chronic carriers can harbor HCV genomic sequences whatever the infecting viral genotype and viral load; second, that such sequences can be found in different hematopoietic cell subsets. Because of the typically low viral loads associated with HCV infections, PCR remains to date the most reproducible and sensitive technique to track the putative presence of virions, passively absorbed or replicating, in cells. We provide evidence that both genomic strands, detected using highly specific assays, are present in specific hematopoietic cell subsets of patients infected with at least three genotypes (1, 2, and 3) and four specific subtypes (1a, 1b, 2a/c, and 3a). This constitutes an original observation compared with previous studies reported in the literature as none of these studies included the systematic characterization of patients' viral genotypes. HCV's well-described genomic diversity (for review see Bukh et al4) makes it particularly difficult to develop experimental conditions equally sensitive and specific for all the existing genotypes. We took particular care in combining conditions providing the most specific amplification of sequences (in particular for the negative strand RNA) together with keeping comparable sensitivity. This could be achieved within a difference of one log factor when templates from the different genotypes (1, 2, 3, and 4) were tested. Both assays used for amplification of the positive and negative strand RNA had an identical sensitivity (102 template copies) for all genotypes tested, except in only three instances for which a sensitivity of 103 template copies was reached: for genotypes 1a and 3a positive strand RNA and for genotype 4c negative strand RNA as evaluated using synthetic transcripts (Fig 2). In our study, there was a trend toward a higher detection rate for both the positive and the negative strand RNA in patients from the genotype 1 (a or b) infected group (ranging from 42% to 78.9%). This difference was statistically significant when compared with the detection rate found in the nongenotype 1 infected group (P < .001). Although a larger number of patients from the nongenotype 1 infected type should be studied, it remains nonetheless that this observation is intriguing. Genotype 1b isolates have been reported to be associated with a more common and active disease on the graft after liver transplant7 and a poorer response to interferon treatment29 (although they are characterized by viral loads equivalent to those found for other genotypes).30 It is tempting to speculate that extrahepatic reservoirs could be favored by viruses from this genotype. Such replication advantage appears independent of the serum viral load because we could not show any influence of such viral load on detection of HCV sequences in PBMCs of the patients (see Fig 3). Our observation further confirms, in that respect, that there is no association between viral load and viral genotype as was previously suggested.30
Although our data are only representative of the conditions and patients used in our study, we observed that in cases of multiple infections, at least with genotypes 1a or b and 2a/c, the genotypes identified in the serum of the patients may not reflect the genotype(s) of viral sequences found in the PBMCs of the same patient. Because the sensitivity of detection of genotype 2 negative strand RNA is as good as the one observed for genotype 1a and 1b sequences (102template copies, see Fig 2), it is difficult to attribute the lack of detection of genotype 2 sequences to a poorer sensitivity of the detection assay. Another explanation could be due to a difference in the viral loads of PBMC-associated HCV sequences depending on the genotype involved. Alternatively, if detection of HCV negative strand RNA is indeed a reflection of active replication, the data would reflect a preferential tropism of genotype 1 isolates for PBMCs compared with genotype 2 isolates. Shimizu et al31 have recently described the existence of a preferential tropism of specific HCV viral quasispecies for PBMCs, an observation that reinforces the concept of PBMCs versus hepatocyte-adapted isolates. Cases of dual infections are not commonly found even in populations with increased risks of infection.32 33 Although additional such cases should be studied, extension of our analysis remains a difficult task to perform.
The detection of HCV sequences, either the positive or the negative strand RNA, does not constitute a sufficient argument to prove replication. The detection of positive strand RNA could simply reflect adsorption of viral particles on the cellular surface membrane. Such adsorption could be due to Fc-receptor–mediated binding of antibody-coated viral particles. This is consistent with our observation that monocytes, granulocytes, and B cells of the patients were indeed found to harbor HCV positive strand RNA. It is also consistent with the observation that immune complexes are typically found in sera of HCV-infected patients.34 35 Detection of HCV negative strand RNA being the theoretical intermediate of replication could be a stronger indication for putative replication in a cell reservoir. However, one should keep in mind that phagocytic cells could take up cellular debris or even entire cells with actively replicating HCV. Nonetheless, because the negative strand RNA was absent from the sera of all patients, trapping of this putative replication intermediate from serum would be unlikely. We observed that clinical features showing liver damage (enzyme levels, histological score) were similar in the group of patients harboring negative strand RNA in their PBMCs compared with the one without evidence of negative strand RNA in PBMCs (Lerat, personal observation). This could suggest that the release of infectious virus particles and/or negative strand RNA from damaged hepatocytes may not be the source of the negative strand RNA detected in PBMCs and that detection of such an intermediate may indeed reflect active replication.
We were able to detect HCV genomic sequences in phagocytic cells (PMNL and M/M) as well as in B lymphocytes. Previous reports had described evidence of HCV RNA in B lymphocytes18-20,33 and monocytes20 of chronic carriers. Examples of viruses replicating in phagocytic cells have been reported, eg, HIV36; Dengue virus37,38; HSV-139; and CMV.40,41 A favored tropism of HCV for B cells may clearly be a factor participating in the unusual high-association rate that has been reported between HCV infection and mixed cryoglobulinemia.42 In our study, both patients harboring HCV negative strand sequences in B cells suffered from severe cryoglobulinemia. Such tropism may also have implications in the development of lymphomas recently described in cases of HCV-infected carriers.43 We could not document HCV RNA in T lymphocytes and platelets. Although T lymphocytes constitute the most abundant population in PBMCs, they thus appear to play no role in the tracking of extrahepatic viral sequences.
Replication of viruses in PBMCs of patients has been proposed and/or shown for members of other hepatitis viruses such as hepatitis A and B viruses (HBV).44-47 A recent study, in particular, clearly describes the presence of all encoded HBV transcripts in PBMCs of chronically infected patients.48 The capacity of HCV to enter and possibly replicate in cells from the immune system and the possible dysregulation of normal cell functions associated with such an invasion could be one factor responsible for the establishment of chronicity characterizing infection by this virus.
ACKNOWLEDGMENT
We thank F. Zoulim (INSERM U271, Lyon) for assisting in the recruitment of patients and JC. Tremisi (Blood Center, Lyon) and C. Biron (Bone Marrow Transplant Unit, Lyon) for providing some patient samples and A. Fatmi for performing the sequencing of samples. We are grateful to C. Bain, S. Lemon, A.M. Prince, and M. Beard for critical review of the manuscript and P. Simmonds and L. Jarvis for help with the genotyping of cloned sequences. The authors are thankful to the Chiron Corporation and J.P. Bonn for providing us with the bDNA Chiron HCV RNA 2.0 assay.
H.L. and S.R. have equally contributed to the study.
Supported in part by l'Association pour la Recherche sur le Cancer; H.L. is supported by a grant from INSERM.
Address correspondence to Geneviève Inchauspé, INSERM U271, 151 Cours Albert Thomas, 69424 LYON Cedex 03, FRANCE; e-mail:inchauspe@lyon151.inserm.fr.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal