ACUTE LYMPHOBLASTIC leukemias (ALL) are characterized by clonal proliferation, accumulation, and tissue infiltration of neoplastic cells. They are mainly regarded as childhood diseases, with an early incidence peak at 2 to 5 years of age, where they represent about 80% of the childhood leukemias in the United States, and occur with an incidence of up to 30 cases per 1 million population per year.1 The age-adjusted incidence of ALL in adults (usually defined as 15 years of age and older) amounts to about one third of that in children.1 However, ALL has a bimodal distribution, with a second peak around age 50 and a low but steady rise in incidence with increasing age.2
Improvements in cytogenetic techniques have yielded significant insight as to the importance of cytogenetic abnormalities in the pathophysiology and prognosis of hematologic malignancies. Heim and Mitelman3 reported an overall increase in the number of reported cases of cancer with cytogenetic alterations from 3,844 in 1983 to more than 22,000 in 1994. Of all neoplasms, leukemias have been by far the most intensively investigated and account for more than 60% of all listed chromosomal aberrations, including more than 3,000 cases of ALL.
The majority of cases of ALL demonstrate an abnormal karyotype, either in chromosome number (ploidy) or as structural changes such as translocations, inversions, or deletions. These changes were detected in only half of ALL patients in the first banding studies.3The scantiness of information gained from chromosomal findings in ALL has been, in large part, due to technical difficulties. Chromosome studies in ALL exhibit poor morphology; chromosomes tend to spread poorly, and appear blurred and fuzzy with indistinct margins, making banding studies challenging or even impossible.4,5Improvements in spreading and banding techniques have resulted in higher rates of detection, and most studies now report chromosomal changes in 60% to 85% of ALL cases.6-10 The Third International Workshop on Chromosomes in Leukemia (TIWCL) found the majority of cytogenetic changes in cases of B-precursor ALL, with only 39% occurring in T-cell ALL.6,9 Williams et al4 used a direct technique of bone marrow (BM) chromosomal analysis developed particularly for studies in ALL, which paid attention to sampling and processing steps using specific flaming techniques and modified G-banding procedures. They identified clonal karyotypic abnormalities in 94% to 98% of cases of ALL. Such improved techniques also detected nonrandomly occurring cytogenetic abnormalities in cases with hyperdiploid chromosome numbers (>50) that had previously been classified as normal in karyotype.11 These results showed a high prevalence of clonal chromosomal abnormalities in ALL, as was shown for acute nonlymphoblastic leukemias by Yunis,12 who used high-resolution banding techniques. These and similar studies underscore the significant yield achieved with thorough cytogenetic studies in ALL.
That cytogenetic abnormalities confer important prognostic information in ALL was first reported by Secker-Walker et al13 in 1978 in a series of childhood ALL. The investigators reported better clinical outcomes in cases with hyperdiploid karyotypes than in those with hypodiploidy or pseudodiploidy, and these findings were confirmed in the follow-up study14 and by other researchers.8,15-17 The TIWCL examined 330 newly diagnosed ALL patients (172 adults and 157 children) and found that chromosomal abnormalities distinguished high-risk from low-risk patients. Complete remission (CR) rates, remission durations, as well as disease-free-survivals (DFS) were significantly affected by the karyotypic abnormalities.6 Among adult patients the highest likelihood of cure (21% to 30%) was projected in patients with chromosome numbers of >50, or 47 to 50, with 6q−, or with a normal karyotype.18 As with children, karyotypes in adults were significant independent predictors of remission duration or DFS, after considering covariates such as age, leukocyte count at presentation, or French-American-British (FAB) morphology.19 Secker-Walker et al10 reported the prognostic effect of certain structural rearrangements, such as t(9;22), to be independent of other single variables.
Most studies on karyotypic abnormalities and their clinical significance have been performed in childhood ALL.15,20-24Adult ALL may show nonrandom chromosomal abnormalities similar to those found in childhood ALL, but their distribution and, possibly, their biological significance are different. Few studies have addressed these issues in adult ALL.6-8,18,25 26 This review focuses on the most important chromosomal abnormalities found in adult ALL and their prognostic and therapeutic implications.
NUMERICAL CHROMOSOME ABNORMALITIES
Numerical chromosome abnormalities, either alone or in association with structural changes, are found in about half of ALL cases. Several ploidy groups have been identified (Table1).15,27 These include low hyperdiploidy (modal number 47 to 50), high or massive hyperdiploidy (>50), hypodiploidy (46 and lower), pseudodiploidy (normal number of chromosomes, but with associated structural changes), as well as gain or loss of a single chromosome as the sole karyotypic change. Sole numerical aberrations are less frequent than the numerical aberrations in combination with structural changes, which occur in about 40% to 70% of cases.5 7
Frequency of Ploidy Groups in Adult ALL Compared With Childhood ALL
| Ploidy Group . | Frequency in: . | |
|---|---|---|
| Adult ALL (%) . | Childhood ALL (%) . | |
| Normal karyotype | 26-34 | 8-56 |
| Hypodiploid <46 | 2-8 | 5-6 |
| Pseudodiploid | 7-59 | 3-42 |
| Hyperdiploid 47-50 | 7-17 | 8-16 |
| Hyperdiploid >50 | 4-9 | 14-27 |
| Near triploidy | 3 | <1 |
| Near tetraploidy | 2 | 1 |
| Ploidy Group . | Frequency in: . | |
|---|---|---|
| Adult ALL (%) . | Childhood ALL (%) . | |
| Normal karyotype | 26-34 | 8-56 |
| Hypodiploid <46 | 2-8 | 5-6 |
| Pseudodiploid | 7-59 | 3-42 |
| Hyperdiploid 47-50 | 7-17 | 8-16 |
| Hyperdiploid >50 | 4-9 | 14-27 |
| Near triploidy | 3 | <1 |
| Near tetraploidy | 2 | 1 |
Data from references 7, 8, 15, 18, 26, 27, 32, and 37.
Hyperdiploidy
Chromosome numbers of 47 or more are found in up to one fourth of adult ALL cases, making the hyperdiploid group one of the most frequent ploidy groups (Table 1). Hyperdiploid chromosome numbers, particularly those with >50 chromosomes, are more frequent in children than in adults. Chromosome numbers cluster around 47 and between 51 to 55 so that a bimodal distribution becomes evident, similar to the distribution in childhood ALL.15 A low hyperdiploidy group with 47 to 50 chromosomes can be distinguished from a massive hyperdiploidy group with >50 chromosomes. Disomies and, particularly, trisomies or tetrasomies, are frequent and are dominant in the group with >50 chromosomes.
Although various chromosomes are involved in hyperdiploidy, certain karyotypes seem to prevail. In a series of 31 patients, Ankathil et al28 found that hyperdiploid karyotypes were mainly characterized by trisomies of chromosomes 8, 18, 19, and 21. The TIWCL study also showed, in both hyperdiploidy groups, gains in chromosomes 4 and 8, as well as 21 and 18, to be more frequent. Additional chromosomes 6, 10, and 14 appeared almost exclusively in the massive hyperdiploidy group.6 The Groupe Français de Cytogénétique Hématologique (GFCH) found in the group of >50 chromosomes mainly chromosomes 4, 6, 8, 10, 14, 17, and 21 and in the 47 to 50 group mainly chromosomes 5, 8, 10, and 21.7About half of all adult ALL cases with hyperdiploidy of a modal number of >50 can be expected to show additional chromosomal rearrangements, with translocation t(9;22) being the most common.7,9 In contrast, the TIWCL found relatively few structural rearrangements in both hyperdiploidy groups. This difference reflects the impact of advances in technology and expertise over little more than a decade on the yield of cytogenetic studies. In addition, the technical quality of hyperdiploid metaphase spreads is frequently substandard, and structural aberrations are still not reliably identified in a significant number of cases.29
In the TIWCL study, almost all cases with >50 chromosomes were of precursor B-cell type and associated with favorable prognostic features such as low white blood cell count (WBC) at presentation, low lactate dehydrogenase (LDH) levels, and FAB L1 or L2 morphology.6
In children with ALL, chromosomal numbers of >50 have been reported to have better response duration as well as median survival time (MST), whereas numbers between 47 and 50 conferred an intermediate prognosis. In this context, the combination of trisomies of both chromosomes 4 and 10 in children with hyperdiploid ALL identified a subgroup of patients with an extremely favorable DFS, and likelihood of cure with antimetabolite-based chemotherapy.30
No such favorable constellation could be identified in adult ALL, where the impact of hyperdiploidy on prognosis appeared less significant (Table 2). Although the TIWCL showed higher CR rates, as well as increased CR duration and MST in this ploidy group for both children and adults, children still survived significantly longer than adults for modal numbers >50 or 47 to 50. The GFCH study on 443 adult ALL patients found a favorable prognosis in patients with hyperdiploidy of >50 chromosomes without the Philadelphia chromosome (Ph), as well as in patients with tetraploidy.7 The UKALL XA trial observed better outcomes with hyperdiploidy which added prognostic significance to age, sex, and WBC, unless these variables were combined.10 Campbell et al31 showed a survival advantage for adult patients with only numerical abnormalities and particularly for those with chromosome numbers more than 50. However, other studies could not confirm the association of hyperdiploidy with good outcome in adult ALL. Fenaux et al8found no significant differences in CR rate or duration between cytogenetic groups in 73 adult patients, except for a slight increase in CR duration with normal karyotypes, and worse outcomes with the Ph abnormality. This contrasted with their findings in pediatric patients, where a significant difference in response to treatment was found between children with hyperdiploidy and those with hypodiploidy. Walters et al,32 in a series of 91 adult patients, could not demonstrate superior outcome with hyperdiploidy.
Characteristics and Clinical Outcome in Various Ploidy Groups
| . | Karyotype . | References . | ||||
|---|---|---|---|---|---|---|
| Normal . | <46 . | 46 abnl . | 47-50 . | >50 . | ||
| FAB category (%) | ||||||
| L1 | 36 | 11 | 14 | 30 | 0 | TIWCL‡ |
| 47 | 100 | 50 | 40 | 20 | Fenaux et al* | |
| 71 | 48 | 53 | 33 | 60 | GFCH | |
| NA | NA | NA | NA | NA | UKALL XA | |
| L2 | 62 | 89 | 86 | 70 | 100 | TIWCL‡ |
| 53 | 0 | 50 | 60 | 80 | Fenaux et al* | |
| NA | NA | NA | NA | NA | GFCH | |
| NA | NA | NA | NA | NA | UKALL XA | |
| LSM (%) | ||||||
| Pre-B | 62 | 75 | 69 | 71 | 100 | TIWCL‡ |
| 77 | 0 | 66 | 075 | 100 | Fenaux et al* | |
| 65 | 92 | 78 | 67 | 100 | GFCH | |
| 75 | 91 | 71 | 75 | 96 | UKALL XA | |
| T | 38 | 25 | 31 | 29 | 0 | TIWCL‡ |
| 23 | 100 | 33 | 25 | 0 | Fenaux et al* | |
| 35 | 5 | 19 | 33 | 0 | GFCH | |
| 25 | 9 | 29 | 25 | 4 | UKALL XA | |
| CR rate (%) | 86 | 50 | 67 | 67 | 78 | TIWCL‡ |
| 63 | 0† | 100 | 75 | 40 | Fenaux et al | |
| 79 | 65 | 71 | 84 | 74 | GFCH | |
| 90 | 57 | 89 | 96 | 96 | UKALL XA | |
| Median CR duration (mo) | 9 | 5 | 11 | 10 | 17 | TIWCL‡ |
| 17 | NA | 6 | 6 | 15.5 | Fenaux et al | |
| NA | NA | NA | NA | NA | GFCH | |
| NA | NA | NA | NA | NA | UKALL XA | |
| Median survival time (mo) | 24 | 7 | 10 | 8 | 21 | TIWCL‡ |
| NA | NA | NA | NA | NA | Fenaux et al | |
| NA | NA | NA | NA | NA | GFCH | |
| NA | NA | NA | NA | NA | UKALL XA | |
| Median DFS (mo) | 8 | 2 | 6 | 6 | 13 | TIWCL‡ |
| NA | NA | NA | NA | NA | Fenaux et al | |
| 24 | 4 | 7 | 10 | 8 | GFCH | |
| 18 | 4 | 13 | 32 | 36 | UKALL XA | |
| . | Karyotype . | References . | ||||
|---|---|---|---|---|---|---|
| Normal . | <46 . | 46 abnl . | 47-50 . | >50 . | ||
| FAB category (%) | ||||||
| L1 | 36 | 11 | 14 | 30 | 0 | TIWCL‡ |
| 47 | 100 | 50 | 40 | 20 | Fenaux et al* | |
| 71 | 48 | 53 | 33 | 60 | GFCH | |
| NA | NA | NA | NA | NA | UKALL XA | |
| L2 | 62 | 89 | 86 | 70 | 100 | TIWCL‡ |
| 53 | 0 | 50 | 60 | 80 | Fenaux et al* | |
| NA | NA | NA | NA | NA | GFCH | |
| NA | NA | NA | NA | NA | UKALL XA | |
| LSM (%) | ||||||
| Pre-B | 62 | 75 | 69 | 71 | 100 | TIWCL‡ |
| 77 | 0 | 66 | 075 | 100 | Fenaux et al* | |
| 65 | 92 | 78 | 67 | 100 | GFCH | |
| 75 | 91 | 71 | 75 | 96 | UKALL XA | |
| T | 38 | 25 | 31 | 29 | 0 | TIWCL‡ |
| 23 | 100 | 33 | 25 | 0 | Fenaux et al* | |
| 35 | 5 | 19 | 33 | 0 | GFCH | |
| 25 | 9 | 29 | 25 | 4 | UKALL XA | |
| CR rate (%) | 86 | 50 | 67 | 67 | 78 | TIWCL‡ |
| 63 | 0† | 100 | 75 | 40 | Fenaux et al | |
| 79 | 65 | 71 | 84 | 74 | GFCH | |
| 90 | 57 | 89 | 96 | 96 | UKALL XA | |
| Median CR duration (mo) | 9 | 5 | 11 | 10 | 17 | TIWCL‡ |
| 17 | NA | 6 | 6 | 15.5 | Fenaux et al | |
| NA | NA | NA | NA | NA | GFCH | |
| NA | NA | NA | NA | NA | UKALL XA | |
| Median survival time (mo) | 24 | 7 | 10 | 8 | 21 | TIWCL‡ |
| NA | NA | NA | NA | NA | Fenaux et al | |
| NA | NA | NA | NA | NA | GFCH | |
| NA | NA | NA | NA | NA | UKALL XA | |
| Median DFS (mo) | 8 | 2 | 6 | 6 | 13 | TIWCL‡ |
| NA | NA | NA | NA | NA | Fenaux et al | |
| 24 | 4 | 7 | 10 | 8 | GFCH | |
| 18 | 4 | 13 | 32 | 36 | UKALL XA | |
For each karyotype group the percentage of patients who had each characteristic is given, as is the response and survival.
Abbreviations: abnl, abnormal; NA, not available; LSM, lymphocyte surface marker; TIWCL, Third International Workshop on Chromosomes in Leukemia (refs 6, 18, 25, 36, 37); Fenaux et al (ref 8); GFCH, Groupe Français de Cytogénétique Hématologique (ref 7); UKALL XA, MRC Adult Leukaemia Working Party (ref 10).
Low patient numbers.
Based on one patient only.
Data included from Bloomfield et al (ref 18, 25).
The less favorable prognosis in adult ALL with hyperdiploid karyotype, compared with children, may be explained by the higher prevalence of associated unfavorable structural changes. The GFCH study, for example, reported the additional presence of Ph in 11 of 30 patients with hyperdiploid (>50) karyotypes.7 The UKALL XA trial showed that the prognostic impact of structural changes overrode the prognostic significance of ploidy groups. Poor-risk structural rearrangements (such as Ph) confer a bad outcome even if they occur in otherwise good-risk ploidy groups.10
Hypodiploidy
Modal chromosome numbers of 45 and less are rare, particularly the nearly haploid numbers of 24 to 36. In most series of adult patients, hypodiploid chromosome numbers were found in 2% to 8% (Table 1). Sandberg33 compiled, from various references, 26 patients with near-haploid or severe hypodiploid leukemias, including seven adults. The most common losses involved chromosomes 1, 5, 6, 10, 11, 18, 19, 21, and 22. Interestingly, these were the same chromosomes involved in the hyperdiploid karyotypes.6,7,33 The reason for this preference is not known. Near-haploid cases almost always have only numerical changes, whereas cases with modal numbers of 30 to 44 show frequent structural aberrations, mainly translocations.6,34 In nearly all hypodiploid cases analyzed by Rieder et al,9 additional structural abnormalities were identified (half of them involving the Ph).
How massive chromosome losses contribute to leukemogenesis is unclear. Oshimura et al35 proposed multipolar mitosis with subsequent misdivision as a possible explanation. Conceivably, a near-haploid karyotype allows expression of recessive genes that would otherwise have been under the dominance of their allelic counterparts, with loss of regulatory control of growth and differentiation of lymphoid cells.33 Many ALL patients also have a second hyperdiploid population with 52 to 56 chromosomes, twice the near-haploid number, at some time in their disease course, possibly arising by secondary endoreduplication.34 In these instances hyperdiploidy may be an expression of clonal evolution from a near-haploid stem line.33
ALL associated with a hypodiploid karyotype is usually of a precursor B-cell type, although in the TIWCL series, 20% of hypodiploid cases had T-cell ALL.6 Median initial WBC and percentage of blasts were higher than with diploid or hyperdiploid karyotype, and morphology was predominantly FAB L2.6 Hypodiploid adult patients had the worst 3-year survival in the UKALL XA trial, and hypodiploidy was shown to have prognostic significance independent of other important variables (including the combination of age, sex, and WBC).10 Like hyperdiploidy, hypodiploidy appeared to have more effect on outcome in children than in adults, where data are still limited.25 Nevertheless, hypodiploidy conferred poor prognosis in most studies of adult ALL, comparable to other poor-risk chromosomal translocations such as t(4;11) and t(1;19) (Table2).7,10 18
Pseudodiploidy
A normal chromosome number with structural changes is the most frequently found abnormal karyotype in adult ALL, and provides for the most heterogeneous group of patients (Table 1). The GFCH showed a pseudodiploid karyotype in 59% of ALL patients, 60% of whom had recurrent translocations (Ph in 40%). This group was also characterized by the highest peripheral WBC counts.7 The TIWCL study found that, compared to other karyotype groups, the group with pseudodiploidy had increased ratios of adults to children, and of L2 to L1 morphology, as well as high initial leukocyte and blast counts.36 The pseudodiploid group was also notable for having the highest percentage of T-cell ALL.37 Most cases had structural changes, with only a few combining structural and numerical aberrations. Chromosomes frequently involved were 1, 6, 9, and 14 but included every autosome with the exception of 4, 16, 18, and 21.36 In the UKALL XA trial, pseudodiploidy was observed in association with translocation t(9;22), with abnormalities of 6q or 9p, or, less often, with translocations t(4;11) and t(1;19).10In some studies, initial leukemic cell burden was high in pseudodiploid cases, which was reflected by increased leukocyte counts and LDH levels.6 14 The poor prognosis associated with pseudodiploidy (Table 2) is likely a reflection of structural rearrangements and other poor-risk features. More accurate identification of underlying specific structural abnormalities may soon make the pseudodiploid group redundant in a prognostically useful classification.
Single Chromosomal Gains or Losses
Nonrandom single chromosome gains or losses occur frequently in ALL,38 although their incidence is lower than in myeloid leukemias.34,39-42 Rarely are they the sole karyotypic abnormality.38 The mechanisms by which such changes contribute to leukemogenesis are unknown. Heim and Mitelman43 offered two possible explanations: a dose effect, with abundance of certain gene products resulting in abnormal proliferation or differentiation, or a duplication of a small genetic defect with oncogenic potential. Most reports on single chromosome gains or losses have been published in childhood ALL, mainly with trisomy 8, monosomy 20, and trisomy 21.34,42 44-46 Only in 10% to 20% of these were the trisomy or monosomy the only karyotypic anomaly, and no particular distinguishing characteristics could be observed.
The TIWCL demonstrated trisomy 21 as the most frequent chromosomal gain in ALL, and further studies confirmed its relatively high incidence, but mainly as part of other cytogenetic changes such as hyperdiploidy.36,47 Trisomy 8 as the only karyotypic change is a frequent occurrence, mainly in myeloid leukemias.48Garipidou et al44 estimated its incidence in ALL to be 1% to 2%. It is indicative of a poor prognosis in acute myeloid leukemias (AML), but its prognostic significance in either adults or children with ALL is not established.49
Trisomy 4 has been observed in a broad range of hematologic malignancies,38 50 but is rare in ALL51-53 and, as the sole abnormality, tends to be associated with myeloid leukemias, while no such association existed when there were additional cytogenetic changes.
The first report of trisomy 5 in ALL, by Sandberg et al,54was of a 26-year-old male patient with B-lineage ALL. Nagesh Rao et al55 described a 24-year-old woman with T-lymphoblastic lymphoma and trisomy 5 as the sole cytogenetic abnormality. Their review of the literature found only five cases in addition to the case reported by Sandberg et al: 3 AML, 1 non-Hodgkin's lymphoma, and 1 case of Hodgkin's disease. Chen et al56 delineated certain regions on chromosome 5, such as 5p13 and 5q11-31, that were supposedly more specific to ALL.
Monosomies of chromosomes 5 and 7 through deletion or losses of chromosomal material are frequently found in AML and myelodysplastic syndromes. However, their occurrence and significance is not well established in ALL. Dabaja et al57 analyzed 468 adult patients with ALL and found abnormalities of chromosome 5 in 3 and of chromosome 7 in 31 patients; Ph was an associated abnormality in one third. This association translated into lower CR and 3-year survival rates compared with patients who did not have abnormalities of chromosomes 5 or 7. However, when patients with −5 and −7, but without Ph were considered, clinical outcome was not different. The GFCH reported 45 cases with monosomy 7 among 443 adult patients. It was the sole abnormality in only one patient with T-cell ALL.7Rieder et al9 reported only 1 case out of 100 with loss of chromosome 7 as the sole cytogenetic abnormality, confirming the rarity of this karyotype.
Monosomy 20 rarely presents as the single anomaly in ALL. Most studies refer to pediatric patients but occasionally describe this finding in adults, where it correlated with FAB L1 morphology, although this association was not consistent.45 58
Overall, the significance of monosomies or trisomies as isolated karyotypic changes is unclear, and no specific disease characteristics have been established. Furthermore, reports of single chromosomal gains or losses in adults were too sporadic to draw conclusions of clinical relevance.
STRUCTURAL CHROMOSOME ABNORMALITIES
More than 30 different nonrandomly occurring rearrangements are presently known in ALL (Table3).34 The GFCH found structural abnormalities in 78% of cases distributed across all ploidy groups.7 Translocations constituted the most common changes. They were found in 30% to 37% of adult cases, with the t(9;22) translocation being the most frequent.7,33,37 The TIWCL showed significant differences between chromosome groups regarding central nervous system (CNS) involvement, leukocyte and blast counts, FAB morphology, and immunophenotype.37 In adults, translocation t(4;11) and the Ph karyotypes were associated with higher leukocyte and blast counts than were other chromosomal rearrangements. Translocations t(8;14) and t(4;11), as well as 14q+, correlated with a higher risk of CNS involvement. Both children and adults with chromosomal translocations had worse survival than those with normal karyotypes. In children the presence of cytogenetic abnormalities other than translocations was associated with outcome similar to normal karyotypes, but in adults all structural abnormalities adversely influenced survival.
Nonrandom Structural Chromosomal Aberrations in Adult ALL
| Chromosome Aberration . | Involved Genes . | Protein Product . | Function of Protein Product . | Frequency . | Phenotype . | FAB-Morphology . | References . |
|---|---|---|---|---|---|---|---|
| del(6q) | ? | ? | ? | <5% | B- or T-lineage | L1, L2 | 6, 7, 10, 18, 25, 28, 33, 240-247 |
| i(6p) | ? | ? | ? | <5% | 34, 248-251 | ||
| i(7q) | ? | ? | ? | <5% | 34, 248-252 | ||
| 7q32-35 | TCR-β | <5% | T-lineage | L1 > L2 | 6, 7, 157, 168-193 | ||
| t(1;7)(p32;q35) | TAL1/TCR-β | Basic HLH protein | Transcription factor | ||||
| t(1;7)(p34;q34) | LCK/TCR-β | Protein kinase | Signal transduction | ||||
| t(1;7)(q11-21; q35-36) | ? | ? | ? | ||||
| t(7;9)(q34;q32) | TAL2/TCR-β | Basic HLH protein | Transcription factor | ||||
| t(7;9)(q34;p34) | TAN1/TCR-β | Drosophila notch | Signal transduction | ||||
| Homologue | |||||||
| t(7;10)(q35;q24) | RHOM2/RCR-β | LIM domain protein | Transcription factor | ||||
| t(7;11)(q35;p13) | RHOM2/TCR-β | LIM domain protein | Transcription factor | ||||
| LYL1/TCR-β | Basic HLH protein | Transcription factor | |||||
| 8q24 | c-myc | Basic HLH protein | Transcription factor | <3-5% | B | L3 | 5-8, 18, 25, 33, 34, 36, 37, 151, 171, 194-208 |
| t(2;8)(p12;q24) | lgκ/c-myc | ||||||
| t(8;14)(q24;q32) | c-myc/lgH | ||||||
| t(8;22)(q24;q11) | c-myclgλ | ||||||
| i(9q) | ? | ? | ? | <5% | Pre-B | L2 | 34, 248-251 |
| t(9;22)(q34;q11) | BCR, ABL e1a2 b2a2/b3a2 e19a2 | p190 p210 p230 | Increased tyrosine kinase activity (? p23) | 20-30% | Pre-B | L1, L2 | 7, 8, 36, 54, 59-87 |
| del(9q)(p21-22) | MTS1/MTS2 | p16INK4A/p15INK4B | Cyclin-dependent kinase inhibitors | 7-15% | B- or T-lineage | L1, L2 | 7, 92-119 |
| t/dic(9;12)(p11-12; p11-13) | ? | ? | ? | <5% | Early-pre-B or pre-B | L1, L2 | 22, 34, 120-123 |
| 11q23 t(1;11)(p32;q23) t(4;11)(q21;q23) t(10;11)(p12-14; q14-23) t(11;19)(q23; p13) | MLL (HRX, ALL-1, HTRX-1) AF1P/MLL AF4/MLL AF10/MLL MLL/ENL | MLL fusion proteins with conservation of 5′-N-terminal sequences including AT-hook and DNA-methyltransferase regions | Probably involved in transcriptional regulation | 3-10% | Early-pre-B T-lineage biphenotypic | L1 L1, L2 L2 L1, ,L2 | 7, 9, 18, 33, 90, 124-143 |
| 12q t(9;12)(q34;p13) t(12;21)(p11-12; q22) | TEL (ETV6) ABL/TEL TEL/AML1 | HLH protein tyrosine kinase runt/transactivating domain | Phosphorylation Transcription regulation | 3-4% | pre-B | L1, L2 | 7, 10, 24, 209-239 |
| 14q11 | TCR-α/δ | T-lineage | L1 > L2 | 6, 7, 157, 168-193 | |||
| t(1;14)(p32-34;q11) | TAL1/TCR-δ | Basic HLH protein | Transcription factor | ∼20% | |||
| t(5;14)(p15;q11) | ? | ? | ? | ? | |||
| t(8;14)(q24;q11) | c-MYC/TCR-α | Basic HLH protein | Transcription factor | <1% | |||
| t(10;14)(q24;q11) | HOX11/TCR-δ | Homeodomain protein | Transcription factor | 3-7% | |||
| t(11;14)(p13;q11) | RHOM2/TCR-δ | LIM domain protein | Transcription factor | 5-7% | |||
| t(11;14)(p15;q11) | RHOM1/TCR-δ | LIM domain protein | Transcription factor | <1% | |||
| inv(14)(q11;q32.1) | TCL1/TCR-α | ? | ? | <1% | |||
| inv(14)(q11;q32.3) | IgH/TCR-α/δ | Ig | ? | <1% | |||
| i(17q) | ? | ? | ? | 7-9% | Pre-B | 34, 248-251, 253 | |
| 19q13 t(1;19)(q23;p13) t(17;19)(q22;p13) | E2A PBX1/E2A HLF/E2A | Fusion proteins with preservation of E2A activation domains | Transcription factor | <5% | Pre-B Pre-B | L1 L1 | 7, 8, 15, 29, 33, 34, 90, 145-166 |
| i(21)q | ? | ? | ? | <5% | B-lineage | 34, 248-251 |
| Chromosome Aberration . | Involved Genes . | Protein Product . | Function of Protein Product . | Frequency . | Phenotype . | FAB-Morphology . | References . |
|---|---|---|---|---|---|---|---|
| del(6q) | ? | ? | ? | <5% | B- or T-lineage | L1, L2 | 6, 7, 10, 18, 25, 28, 33, 240-247 |
| i(6p) | ? | ? | ? | <5% | 34, 248-251 | ||
| i(7q) | ? | ? | ? | <5% | 34, 248-252 | ||
| 7q32-35 | TCR-β | <5% | T-lineage | L1 > L2 | 6, 7, 157, 168-193 | ||
| t(1;7)(p32;q35) | TAL1/TCR-β | Basic HLH protein | Transcription factor | ||||
| t(1;7)(p34;q34) | LCK/TCR-β | Protein kinase | Signal transduction | ||||
| t(1;7)(q11-21; q35-36) | ? | ? | ? | ||||
| t(7;9)(q34;q32) | TAL2/TCR-β | Basic HLH protein | Transcription factor | ||||
| t(7;9)(q34;p34) | TAN1/TCR-β | Drosophila notch | Signal transduction | ||||
| Homologue | |||||||
| t(7;10)(q35;q24) | RHOM2/RCR-β | LIM domain protein | Transcription factor | ||||
| t(7;11)(q35;p13) | RHOM2/TCR-β | LIM domain protein | Transcription factor | ||||
| LYL1/TCR-β | Basic HLH protein | Transcription factor | |||||
| 8q24 | c-myc | Basic HLH protein | Transcription factor | <3-5% | B | L3 | 5-8, 18, 25, 33, 34, 36, 37, 151, 171, 194-208 |
| t(2;8)(p12;q24) | lgκ/c-myc | ||||||
| t(8;14)(q24;q32) | c-myc/lgH | ||||||
| t(8;22)(q24;q11) | c-myclgλ | ||||||
| i(9q) | ? | ? | ? | <5% | Pre-B | L2 | 34, 248-251 |
| t(9;22)(q34;q11) | BCR, ABL e1a2 b2a2/b3a2 e19a2 | p190 p210 p230 | Increased tyrosine kinase activity (? p23) | 20-30% | Pre-B | L1, L2 | 7, 8, 36, 54, 59-87 |
| del(9q)(p21-22) | MTS1/MTS2 | p16INK4A/p15INK4B | Cyclin-dependent kinase inhibitors | 7-15% | B- or T-lineage | L1, L2 | 7, 92-119 |
| t/dic(9;12)(p11-12; p11-13) | ? | ? | ? | <5% | Early-pre-B or pre-B | L1, L2 | 22, 34, 120-123 |
| 11q23 t(1;11)(p32;q23) t(4;11)(q21;q23) t(10;11)(p12-14; q14-23) t(11;19)(q23; p13) | MLL (HRX, ALL-1, HTRX-1) AF1P/MLL AF4/MLL AF10/MLL MLL/ENL | MLL fusion proteins with conservation of 5′-N-terminal sequences including AT-hook and DNA-methyltransferase regions | Probably involved in transcriptional regulation | 3-10% | Early-pre-B T-lineage biphenotypic | L1 L1, L2 L2 L1, ,L2 | 7, 9, 18, 33, 90, 124-143 |
| 12q t(9;12)(q34;p13) t(12;21)(p11-12; q22) | TEL (ETV6) ABL/TEL TEL/AML1 | HLH protein tyrosine kinase runt/transactivating domain | Phosphorylation Transcription regulation | 3-4% | pre-B | L1, L2 | 7, 10, 24, 209-239 |
| 14q11 | TCR-α/δ | T-lineage | L1 > L2 | 6, 7, 157, 168-193 | |||
| t(1;14)(p32-34;q11) | TAL1/TCR-δ | Basic HLH protein | Transcription factor | ∼20% | |||
| t(5;14)(p15;q11) | ? | ? | ? | ? | |||
| t(8;14)(q24;q11) | c-MYC/TCR-α | Basic HLH protein | Transcription factor | <1% | |||
| t(10;14)(q24;q11) | HOX11/TCR-δ | Homeodomain protein | Transcription factor | 3-7% | |||
| t(11;14)(p13;q11) | RHOM2/TCR-δ | LIM domain protein | Transcription factor | 5-7% | |||
| t(11;14)(p15;q11) | RHOM1/TCR-δ | LIM domain protein | Transcription factor | <1% | |||
| inv(14)(q11;q32.1) | TCL1/TCR-α | ? | ? | <1% | |||
| inv(14)(q11;q32.3) | IgH/TCR-α/δ | Ig | ? | <1% | |||
| i(17q) | ? | ? | ? | 7-9% | Pre-B | 34, 248-251, 253 | |
| 19q13 t(1;19)(q23;p13) t(17;19)(q22;p13) | E2A PBX1/E2A HLF/E2A | Fusion proteins with preservation of E2A activation domains | Transcription factor | <5% | Pre-B Pre-B | L1 L1 | 7, 8, 15, 29, 33, 34, 90, 145-166 |
| i(21)q | ? | ? | ? | <5% | B-lineage | 34, 248-251 |
Translocation t(9;22)(q34;q11)
In 1960 Nowell and Hungerford59 discovered the Ph as a distinct chromosomal abnormality in chronic myeloid leukemia (CML). It was the first chromosomal abnormality to be associated with a specific malignant disease in humans, and became a karyotypic hallmark of CML. In 1970 Propp and Lizzi60 reported a 53-year-old patient with ALL who had the classic Ph in a high percentage of marrow cells. It is now well established that a t(9;22) translocation can be observed in up to 95% of patients with CML, in about 1% to 2% of patients with AML, as well as in up to 5% of children and 15% to 30% of adults with ALL, making it the most common ALL-associated chromosomal abnormality in the latter group.61-64
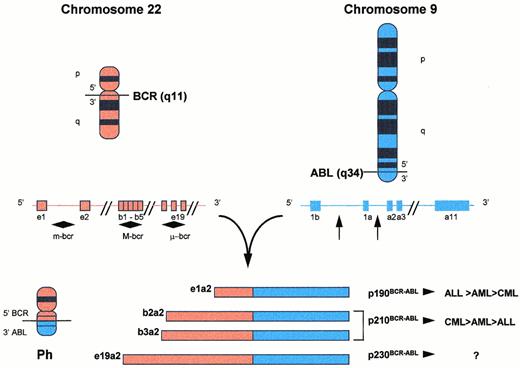
The Ph is a shortened chromosome 22 that results from a reciprocal translocation between the long arms of chromosomes 9 and 22 (Fig 1).62 This transposes the large 3′ segment of the c-ABL gene from chromosome 9 to the 5′ part of the BCR gene on chromosome 22, creating a hybridBCR-ABL gene that is transcribed into a chimericBCR-ABL mRNA.65,66 The breakpoint in theABL region can occur anywhere in a 300-kb intron, but is usually 5′ of ABL exon a2. Deletions of ABL exon a2 and in-frame joining at the mRNA level of 5′ BCR sequences to theABL exon a3 have also been described.67 In almost all cases of CML, as well as in about half of adult Ph+ALL, the ABL gene from chromosome 9 transposes into the major breakpoint cluster region (M-bcr) on chromosome 22, spanning exons 12 to 16 (historically named b1 to b5), giving rise to BCR-ABLfusion transcript mRNAs with a b2a2 or b3a2 junction. The fusion mRNAs translate into a fusion protein of 210 kD called p210BCR-ABL.68 In very rare cases of CML or AML, but in about 50% of adult Ph+ ALL and in 80% of childhood Ph+ ALL, the breakpoint on chromosome 22 falls 5′ of the M-bcr, within a long intron segment separating the alternative exon e2′ from e2 called the minor breakpoint cluster region (m-bcr).69 Splicing out exons e1′ and e2′ creates an e1a2 junction in the BCR-ABL transcript, translating into a smaller BCR-ABL fusion protein called p190BCR-ABL. Both p210BCR-ABL and p190BCR-ABL have significantly increased tyrosine phosphokinase activity compared to the normal human c-ABL protein, p145. Saglio et al70 described a novel position for a breakpoint in the BCR gene that is located in a 3′ direction from the M-bcr region, between exons e19 and e20 (historically c3 and c4), creating a new fusion transcript with an e19a2 junction. This transcript contains a significantly larger part of the BCR gene and codes for the p230BCR-ABL fusion protein. In other cases, complex variant translocations involve three or more chromosomes, mask the Ph marker, or a fusion gene is created by the insertion of ABL sequences into a normal appearing chromosome 22.
Translocation t(9;22)(q34;q11) and fusion products. The Ph is a shortened chromosome 22 that results from the transposition of 3′-ABL segments on chromosome 9 to 5′-BCR segments on chromosome 22. Whereas breakpoint locations on chromosome 9 appear rather constant 5′ of ABL exon a2, several breakpoint cluster regions have been identified along the BCR gene on chromosome 22. Depending on which breakpoints are involved on chromosome 22, differently sized segments from the BCR gene are joined together with the 3′-sequences of the ABL gene. The translocation thus results in fusion mRNA molecules of different length (e1a2, b2a2, b3a2, e19a2) and, subsequently, chimeric protein products with variable molecular weights and presumably function (p190, p210, p230) (see text for details).
Translocation t(9;22)(q34;q11) and fusion products. The Ph is a shortened chromosome 22 that results from the transposition of 3′-ABL segments on chromosome 9 to 5′-BCR segments on chromosome 22. Whereas breakpoint locations on chromosome 9 appear rather constant 5′ of ABL exon a2, several breakpoint cluster regions have been identified along the BCR gene on chromosome 22. Depending on which breakpoints are involved on chromosome 22, differently sized segments from the BCR gene are joined together with the 3′-sequences of the ABL gene. The translocation thus results in fusion mRNA molecules of different length (e1a2, b2a2, b3a2, e19a2) and, subsequently, chimeric protein products with variable molecular weights and presumably function (p190, p210, p230) (see text for details).
Traditional karyotypic studies underestimate the incidence of theBCR-ABL fusion gene, and Ph− cytogenetics with positive molecular tests for the BCR-ABL fusion gene have been documented.62,71-73 Molecular tools for the detection of the BCR-ABL fusion gene include fluorescent in situ hybridization (FISH),74 polymerase chain reaction (PCR),75 and pulsed-field gel electrophoresis,76 which are complementary to cytogenetic analysis.73 Southern blot analysis, as it is applied in CML, is inadequate as a single diagnostic tool for Ph+ ALL, since it detects p210BCR-ABL but not p190BCR-ABL which is found in 50% to 80% of Ph+ ALL, because breakpoints on the BCR gene occur outside the M-bcr region in an area too large to be reliably recognized.71
The variability of the breakpoint locations within the BCRgene, and the fact that p190BCR-ABL is predominantly associated with ALL, while p210BCR-ABL is most consistently associated with CML suggests a distinction between true de novo ALL (m-bcr, p190BCR-ABL) from CML in lymphoid blastic phase (M-bcr, p210BCR-ABL). There is also a difference in the frequency of expression of p190BCR-ABL between pediatric and adult ALL (80% v 50% of Ph+ ALL cases).77,78 In vitro studies showing that p190BCR-ABL is a more active tyrosine kinase than is p210BCR-ABL also suggest that these entities are different on the basis of their breakpoint location.77,79,80 The relation among these disease groups is still not well understood.81 Melo61 reported that the location of the breakpoint in the BCR gene (and/or possibly in the ABL gene) determined the disease phenotype. The hybrid gene product p190BCR-ABL may function preferentially during lymphoid and monocytic differentiation, while p210BCR-ABLcontains additional BCR-sequences that appear to affect pathways common to both ALL and CML precursors. However, p190BCR-ABLtranscripts are not confined to primary acute leukemias. Using quantitative reverse transcription-PCR (RT-PCR) for p210BCR-ABL and p190BCR-ABL mRNA, Van Rhee et al,82 in a study of adult leukemias, detected p190BCR-ABL mRNA in 88% (14 of 16) of patients with CML in chronic phase, 100% (10 of 10) of patients with CML in lymphoid blastic phase, and 100% (10 of 10) of cases with p210BCR-ABL-positive ALL. However, the p190BCR-ABL/ p210BCR-ABL ratio was 10 times greater in ALL than CML, whereas the ratio was similar in CML at diagnosis, in chronic phase, and in lymphoid blastic phase.
Several studies reported on the association between breakpoint location and clinical features and prognosis in Ph+ALL.79,80 Secker-Walker et al80 studied 113 adult patients with Ph+ ALL and found no significant difference between those with an M-bcr breakpoint and those with a different breakpoint location for age, immunophenotype, or outcome. Only the peripheral WBC count was significantly higher with an M-bcr breakpoint. Likewise, Kantarjian et al77 found no significant clinical, laboratory, or karyotypic differences in patients with p210BCR-ABL versus p190BCR-ABL ALL. Patients with either abnormality had similar incidences of older age, organomegaly, anemia, leukocytosis, thrombocytopenia, and blastosis. Although patients with p190BCR-ABL disease showed a trend for longer CR duration, overall survival was not influenced by the expression of either p190BCR-ABL or p210BCR-ABL. When maintenance therapy was not given after CR, some patients with p210BCR-ABL disease demonstrated a “second chronic phase” CML-like hematologic picture in the blood.77
In patients treated with bone marrow transplantation (BMT) for Ph+ ALL, a different clinical behavior was reported for Ph+ ALL expressing p190BCR-ABL compared with p210BCR-ABL. In a study of 36 patients (29 adults) with Ph+ ALL, Radich et al83 showed that detection of BCR-ABL by PCR after BMT correlated with a high risk of relapse, and that the expression of p190BCR-ABL was associated with a higher risk of relapse than was the expression of p210BCR-ABL. All of the relapsed patients were adults.
Adult patients with Ph+ ALL tended to be older, had higher WBC and blast counts and, in some studies, more frequent lymphadenopathy and splenomegaly than Ph−patients.84,85 Secker-Walker et al80 found that 44% of ALL patients older than 50 years were Ph+. Almost all cases of Ph+ ALL had a pre-B phenotype,7,8,36,84 and other immunophenotypes were rare.7,36,86,87 Increased expression of CD10 (“common ALL antigen” or CALLA) was found with Ph+ ALL, and in CALLA+ cases BCR-ABL identified a group of patients with short remission duration and poor DFS.73,84 Myeloid markers were present in 40% to 65% of Ph+ ALL cases.6 86
In children, Ph+ ALL has a dismal prognosis even with intensive chemotherapy programs that have improved survival in other cytogenetic subgroups.15,18,88,89 A similarly poor prognosis was reported in adults where, despite high remission rates comparable to those of Ph− ALL, remission duration and survival times were short (Table4).25,54,73,84,90 Therefore, patients with Ph+ALL are suitable candidates for innovative and intensified strategies. In younger patients, allogeneic BMT with a related or unrelated donor, or peripheral blood stem cell transplantation during first CR are indicated, although relapse rates tend to be high ranging from 40% to 80%, and disease recurrence after BMT has a particularly devastating prognosis in adults.83,91 In cases where no match can be found or marrow transplantation is not an option for other reasons, high-dose ara-C containing regimens can achieve considerable response rates.84 Other treatment alternatives include interferon, adoptive immunotherapy, and antibody-based therapies.83Unfortunately, to date, no satisfying track-record of durable responses has been established with any of these approaches.
Characteristics and Clinical Outcomes of Ph+ Adult ALL
| No. . | Ph (%) . | Median Age (yr) . | Median WBC Count . | CR (%) . | CRD (mo) . | MST (mo) . | References . |
|---|---|---|---|---|---|---|---|
| 334 | 12 | 46% > 50 | 34% > 50 | 56 | 9 | 11 | Preti et al84 |
| 15 | 50 | 26.5 | 89.5 | 50 | 5-10 | 7.5 | Bloomfield et al85 |
| 113 | 20 | 39.7 | 73.3 | 63.6 | NA | 7 | Secker-Walker et al80 |
| 56 | 30 | 39 | 26.3 | 71 | 10 | 11.2 | Westbrook et al73 |
| 56 | 26.8 | 39 | 123-150 | 73.33-150 | 6 | NA | Specchia et al86 |
| 443 | 29 | 45 | 30 | 59 | NA3-151 | NA | GFCH7 |
| 58 | 21 | 51 | 57 | 75 | 8.5 | NA | Fenaux et al8 |
| 350 | 11 | NA | NA | 83 | NA | NA3-152 | UKALL XA10 |
| 172 | 17 | 35 | 24 | 46 | 7 | 11 | TIWCL18,373-153 |
| No. . | Ph (%) . | Median Age (yr) . | Median WBC Count . | CR (%) . | CRD (mo) . | MST (mo) . | References . |
|---|---|---|---|---|---|---|---|
| 334 | 12 | 46% > 50 | 34% > 50 | 56 | 9 | 11 | Preti et al84 |
| 15 | 50 | 26.5 | 89.5 | 50 | 5-10 | 7.5 | Bloomfield et al85 |
| 113 | 20 | 39.7 | 73.3 | 63.6 | NA | 7 | Secker-Walker et al80 |
| 56 | 30 | 39 | 26.3 | 71 | 10 | 11.2 | Westbrook et al73 |
| 56 | 26.8 | 39 | 123-150 | 73.33-150 | 6 | NA | Specchia et al86 |
| 443 | 29 | 45 | 30 | 59 | NA3-151 | NA | GFCH7 |
| 58 | 21 | 51 | 57 | 75 | 8.5 | NA | Fenaux et al8 |
| 350 | 11 | NA | NA | 83 | NA | NA3-152 | UKALL XA10 |
| 172 | 17 | 35 | 24 | 46 | 7 | 11 | TIWCL18,373-153 |
Abbreviation: NA, not available.
Not significantly different from Ph− ALL.
Five months median DFS.
13% DFS at 3 years.
Includes data from Bloomfield et al.18
Abnormalities of the Short Arm of Chromosome 9
Loss or mutation of 9p21-22.
In 1983 Kowalczyk and Sandberg92 described 7 children with ALL, 5 with deletions of the short arm of chromosome 9 (9p−) and 2 with total loss of chromosome 9. In all cases segment 9p21-pter was missing. Compared with other children with ALL, the group had distinct clinical features such as older age, prominent lymphadenopathy and splenomegaly, and high WBC count and percentage of blasts. Four of the seven children had T-cell ALL. Median survival time for children with 9p− abnormalities was 1 year shorter than for children without these anomalies. In 1985 Chilcote et al93reported 65 patients with ALL (41 children and 24 adults). They singled out 8 patients (4 children and 4 adults) with “lymphomatous” features and showed that 6 of them (3 children and 3 adults) had loss of bands p21-p22 on the short arm of chromosome 9, whereas these bands were missing in only 1 out of the remaining 57 cases. The mechanisms involved included deletions, unbalanced translocations, or loss of the entire chromosome. The outcome for these patients was poor, as in the Kowalczyk and Sandberg study. The investigators hypothesized a possible suppressor gene in or near 9p21-22 that is involved in the control of proliferation of lymphoid precursors. Finally, in 1987 Pollack and Hagemeijer94described 32 patients (18 adults and 14 children) with a 9p− karyotype, including 20 with ALL (10 adults and 10 children). Among these, 3 adults showed 9p− as the sole karyotypic anomaly, whereas none of the children had 9p− without other chromosomal changes. On average, the karyotypes in children appeared more complex than in adults. However, no association of 9p− with T-cell ALL or “lymphomatous” features was seen. Other studies have confirmed that 9p anomalies are more likely in ALL than in other hematological malignancies, with a reported frequency of 7% to 13%, and no apparent difference in childhood versus adult ALL.92,93 95-97 An association with high-risk clinical features such as older age, higher leukocyte and blast counts, bulky disease, increased incidence of CNS disease, or T-cell immunophenotype was found in some, but not in all studies.
The smallest segment lost included 9p21. Specific deletions of 9p21 have also been described in a variety of human, rodent, and simian cancer cell lines, pointing at 9p21 as a possible location for a tumor suppressor gene.98,99 Detailed molecular analyses of this location, searching for a possible tumor suppressor gene, have been performed with a multitude of different probes.100 Trent et al101 have localized the interferon gene cluster to 9p21, and the assignment by Carrera et al102 of the methyladenosine phosphorylase (MTAPase) gene, coding for an essential enzyme in the purine salvage pathways, to the same region assumed new clinical relevance. Using molecular analysis, the interferon gene cluster was shown to be deleted in 43% of leukemia-derived cell lines and in 29% of primary leukemia samples,95,103 whereas MTAPase deficiency could be demonstrated in 10% of leukemias and, in particular, in 38% of cases of T-cell ALL.104 Kamb et al105 and Nobori et al106 analyzed 9p21 deletions in detail and localized two previously cloned genes to the 9p21 segment: p16INK4A and the structurally homologous gene, p15INK4B, located only 25 kb centromeric to p16INK4A. They named these genes Multiple Tumor Suppressor Gene 1 (MTS1) and Multiple Tumor Suppressor Gene 2 (MTS2), respectively, as they could demonstrate high frequencies of homozygous deletions in a variety of human tumor cell lines. Kamb et al105 also studied four leukemia cell lines and found deletions within 9p21 in one. Nobori et al106 analyzed 14 leukemia cell lines by PCR and foundMTS1 deletions in 9 of them (64%), with deletions of the MTAPase and interferon genes in 29% and 50% of cases, respectively. Both MTS1 and MTS2 encode for proteins that inhibit the cyclin-dependent kinases CDK4 and CDK6 and play a crucial role in cell cycle progression,100,107 108 which makes them ideal candidates for a putative tumor suppressor gene at 9p21.
The main mechanism for p16INK4A inactivation is biallelic deletions, with p15INK4B gene codeletions in most but not all cases (Table5).108 Current data suggest thatp16INK4A is the primary target of such deletions. This is underscored by the fact that in most studiesp15INK4B deletions occur only inp16INK4A-deleted cases, whereas the converse does not.109-112 This has also been shown forinterferon-α gene deletions.113 The frequency of homozygous deletions of theinterferon-α gene is lower than that forp16INK4A in one published report.103
p16INK4A/p15INK4B Deletions (in %) in ALL
| References . | p16 Ho . | p16 He . | p16 Mu . | p15 Del . | Comments . |
|---|---|---|---|---|---|
| Hebert et al109 and Cayuela et al110 | 45 | NA | NA | 36 | Adults and children Strong correlation of p16 and p15 deletion with T-cell ALL |
| Cayuela et al97 | 76 | 10 | 50 (2/4 He) | NA | Adults and children |
| Only T-cell samples analyzed | |||||
| Quesnel | 14 | NA | 1.1 (1/88) | NA | Mainly adults |
| et al113 | No association with T-cell ALL | ||||
| Associated with poor prognostic features | |||||
| 33% IFN-α gene deletion in p16-deleted cases | |||||
| Rasool et al114 | 27 | NA | 11 (1/9) | 4 | Children |
| 9 hemizygous deletions for 9q21 markers | |||||
| No association with T-cell ALL | |||||
| Haidar et al111 | 22 | 0 | NA | 11 | Adults |
| No association with T-cell ALL | |||||
| Ogawa et al112 | 27 | 16 | 0 | 34 | Mainly adults |
| p16 deletions more frequent in pre-B ALL | |||||
| All patients with loss of p15 also had loss of p16 | |||||
| IFN-α gene loss only with loss of p16 | |||||
| Dreyling et al115 | 45 | 45 | 0 | 45 | Only adult data presented, children omitted |
| Schröder et al118 | 17 | 10 | NA | NA | Adults and children |
| p16 deletions in 1/23 B-lineage and 6/7 T-cell ALL |
| References . | p16 Ho . | p16 He . | p16 Mu . | p15 Del . | Comments . |
|---|---|---|---|---|---|
| Hebert et al109 and Cayuela et al110 | 45 | NA | NA | 36 | Adults and children Strong correlation of p16 and p15 deletion with T-cell ALL |
| Cayuela et al97 | 76 | 10 | 50 (2/4 He) | NA | Adults and children |
| Only T-cell samples analyzed | |||||
| Quesnel | 14 | NA | 1.1 (1/88) | NA | Mainly adults |
| et al113 | No association with T-cell ALL | ||||
| Associated with poor prognostic features | |||||
| 33% IFN-α gene deletion in p16-deleted cases | |||||
| Rasool et al114 | 27 | NA | 11 (1/9) | 4 | Children |
| 9 hemizygous deletions for 9q21 markers | |||||
| No association with T-cell ALL | |||||
| Haidar et al111 | 22 | 0 | NA | 11 | Adults |
| No association with T-cell ALL | |||||
| Ogawa et al112 | 27 | 16 | 0 | 34 | Mainly adults |
| p16 deletions more frequent in pre-B ALL | |||||
| All patients with loss of p15 also had loss of p16 | |||||
| IFN-α gene loss only with loss of p16 | |||||
| Dreyling et al115 | 45 | 45 | 0 | 45 | Only adult data presented, children omitted |
| Schröder et al118 | 17 | 10 | NA | NA | Adults and children |
| p16 deletions in 1/23 B-lineage and 6/7 T-cell ALL |
Abbreviations: IFN-α, interferon-α; Ho, homozygous deletion; He, hemizygous deletion; Mu, point mutation; del, deletion; NA, not available.
Data analysis similar to Quesnel et al.108
Point mutations within p16INK4A are rare (Table 5). Quesnel et al113 analyzed p16INK4A gene deletions by Southern blot in 63 ALL patients (61 adults) and found 9 cases with homozygous deletions of p16INK4A (3 T-cell ALL, 6 precursor B-cell ALL). Single-stranded conformation polymorphism (SSCP) analysis of exons 1 and 2 ofp16INK4A was performed in 88 cases of ALL, including the 63 cases mentioned. Only one missense mutation at codon 49 was detected. Inactivation of p16INK4A occurred mainly through deletions of both copies of the gene, whereas deletion of one allele and point mutation of the other (as is frequently observed with the tumor suppressor gene p53) were rare.97,112,114 115
More recently Herman et al116 and Batova et al117 presented evidence for an additional mechanism of gene inactivation. They showed that the p15INK4Bgene was preferentially hypermethylated at a 5′-CpG island. This was demonstrated in 17 of 45 children with T-cell ALL at diagnosis and in 7 of 32 at relapse. Methylation of the p16INK4A gene was rare; it occurred in only 2 of 49 patients at diagnosis and none at relapse. Methylation correlated with loss of transcription, and the preferred methylation site at p15INK4B in ALL lended strong support for a role of p15INK4Binactivation in the pathogenesis of ALL. Whether a similar mechanism is operative in adult ALL patients remains to be investigated.
Disagreement still exists about whether 9p21 abnormalities correlate with immunophenotypic or prognostic features. Strong correlations with T-cell ALL have been reported by some investigators96,109,110,118 but not by others.111,112 Quesnel et al113 found at least one poor prognostic factor, such as bulky disease or high WBC count, in patients with ALL and p16INK4A gene deletions, and most of these patients relapsed. However, no strong association with T-cell lineage was evident. Likewise, Fizzotti et al119reported a significant relationship betweenp16INK4A/p15INK4B gene deletions and leukemic cell mass and WBC count, but no difference by immunophenotype. The GFCH found 9p21 abnormalities in 15% of ALL patients.7The abnormalities had no effect on prognosis and no significant correlation with T or B lineage. Nevertheless, abnormalities of chromosome 9p21 with deletions of thep16INK4A/p15INK4B region are highly specific to lymphoid tumors and have been among the most consistent genetic defects found in ALL to date.
t/dic(9;12)(p11-12)(p11-13).
This rare group of chromosomal abnormalities represents, for the most part, unbalanced translocations with loss of part of the short arms of chromosomes 9 and 12. Considerable breakpoint variation exists on 9p11, where the translocation can fall anywhere in a 300-kb segment.34 This abnormality was first discovered in 1985 by Heerema et al,22 who described a child with the karyotype 45, XY, −9, −12, +der(12)t(9;12)(q1?;p13). In 1987 Carroll et al120 reported eight children with precursor-B-cell ALL and t/dic(9;12)(p11-13;p11-12); seven were hypodiploid, with loss of both chromosomes 9 and 12 and a remaining der(12)t/dic(9;12), whereas the eighth child retained two normal chromosomes 9, with one normal chromosome 12 and a der(12)t/dic(9;12). In four children this abnormality was the only cytogenetic change. Since then other dicentric translocations involving the short arms of chromosomes 9 and 12 have been described. Larger series were published in 1992 by the United Kingdom Cancer Cytogenetics Group121 and by Mahmoud et al.122 In the latter series 15 of 2,303 children carried a t/dic(9;12). There was a striking association with a pre-B/early-pre-B-cell ALL phenotype, male gender, and an excellent prognosis. In 1995 a dic(9;12) study group analyzed additional 14 cases of dic(9;12)(p11-13;p11-12),123 including 11 cases of ALL with a precursor-B phenotype (5 children and 6 adults). The ALL patients were characterized by young age (median age 15 years, all but one under 25 years), a median WBC count of 6.3 × 109/L, no CNS involvement, predominantly FAB L1 morphology, early pre-B-cell phenotype, positivity for CD10, and an excellent prognosis. Almost all achieved CR, with DFS and overall survivals (OS) of 93% and 97%, respectively, at a median follow-up of 61 months. In 41% of cases dic(9;12) was the sole cytogenetic abnormality, in which cases the dicentric chromosomes resulted in hypodiploidy. Additional numerical abnormalities were found in 34%, with trisomy 8 being the most common.
Behrendt et al123 described an interesting case of relapsed ALL with both dic(9;12) and t(21;21)(q21;q22). Translocations involving 21q22 occur in secondary leukemia patients after treatment with topoisomerase II inhibitors, and the patient had been previously treated with VP16.
Comparable data for adult patients have to be largely extrapolated from the available information in children or adolescents. This chromosomal abnormality is rare in adults, and little is known about its effect on prognosis.
Abnormalities Involving 11q23
Abnormalities of 11q23 are among the most frequent cytogenetic abnormalities in a variety of adult hematopoietic malignancies. They also occur in 60% to 70% of acute leukemias in infants.124-126 Their frequency in older children and adults with ALL is lower (up to 10%).124-126 When there has been previous therapy with topoisomerase II inhibitors, frequencies as high as 80% can be observed.127 128
The common molecular denominator is the disruption of a gene located at band q23 of chromosome 11. In 1991 Ziemin-van der Poel et al129 and others identified this gene at 11q23 as the “mixed-lineage leukemia,” or “myeloid-lymphoid leukemia,” gene (MLL, also referred to asALL-1, HRX, or HTRX1).129-131 The gene contains at least 21 exons and spans about 100 kb. It encodes a protein of 3,968 amino acids with a molecular weight of 431 kD. It contains two central zinc-finger domains and a 210-amino acid C-terminal and shows significant homology with the Drosophila trithoraxprotein.130,131 At the N-terminal are a methyltransferase domain and three AT-hook motifs through which the protein can bind to AT-rich regions of the minor groove of the DNA double helix.132
The function of the MLL protein is not entirely clear, but it appears to act as a transcription factor in the regulation of differentiation pathways by direct interaction with DNA or with other DNA-binding proteins.133,134 In 1993 Thirman et al135identified a single complementary DNA (cDNA) probe from the MLLgene that could detect rearrangements with a breakpoint at band 11q23 when DNA from leukemia patients was digested with a single enzyme. The breakpoints cluster within an 8.3 kb-region between exons 5 and 11. This is a comparatively restricted area that can be identified by Southern blot analysis of genomic DNA probed with a small cDNA fragment that overlaps the cluster region.
More than 20 reciprocal chromosomal loci are known to participate in 11q23 translocations, the most common are 4q21, 9p22, 19p13, and 1p32, and many partner genes have been identified (Table 6).133,136 With the exception of AF10 (10p12-14) and AF17 (17q21), as well as ENL (19p13) and AF9 (9p22), no close relation exists among the various partner genes on a structural or functional basis.134 The breakpoints on the reciprocal chromosomes are also scattered over larger segments (in the case of 4q21, over at least 38 kb) than is the case with MLL, giving rise to fusion transcripts that vary in length and exon composition.137Janssen et al125 used RT-PCR to show fusion transcripts containing MLL and AF4, the gene located at 4q21. They demonstrated that a large number of differently sized fusion transcripts was probably resulting from alternative splicing events. Overall they identified eight different MLL-AF4 versions. The fusion genes contain nearly identical 5′ portions of MLLtransposed to various partner genes. The resultant chimeric proteins contain the N-terminal portions of the MLL protein, including the AT-hook motifs and methyltransferase domain, but not the C-terminal segments including the central zinc-finger motifs.133 138
11q23 Partner Chromosome Breakpoints and Fusion Transcripts in ALL
| Chromosome Breakpoint . | Gene Involved . | Fusion Transcript . | Comments . |
|---|---|---|---|
| 1p32 | AF1p | AF1p-MLL | Normally expressed as 4.4 kb transcript in various tissues |
| No known similarity with other genes known to fuse withMLL, but 88% homology with eps15 (refs 133, 134) | |||
| 4q21 | AF4 (FEL, MLLT2) | AF4-MLL | Expressed as two 12-kb and 10.5-kb transcripts in various tissues |
| Encodes protein of 140 kD with putative nuclear targeting sequence and consensus sequence for ATP/GTP binding | |||
| Proline/serine-rich protein | |||
| Most common 11q23 fusion gene (refs 133, 134) | |||
| 10p12-14 | AF10 | AF10-MLL | 5.5-kb mRNA encoding for protein of 1,027 amino acids |
| Contains zinc-finger motif near N-terminus and leucine-zipper motif near C-terminus | |||
| High degree of similarity with AF17 | |||
| IL-2 gene maps to breakpoint region (refs 133, 134) | |||
| 19p13 | ENL (MLLT1, LTG19) | MLL-ENL | 4.7-kb mRNA encoding for protein of 559 amino acids |
| Contains putative nuclear targeting sequence and consensus sequence for ATP/GTP binding | |||
| Proline/serine-rich protein | |||
| 56% identical with AF9 (refs 133, 134) |
| Chromosome Breakpoint . | Gene Involved . | Fusion Transcript . | Comments . |
|---|---|---|---|
| 1p32 | AF1p | AF1p-MLL | Normally expressed as 4.4 kb transcript in various tissues |
| No known similarity with other genes known to fuse withMLL, but 88% homology with eps15 (refs 133, 134) | |||
| 4q21 | AF4 (FEL, MLLT2) | AF4-MLL | Expressed as two 12-kb and 10.5-kb transcripts in various tissues |
| Encodes protein of 140 kD with putative nuclear targeting sequence and consensus sequence for ATP/GTP binding | |||
| Proline/serine-rich protein | |||
| Most common 11q23 fusion gene (refs 133, 134) | |||
| 10p12-14 | AF10 | AF10-MLL | 5.5-kb mRNA encoding for protein of 1,027 amino acids |
| Contains zinc-finger motif near N-terminus and leucine-zipper motif near C-terminus | |||
| High degree of similarity with AF17 | |||
| IL-2 gene maps to breakpoint region (refs 133, 134) | |||
| 19p13 | ENL (MLLT1, LTG19) | MLL-ENL | 4.7-kb mRNA encoding for protein of 559 amino acids |
| Contains putative nuclear targeting sequence and consensus sequence for ATP/GTP binding | |||
| Proline/serine-rich protein | |||
| 56% identical with AF9 (refs 133, 134) |
Several mechanisms for MLL gene rearrangements are possible including aberrant V-D-J recombination, homologous recombination between Alu-repeats, and topoisomerase-II–mediated nonhomologous recombinations.133 Truncation or loss of function of theMLL gene alone may be the crucial event in leukemogenesis, with partner genes assuming a minor role and being interchangeable. However, Rogaia et al139 recently showed that fusion of theMLL gene with eps15 (on chromosome 1, band p32) in AML altered the cellular compartmentalization of the fusion protein, providing a putative mechanism for activation of MLL in 11q23 recombinations and suggesting a more active role of the fusion partner genes in leukemogenesis.
t(4;11)(q21;q23).
The most common translocation involving 11q23 is translocation t(4;11)(q21;q23), first described by Oshimura et al140 in 1977. It is observed in more than 60% of infants with ALL, 2% of children with ALL, and 3% to 6% of adults with ALL.7,9,18,37 It is invariably associated with young age (generally under 2 years), female sex, and high WBC counts, and is frequently associated with organomegaly and involvement of sanctuary sites such as the CNS.33,90,125,141 The immunophenotype is of the early pre-B or pre-B-cell type, is positive for TdT, HLA-DR, and CD19 with rearrangements of the Ig heavy-chain (IgH) genes, and is variably CD10−. Cells frequently coexpress myeloid antigens, such as CD13, CD15, or CD33. Janssen et al125found a significant association of MLL-AF4 recombinations in particular with CDw65, compared with early pre-B-cell ALL without this genetic translocation. This association emphasizes an important characteristic of 11q23 abnormalities suggesting that the transforming event originates at the stage of a pluripotential progenitor cell with the capacity to differentiate to both lymphoid and myeloid lineages.142 In fact, 11q23 abnormalities can frequently be detected in biphenotypic or acute myeloid leukemias.128 143
The clinical outcome for both adults and children with the t(4;11)(q21;q23) translocation is poor (Table7).33
Clinical Outcomes for Patients With t(4;11)(q21;q23)
| Proportion With t(4;11)(q21;q23) . | UKALL XA Adults . | TIWCL Adults . | TIWCL Children . |
|---|---|---|---|
| 10/350 . | 9/172 . | 9/157 . | |
| CR (%) | 70 | 50 | 88 |
| MCRD (mo) | NA | NA | 3 |
| MST (mo) | NA | 7 | 9 |
| MDFS (mo) | 4 | 2 | 5 |
| DFS | 24% at 3 yr | 0% at 5 yr | 0% at 5 yr |
| Proportion With t(4;11)(q21;q23) . | UKALL XA Adults . | TIWCL Adults . | TIWCL Children . |
|---|---|---|---|
| 10/350 . | 9/172 . | 9/157 . | |
| CR (%) | 70 | 50 | 88 |
| MCRD (mo) | NA | NA | 3 |
| MST (mo) | NA | 7 | 9 |
| MDFS (mo) | 4 | 2 | 5 |
| DFS | 24% at 3 yr | 0% at 5 yr | 0% at 5 yr |
Abbreviations: MCRD, median complete remission duration; MDFS, median disease-free survival.
The GFCH demonstrated 11q23 abnormalities in 7% of adult patients with ALL.7 Half of these patients had a t(4;11), 50% of whom had coexpression of myeloid markers: their CR rate was 75%, with a median DFS of 7 months and no DFS at 3 years. This was slightly worse than for the whole 11q23 group, in whom the CR rate was 72% and the median DFS was 8 months, but the 3-year DFS was 26%. No differences in DFS were noted with translocations as opposed to deletions of 11q23.
Janssen et al125 analyzed MLL-AF4 rearrangements by PCR in a series of 46 patients with early pre-B-ALL (34 adults, 12 children). They demonstrated fusion transcripts in 39% of the patients (14 adults, 4 children). Using intensified treatment strategies which included BMT in 4 adult cases, they reported 9 of 19 ALL patients withMLL-AF4 to be in remission for up to 54 months (median CR duration [CRD], 26 months), with 7 of the 9 showing PCR negativity for minimal residual disease.
These results emphasize that survival can be improved substantially by applying intensive therapy to cytogenetic groups that had been previously defined as poor risk by their response to standard treatments. Although the data in adult ALL with translocation t(4;11) are scarce, risk-adapted therapy might benefit this group as it did in children.33
t(11;19)(q23;p13).
Translocation t(11;19)(q23;p13) shares similar clinical and prognostic features with t(4;11). It results in a fusion of the ENL gene (also called LTG19 or MLLT1) from chromosome 19 to the 5′ end of MLL on 11q23.144 The translocation is associated with younger age and high WBC count, as well as IgHgene rearrangements in blast cells and coexpression of lymphoid and myelomonocytoid antigens.
Abnormalities Involving 19p13
t(1;19)(q23;p13).
The two known translocations involving band p13 of chromosome 19 are t(1;19)(q23;p13) and its rarer variant t(17;19)(q21-22;p13). Translocation t(1;19) was first reported in 1984 by Carroll et al,145 who found that some leukemias with a pre-B-cell ALL phenotype (cytoplasmic Ig [clg]-positive and surface Ig [slg]-negative) carried this abnormality. It was confirmed in subsequent studies as one of the most common recurring translocations in childhood ALL, with a frequency of 5% to 6% overall, and of 25% in pre-B-cell ALL.15,90,146 It is also seen in 1% of childhood cases of early pre-B-cell ALL (clg−, slg−) and occasionally in ALL patients with a transitional pre-B phenotype (clg+, μ heavy-chain Igs detectable on cell surface).147-150 It is present in less than 5% of adult ALL cases.7,8 151
The translocation can be observed in two principal forms. In its unbalanced form, −19, +der(19)t(1;19), two normal chromosomes 1 are present. Shikano et al152 suggested that this anomaly may result from loss of the der(1)t(1;19)(q23;p13.3) by nondisjunction or asynchronous replication, with replacement of the normal chromosome 1 by a second copy, which ultimately resulted in trisomy for 1q23 → 1qter. Abnormalities of chromosome 1, such as complete or partial trisomies for the long arm, are known to arise during clonal evolution and can be observed in recurrent hematologic malignancies.153-155 The other form of the translocation is a balanced rearrangement, a simple reciprocal translocation without net loss or gain of genetic material. Unbalanced translocations appear to be more common than balanced rearrangements (about 75% are unbalanced). No differences in clinical presentation or prognosis exist.34 153
At a molecular level, the breakpoint on chromosome 19 has been mapped to a tightly clustered region on band p13.2-p13.3. This segment contains a gene, E2A, which encodes the two transcription factors E12 and E47 via alternative splicing of precursor mRNA.156 E12 and E47 are ubiquitous helix-loop-helix (HLH)-containing proteins that bind to the E-box element in the κ light-chain DNA-enhancer region.157 They are considered essential for normal lymphopoiesis and regulation of B-cell development.158 Breakpoints in the E2A gene occur almost exclusively in a 3.5-kb intron segment between exons 13 and 14.159 The breakpoint region on chromosome 1q23 appears to be more dispersed and lies within an intron of at least 50 kb in size.159 It disrupts the homeobox-containing “pre-B-cell leukemia” gene (PRL, also referred to as PBX1), which is transcriptionally silent in lymphoid cells. The genomic organization of PBX1 has not yet been detailed in entirety.158 The chromosomal rearrangement fuses the 5′ sequences of the E2A gene with 3′ sequences of PBX1.The resulting fusion transcript is a chimeric transcription factor which preserves the activation domain of E2A but has its DNA-binding domain and HLH dimerization domain replaced byPBX1.159E2A-PBX1 appears to function as a potent transcriptional activator.160,161 Despite the size of the breakpoint location on 1q23, an unvarying portion ofPBX1 is fused with E2A as most mRNAs generated by the translocation have the same sequence, suggesting that the site-specific fusion between E2A and PBX1 is pathogenetically important.157 However, differential splicing seems to occur at the mRNA level, leading to the production of two E2A-PBX1 chimeric proteins that differ in their extreme carboxyl-terminal end.158 Several groups have detected E2A-PBX1fusion transcripts that differ in their mRNA sequences.162,163 Their exact derivation, as well as their clinical significance, are unknown. Other members of the PBXfamily with homeodomains that are nearly identical to that ofPBX1, namely PBX2 and PBX3, have been identified, but their role in lymphoid cell transformation is unknown.164
A strong association exists between t(1;19) and pre-B-ALL, especially in children,145 where the translocation is present in 25% to 30% of cases.15,33,90 Among 73 adults and 101 children with ALL, Fenaux et al8 found that all patients with t(1;19) (1 adult and 7 children) had a pre-B-ALL phenotype. No other translocation demonstrated a comparably close relationship to a specific immunophenotype in this study. Raimondi et al147found that most cases with pre-B-ALL and t(1;19) were pseudodiploid, and hyperdiploidy with more than 50 chromosomes was virtually absent. The immunophenotype in the majority of cases was positive for CD10, CD19, and clg. Borowitz et al165 analyzed the surface marker expression of t(1;19) ALL in more detail. They found 22 cases with t(1;19) among 697 patients with pre-B-ALL. Twenty of them were characterized by an identical, complex phenotype with homogenous expression of CD10, CD19, and CD9, complete absence of CD34, and partial absence of CD20. All 12 cases analyzed with molecular studies showed E2A-PBX1 abnormalities. The same complex phenotype was seen in only 8% of children with pre-B-ALL without t(1;19). Pui et al153 later added positivity for CD22 and negativity for CD21 to the characteristic surface-marker profile.
In rare cases, t(1;19) is seen in clg−, early-pre-B-ALL. Involvement of either E2A or PBX1 in these cases could not be consistently shown.150,162 In contrast to the adverse clinical prognosis of patients with pre-B-ALL and translocation t(1;19) (see below), patients with early pre-B-ALL and t(1;19) appear to fall into a standard-risk category.148 162
Clinical characteristics of pre-B-ALL with t(1;19) include presentation with high WBC counts, high LDH levels, and a DNA index of less than 1.6, which underscores that this type of leukemia is usually associated with a pseudodiploid karyotype. It frequently occurs in black people. The GFCH7 observed t(1;19)-positive leukemias in 11 (3%) of its adult ALL patients. Nine had a pseudodiploid karyotype, whereas two were hyperdiploid, with 47 to 50 chromosomes. Contrary to most other studies with t(1;19), patients tended to be younger and presented with lower WBC counts. Although all patients achieved a CR, the median DFS was only 6 months, and the 3-year DFS was 20%.
Pediatric Oncology Group studies showed that children with ALL who express E2A-PBX1 and t(1;19) and do poorly with standard or less aggressive therapy, have good responses to more intensified treatment.148,162 Therefore, to plan appropriate therapy, adequate and thorough cytogenetic studies, including molecular analysis, assume a critical role in diagnosis.148Comparable data for adult patients with ALL and expression ofE2A-PBX1 who share equally dismal outcomes with standard therapy as do children are not available.
t(17;19)(q21-q22;p13).
This less well-recognized translocation fuses the E2A gene to the “hepatic leukemia factor” (HLF) gene, a basic leucine zipper transcription factor gene, on chromosome 17.166 Whereas HLF breakpoints on chromosome 17 appear to lie consistently in intron 3, E2A breakpoints differ with respect to the inclusion of exon 13 and the insertion of a cryptic exon that contains E2A intronic sequences at its 5′ end andHLF intronic sequences at its 3′ end. These translocations involving the E2A gene have been reviewed by Hunger.158
Abnormalities Involving the T-Cell Receptor (TCR) Genes
Translocations involving the TCR genes are among the most common abnormalities in T-cell ALL,90 whereas TCR gene rearrangements are found at a lower frequency in patients with B-lymphoid markers.167 T-cell ALL is somewhat peculiar in that the majority of patients do not have a cytogenetically detectable chromosomal abnormality, more patients have pseudodiploid karyotypes, and only few have hyperdiploidy.6,168 Raimondi et al169 described an overall frequency of 40% to 45% of translocations in childhood T-cell ALL, half having breakpoints mapped to chromosomal regions encoding for TCR genes. These loci include chromosome 14 band q11 (TCR-α and -δ genes), 7q32-36 (TCR-β gene), as well as 7p15 (TCR-γ gene).170 The β-chain locus on 7q32-36 is less frequently rearranged than the α-δ chain loci proximally on 14q. Rearrangements of theTCR-γ gene on 7p15 are extremely rare. Although no specific karyotypic abnormality can be associated with a distinct clinical subtype of T-cell ALL (as in B-cell ALL [eg, Burkitt's lymphoma and t(8;14)]),171 a number of distinct chromosomal translocations have been identified.
A common theme is the juxtaposition of TCR promotor/enhancer elements to a variety of putative or proven transcription factors located at or near breakpoints on the partner chromosomes. Several families of involved transcription factors can be distinguished on the basis of structural motifs that characterize their DNA binding or dimerization domains (Table 8).157 For example, the TAL1 (also referred to as SCL, TCL5),TAL2, LYL1, and c-myc genes, encode for transcription factors that have a characteristic HLH motif. This structure consists of two amphipathic α helices that are separated by an intervening nonhelical loop and which stretch over 50 to 60 amino acids.157,172 The HLH motif allows for specific protein-protein interactions such as homodimerization or heterodimerization. A stretch of basic residues at the N terminus seems to mediate sequence-specific DNA-binding.172TAL1, TAL2, and LYL1 share more than 85% homology in their basic HLH domains. A second class of transcription factors are encoded for by the rhombotin gene family located on chromosomes 11 and 12.173 The two genes RBTN1 (or Lmo1, Ttg1, Rhom1) and RBTN2 (or Lmo2, Ttg2, Rhom2) are both involved in translocations associated with T-cell ALL. Their protein products contain two cysteine-rich regions that are referred to as LIM domains and, through stabilization of their tertiary folds by zinc, are able to interact and bind with nucleic acids or with other proteins. Finally, HOX11 is a homeobox-containing gene that is translated into a helix-turn-helix structure that shows remarkable conservation across species of a region containing about 60 amino acids that is responsible for DNA binding.157
Protein Domains and Transcription Factor Families Involved in Rearrangements With TCR Gene Loci
| Protein Domain . | Involved Gene . | Location . | Translocation . |
|---|---|---|---|
| Transcription factors | |||
| Basic HLH proteins | TAL1 (SCL, TCL5) | 1p32 | t(1;14)(p32-34;q11) |
| t(1;7)(p32;q35) | |||
| TAL2 | 9q32 | t(7;9)(q34;q32) | |
| LYL1 | 19p13 | t(7;19)(q34;p13) | |
| c-myc | 8q24 | t(8;14)(q24;q11) | |
| LIM domain proteins | RBTN1 (RHOM1, Ttg1, Lmo1) | 11p15 | t(11;14)(p15;q11) |
| RBTN2 (RHOM2, Ttg2, Lmo2) | 11p13 | t(11;14)(p13;q11) t(7;11)(q35;p13) | |
| RHOM3 | 12p12-13 | None known | |
| Homeodomain | HOX11 (TCL3) | 10q24 | t(10;14)(q24;q11) |
| proteins | t(7;10)(q35;q24) | ||
| Protein kinases | LCK | 1p34 | t(1;7)(p34;q34) |
| Notch homologue | TAN1 | 9p34 | t(7;9)(q34;p34) |
| Chimeric Ig-TCR gene | IgH | 14q32.3 | inv(14)(q11;q32.3) |
| Unknown | TCL1 | 14q32.1 | inv(14)(q11;q32.1) |
| t(14;14)(q11;q32.1) |
| Protein Domain . | Involved Gene . | Location . | Translocation . |
|---|---|---|---|
| Transcription factors | |||
| Basic HLH proteins | TAL1 (SCL, TCL5) | 1p32 | t(1;14)(p32-34;q11) |
| t(1;7)(p32;q35) | |||
| TAL2 | 9q32 | t(7;9)(q34;q32) | |
| LYL1 | 19p13 | t(7;19)(q34;p13) | |
| c-myc | 8q24 | t(8;14)(q24;q11) | |
| LIM domain proteins | RBTN1 (RHOM1, Ttg1, Lmo1) | 11p15 | t(11;14)(p15;q11) |
| RBTN2 (RHOM2, Ttg2, Lmo2) | 11p13 | t(11;14)(p13;q11) t(7;11)(q35;p13) | |
| RHOM3 | 12p12-13 | None known | |
| Homeodomain | HOX11 (TCL3) | 10q24 | t(10;14)(q24;q11) |
| proteins | t(7;10)(q35;q24) | ||
| Protein kinases | LCK | 1p34 | t(1;7)(p34;q34) |
| Notch homologue | TAN1 | 9p34 | t(7;9)(q34;p34) |
| Chimeric Ig-TCR gene | IgH | 14q32.3 | inv(14)(q11;q32.3) |
| Unknown | TCL1 | 14q32.1 | inv(14)(q11;q32.1) |
| t(14;14)(q11;q32.1) |
Abbreviation: HLH, helix-loop-helix.
Data from refs 164, 167, 172, 173, and 194.
TAL1 gene rearrangements.
TAL1 gene rearrangements are the most common abnormalities identified in children with T-cell ALL. They are associated with CD3− leukemias with commitment to the α/β lineage.174 Two mechanisms of rearrangement exist. About 3% of cases occur via a chromosomal translocation that transposes the TAL1 gene on chromosome 1 to either theTCR-α/δ locus on chromosome 14q11, as in translocation t(1;14)(p33;q11), or to theTCR-β location on 7q35, as in translocation t(1;7)(p33;q35). The latter translocation is less frequent. Another translocation, t(1;3)(p32;p21) that involves an uncharacterized segment on chromosome 3 and a segment 5′ to TAL1 has been described by Aplan et al.175 In 25% to 30% of pediatric patients withTAL1 alterations, the tumor-specific rearrangement ofTAL1 is not detectable by routine cytogenetic analysis.157,172,176,177 In these patients site-specific, submicroscopic deletion of a 90-kb to 100-kb segment fuses the coding exons of TAL1 to the first noncoding exon of the “SCL interrupting locus” (SIL) gene centromeric in a 5′ orientation on chromosome 1p33.176,178 Either translocation or interstitial deletion results in the juxtaposition of the TAL1gene to regulatory elements of either the SIL gene or the TCR genes. Although the breakpoint on the SIL gene remains remarkably constant, several breakpoints of the TAL1 gene have been identified (tald1-d3), of which the most frequent, tald1 and tald2, leave all coding exons ofTAL1 intact. The site specificity and precision of the gene rearrangement process, as well as the fact that the deletions are mediated by specific heptamer and nonamer flanking sequences that share homology to the heptamers in the recombination sequences of Ig and TCR genes, suggest that abnormal V-D-J recombination is effective in activating the TAL1 gene locus.157,179 AlthoughSIL and TCR genes are normally expressed in T cells,TAL1 is transcriptionally silent in normal T cells although it is expressed in megakaryocytes, basophilic granulocytes, and erythroblasts.172 Alternative splicing produces a variety of TAL1 transcripts.172 The two most relevant are the full-length, 42-kD TAL1α polypeptide (pp42TAL1, amino acids 1-331), which contains all three TAL1 coding exons (exons 4 to 6), and the 22-kD, N-terminally truncated polypeptide, TAL1β (pp22TAL1, amino acids 176-331), which lacks coding exon 4. Both polypeptides can be found in T-cell ALL, and both are able to form heterodimers with other basic HLH proteins such as E12 or E47, thus specifically recognizing E-box DNA transcriptional enhancer motifs.157 172
A subset of T-cell ALL patients displays cytogenetic alterations of both the TAL1 gene and RBTN2 (see below), and theTAL1 and RBTN2 polypeptides seem to dimerize with each other easily in vivo.180 Both genes appear to play a crucial role in normal hematopoiesis as severe defects in erythropoiesis occur in mice that are deficient in either gene.181 Ono et al182 recently showed in T-cell ALL cell lines that aberrant expression of TAL1 was invariably accompanied by aberrant expression of either RBTN1 orRBTN2. The coexpression also strongly induced TALLA1, a highly specific T-cell marker. It is conceivable that the TAL1and RBTN gene products act synergistically in the genesis of T-cell ALL, and that their influence on the expression of downstream genes, such as TALLA1 or other unknown genes, is part of the mechanism of leukemogenesis.
Disruptions of the TAL1 gene locus are less frequent in adults than in children, where they occur with a frequency of up to 30% of T-cell ALL cases. Stock et al183 performed Southern Blot and PCR analysis in 33 newly diagnosed adult patients with T-cell ALL and detected only 1 case with a t(1;14)(q34;q11) molecular rearrangement, but no submicroscopic deletions. No comprehensive data exist on the clinical significance of TAL1 gene rearrangements in adults with ALL. In one of the largest studies to date, Bash et al184 found that 26% of 182 children with newly diagnosed T-cell ALL had local rearrangements of the TAL1 locus. No significant clinical correlations were obvious except that WBC counts and hemoglobin were higher at diagnosis in the group with theTAL1 rearrangements. The immunophenotype was characterized by positivity for CD2 and absence of CD10 expression. This marker profile was the same that Stock et al183 had found in their single adult patient with TAL1 gene rearrangement. Response to treatment was not significantly different with or without the rearrangement, although a trend for better DFS was observed with expression of TAL1. The latter finding concurs with results of a smaller study in children by Kikuchi et al,185 who suggested that TAL1 gene rearrangements may identify a subgroup of T-cell ALL with a favorable prognosis.
The closely related genes, LYL1 and TAL2, are rearranged in chromosomal translocations t(7;19)(q34;p13) and t(7;9)(q34q32), respectively. The result is a juxtaposition of both genes to promoter/enhancer elements of the TCR-β gene. These translocations occur infrequently.172
t(10;14)(q24;q11).
Translocation t(10;14)(q24;q11) rearranges the homeobox-containing geneHOX11 (TCL3) with the TCR-δ gene; it is seen in 4% to 7% of T-cell acute leukemias.169,186 As in the TAL1 rearrangements, the coding regions of HOX11 are not disrupted by the translocation.187 The breakpoints on chromosome 10 cluster within a 15-kb region. Immediately telomeric to the breakpoint location lies the coding sequence of HOX11.188HOX11transcripts occur abundantly in leukemic blasts.187 The variant translocation t(7;10)(q35;q24) juxtaposes HOX11 to theTCR-β gene region. Both translocations result in overexpression of normal HOX11 mRNA by bringing HOX11under the influence of TCR promoter sequences.187 188
The GFCH7 found t(10;14)(q24;q11) to be the most frequent chromosomal rearrangement in patients with T-cell ALL. This translocation was associated with a favorable outcome: the CR rate was 100%, with a median DFS of 46 months and a 3-year DFS rate of 75%.7 If other 14q11 abnormalities, such as t(11;14)(p13;q11), t(1;14)(p32;q11), inv(14)(q11;q32), and del(14)(q11) were included, the clinical outcomes were less favorable, with a median DFS of 24 months and a 3-year DFS rate of 42%.
t(11;14)(p15;p11) and t(11;14)(p13;q11).
Translocation t(11;14)(p15;p11) recombines the RBTN1 gene with the TCR-δ chain gene. RBTN1 encodes an 18-kD protein characterized by the presence of two cysteine-rich structural motifs that establish it in the LIM family of transcription factors.173,189 It is a rare but consistent translocation in T-cell ALL. The RBTN2 gene is rearranged in the closely related chromosomal translocation t(11;14)(p13;q11), as well as in t(7;11)(q35;p13). The breakpoint region (T-ALLbcr) on 11p13 lies within a 26-kb segment upstream of the RBTN2promoter.190 Neither RBTN1 nor RBTN2 is expressed in normal T cells, an attribute they share with TAL1.Their increased expression appears to contribute to the pathogenesis of T-cell lymphocytic leukemia by as yet undefined mechanisms.
t(8;14)(q24;q11).
Translocation t(8;14)(q24;q11) is not specific for T-cell ALL, and has been described in pre-B-ALL.191 The rearrangements are similar to those between Ig gene loci and c-myc in B-cell malignancies such as Burkitt's lymphoma (BL), but in the translocations mentioned here, c-myc recombines with theTCR-α gene in 14q11. Breakpoints in theTCR-α gene are located 5′ of one of the joining (J) segments, and breakpoints in c-myc are located distal to coding sequences in a 3′ orientation of the third exon.191-193 These chromosomal rearrangements are too infrequent both in adults and children to provide definite correlation with clinical characteristics and with prognosis.
Translocation t(8;14)(q24;q32) and Its Variants
t(8;14)(q24;q32) is a highly specific and consistent chromosomal rearrangement in mature B-cell neoplasms.194 Mature B-cell leukemias, which account for less than 5% of all cases of ALL in children or adults, represent the leukemic phase of BL.195The chromosome rearrangement does not appear to be different in BL and in its leukemic counterpart. It is present in both the sporadic, nonendemic form of BL and the endemic African tumor-type presentation.34 Patients in the leukemic phase present with bulky extramedullary disease, frequent and early CNS involvement, and a rapidly progressive clinical course due to an unusually high blast cell proliferative rate.33 151 The malignant cells are characterized by strong expression of surface Igs, and cell morphology is almost always L3.
B-cell ALL typically contains one of three specific chromosomal translocations. In about 85% of cases t(8;14)(q24;q32) can be detected. Less frequent are the variants t(2;8)(p12;q24), with 5% prevalence, and t(8;22)(q24;q11), with about 15% prevalence.34 However, association with these chromosomal changes is not absolute. There are cases in which the typical translocations are not found, and additional karyotypic abnormalities such as dup(1q), t(1;19)(q23;p13), del(6q), 13q34 abnormalities, trisomy 20, t(X;8)(p23;q24), the Ph chromosome, or complex three-way translocations have been detected.196-198
The common denominator behind the classic translocations is a break in 8q24. This band contains the c-myc proto-ongene, which in t(8;14) is translocated to the IgH gene locus on chromosome 14q32.34,199 In the variant translocations t(2;8) and t(8;22), c-myc recombines with the Ig kappa light chain (Igκ) gene locus on chromosome 2p12 or the Ig lambda light chain (lgλ) gene locus on chromosome 22q11, respectively.34,199 These rearrangements bring c-myc under the influence of transcription-stimulating regulatory sequences in the proximity of constitutively active Ig loci.34
Of the three exons in c-myc, the first most likely remains untranslated. The protein products encoded in exons 2 and 3 share structural motifs, such as a basic domain with an HLH-segment and leucine zipper regions. Other nuclear transcription factors also share these motifs. C-myc is able to form dimers with another nuclear protein called MAX, and together they show sequence-specific binding to DNA.200
In t(8;14) the breakpoint almost always falls proximal on the centromeric side of exon 2 of c-myc, either in the 5′ flanking region or in the first exon or intron, so that the entire coding segment of c-myc becomes translocated.201 The translocation to 14q32 happens in a head-to-head (5′ to 5′) direction, either in the switch (S) or joining (J) regions on 14q32. The breakpoints at the IgH gene locus may be dispersed over a large area of approximately 170 to 190 kb.202 The 8q24 breakpoints in the variant translocations differ. They are located 3′ on the telomeric side of the c-myc locus, which means that c-myc remains on the rearranged chromosome 8, while the Ig light chain (IgL) gene loci on 2p12 and 22q11 are translocated to chromosome 8 and rearranged with c-myc. TheIgL breakpoints can fall within either the J, diversity (D), or S regions of the gene. Because of the different orientation of the variable regions of the IgL genes the rearrangements happen in a 5′ to 3′ (head-to-tail) fashion.34,203,204 The mechanisms by which c-myc becomes leukemogenic are not entirely clear yet. The dominant hypothesis is that c-myc is brought under the influence of highly active regulatory sequences of Ig genes, and thus itself becomes inappropriately expressed.34 However, other possible mechanisms are submicroscopic mutations, such as deletions of c-myc, or the utilization of different transcriptional promotors.205-207 The normal allele of c-mycremains silent.
The TIWCL found abnormalities of 8q24 (mostly 8q−, 14q+) in 6% of adult ALL patients. All patients had typical L3 morphology. Adults were affected more frequently than children, and males more than females. The majority (15 of 16 patients, including 6 children) had pseudodiploid karyotypes. The CR rate was 44%, the median CRD and survival time 5 months each, and 5-year DFS was 18%. Adult patients had a lower CR rate.5,6,18,25,36,37 The GFCH found t(8;14)(q24;q32) in 5% of patients. Two of these 21 patients showed variant translocations, all were of B-cell type, and 20 of 21 had L3 morphology. Sixty-two percent achieved CR, the median DFS was 2 months, and the 3-year DFS rate was 17%.7 Fenaux et al8 detected only 1 t(8;14) among 73 adult patients with ALL; it was slg+ but of L2 morphology. No survival data are available. Ankathil et al28 detected one t(8;14) in 46 adults and reported a survival time of 10 months.
Although mature B-cell ALL was long believed to be associated with a poor prognosis, the introduction of short-term dose-intensive regimens such as hyperfractionated cyclophosphamide, high-dose methotrexate, and cytarabine have significantly improved the clinical outcomes.151,171,208 This provided additional evidence that a change in therapy can have a beneficial impact on a disease category that has traditionally been considered “poor-risk.” This effect can also be shown in adult patients.145
Abnormalities of the Short Arm of Chromosome 12
Prigogina et al209 first described abnormalities of the short arm of chromosome 12 in ALL. Raimondi et al210 showed that rearrangements of 12p, including deletions and translocations with various partner chromosomes such as 1, 3, 9, 10, and 17, were frequent events in childhood ALL, occurring in about 10% of patients. Most cases were characterized by a pre-B-phenotype and L1 morphology. As the common denominator of these rearrangements was a breakage in 12p, it could be suspected that in these instances a location on the short arm of chromosome 12 was critical to the process of leukemogenesis. The UKALL XA trial10 showed 12p abnormalities in 13 (4%) of 350 adult patients. In 11 cases, translocations involving various partner chromosomes were identified [14q11, 14q13, 10q11, 12q11, 17q12, 22q11 and 3 cases of dic(9;12)(p11;p11)]. The whole group with 12p abnormalities was characterized by a favorable outcome, with a CR rate of 92%, a median DFS of 36 months, and a 3-year DFS rate of 74% (for the total cohort of adult ALL patients: CR rate 88%, median DFS 19 months, 3 year-DFS rate 36%). The GFCH study7 identified 12p abnormalities in 23 (5%) of 443 adults. Twenty of them had monosomy 12 (8 deletions, 12 unbalanced translocations). The 12p abnormalities were most often observed in complex karyotypes and, overall, had no impact on prognosis.
Deletions of 12p13.
Several investigators have identified 12p13 as a hot spot frequently deleted in childhood ALL211,212 but rarely involved in adult ALL.213 Fine mapping of this segment showed that the deleted region lies between two genes, TEL (for “translocation, Ets, leukemia,” or ETV6), andCDKN1B (or KIP1). TEL is located at the telomeric end and encodes for a transcription-regulating protein of the Ets family of transcription factors. CDKN1B has a more centromeric location and encodes for p27kip1, an inhibitor of the cyclin E-cdk2 complex.214 The TEL gene was initially cloned by virtue of its involvement in translocation t(5;12)(q33;p13) in chronic myelomonocytic leukemia, where it is fused with the platelet-derived growth factor-β gene.215 Other chimeric TEL transcripts have been identified in both myeloid and lymphocytic leukemias, as well as in acute and chronic leukemias. Examples are TEL-MN1 in t(12;22)(p13;q11),216TEL-ABL in t(9;12)(q34;p13),217,218 or TEL-10q24 in t(10;12)(q24;p13), a variant of t(5;12).219 More recentlyTEL was shown to be fused to the AML1 (orCBFA2) gene on chromosome 21q22 in childhood B-lineage ALL.220,221 It is noteworthy that the region of loss of heterozygosity on 12p13 not only comprises TEL but also extends centromeric into the KIP1 gene, raising the possibility that loss of function of KIP1, in association with deletions and/or rearrangements of TEL, may play a crucial role in ALL.222-225
t(12;21)(p11-12;q22).
The TEL protein is characterized by two distinct regions. The Ets motif consists of a 70-amino acid, sequence-specific DNA-binding domain that is located at the C-terminal end of the protein. The N-terminus contains an HLH motif, which is able to mediate interactions with other proteins. The fusion partner in t(12;21) is AML1, a heterodimer consisting of an α- and β-subunit. The α-chain possesses a DNA-binding and transactivating motif called “runt” (in homology to the Drosophila runt gene). In translocation t(12;21) the HLH domain of TEL is fused to intron 2 of the AML1 gene in such a way that both the DNA-binding and runt domain remain intact and are expressed.225-227 Of interest, the nonrearrangedTEL allele seems to be deleted in almost all t(12;21) cases.228
Translocation t(12;21) was long considered a rarity, as it was detected in less than 0.05% of patients analyzed with cytogenetic studies.24 Therefore, it was thought to be of no significant prognostic value. However, t(12;21) is difficult to detect, and routine cytogenetic studies grossly underestimated its frequency. In several studies, the application of molecular tools such as FISH, Southern blot analysis, and RT-PCR have shown loss of heterozygosity and TEL-AML1 fusion transcripts in up to 27% of children with B-lineage ALL, making it the most common karyotypic-molecular abnormality in pediatric ALL.222,229-236 TheTEL-AML1 rearrangement conferred an exceptionally good outcome in childhood B-precursor ALL and provided prognostic information independent of the consensus workshop risk groups, age, and WBC count at presentation.231,233,234,236 Lanza et al237described a more heterogeneous behavior of t(12;21) in childhood ALL with regard to lineage involvement and outcome. They studied 51 children with ALL by RT-PCR and found a TEL-AML1 rearrangement in 11 samples. Three of these children showed a hybrid phenotype, coexpressing both lymphoid and myeloid markers. Two of the 11 children (one with hybrid phenotype) died in relapse, in the absence of other poor-risk factors.
Available data are more limited in adult ALL. Raynaud et al238 analyzed 118 adult ALL patients (30 with T-cell ALL, 88 with B-lineage ALL) by RT-PCR for the TEL-AML1 fusion transcript and found only one positive case among the B-cell samples. Aguiar et al239 studied 101 patients by RT-PCR followed by Southern blotting and direct sequencing and detected only 3 cases of common ALL that were positive for TEL-AML1 rearrangements. Overall, the frequency of TEL-AML1+ ALL was low in adult B-lineage ALL (3% to 4%), and little is known about its prognostic significance.236,238 239
Abnormalities of the Long Arm of Chromosome 6
In 1977 Oshimura et al35 described deletion of 6q as a recurrent karyotypic event in leukemias. Del(6q) occurs in about 4% to 6% of childhood ALL and probably somewhat less frequently in adults. Abnormalities of 6q are associated with other karyotypic abnormalities, although in one third of cases they occur as the sole change.33,34 The TIWCL found abnormalities of 6q in 1.7% of adults and 6.4% of children.6,18,25,37 Most deletions involved 6q21. The most frequently associated karyotypic abnormality was +21, followed by +4, +10, +11, and +14. For children, the prognosis was similar to that with no detectable cytogenetic abnormality; for adults, there were too few cases to make definite conclusions.6,18,25,37 Studies have either confirmed no difference in prognosis or reported improved outcomes for 6q−, with longer DFS or prolonged median survival times.7,10 28
Although 6q deletions can be either interstitial or terminal, most seem to be interstitial. Menasce et al240 showed that even if conventional cytogenetic analysis showed a terminal deletion, FISH study revealed it to be interstitial. Although the breakpoint locations vary, they center around a proximal region flanked by 6q14-15 and 6q21 and a more telomeric region distal to 6q23.241-243 This raises the possibility of a putative tumor suppressor gene in that area. Indeed, the c-myb oncogene is localized to 6q22-24; however, it does not appear to be involved in the 6q abnormalities of lymphoid malignancies.244
The estrogen receptor (ER) gene is located on 6q25.1.245 It has been shown to have suppressive activity on growth and metastasis in many different cell lines.246Issa et al247 demonstrated that the promoter region of theER gene is aberrantly methylated at CpG islands in leukemia cell lines, pediatric and adult acute lymphocytic as well as myeloid leukemias, and most hematopoietic tumors, including lymphomas, in which it was associated with a very low or even absent expression of theER gene. The methylation of specific gene loci has not been fully investigated but may possibly contribute or even be an important step in human leukemogenesis.
Isochromosomes
Isochromosomes are common tumor-associated cytogenetic abnormalities. They can be either monomeric or dicentric, in which case they have two homologous arms that are mirror images of one another. They result from aberrant transverse instead of longitudinal chromosome division or, alternatively, by chromatid exchange between the two homologous chromosome arms.248,249 The consequence is gain and/or loss of genetic material of the affected chromosome segments. Isochromosomes have been observed in 7% to 9% of ALL cases.250,251 The most frequent chromosomal changes are i(7q), i(9p), and i(17q). Recently i(21q) has been described in ALL.250 Isochromosomes have been detected more frequently in adults than in children (7.8% v 4.5%).250
i(7q).
i(9q).
Like i(7q), i(9q) is usually not the sole anomaly. It is associated with L2 morphology and a pre-B-cell phenotype.
i(17q).
The isochromosome i(17q) is one of the most frequent isochromosomes in ALL. It has a tendency to occur with early pre-B- and pre-B-cell leukemias and has a well-described association with primary aberrations t(9;22) and t(4;11).251 In t(9;22), i(17q) is rarer in Ph+ ALL than in Ph+ CML, where it serves as a paradigmatic marker for disease transformation. Frequently, i(17q) is observed with hyperdiploid karyotypes.34 Although hyperdiploidy is an indicator of good prognosis, Pui et al253 reported poor remission duration in children with hyperdiploidy and i(17q).
i(21q).
Martineau et al250 recently described i(21q) in ALL at a higher frequency than was found in previous studies. It identified patients with B-lineage disease and low WBC counts at presentation, was more frequent with male gender, and was associated with pseudodiploidy or low hyperdiploidy. The study included both children and adults. Follow-up was too short to permit conclusions regarding prognosis.
TUMOR SUPRESSOR GENES
Tumor suppressor genes define a group of genes whose expression can block the development of a tumorigenic phenotype and, upon inactivation, will facilitate the malignant process. The presence of these genes can be suspected in certain chromosomal locations by identifying loss of material from specific chromosomes. However, tumor suppressor genes are frequently altered by mechanisms that are operational at a submicroscopic level such as point mutations, deletions of coding or regulatory gene sequences, transcriptional silencing of genes by hypermethylation, altered protein expression, or expression of inhibiting proteins. Thus, most tumor suppressor gene abnormalities are detected not by conventional cytogenetics, but by molecular techniques such as Southern blot analysis, PCR, SSCP, nucleotide sequencing, or pulsed-field gel electrophoresis. Although tumor suppressor genes are frequently deleted in solid tumors,254-257 they appear to be present in only a fraction of cases of human leukemias and may not be major contributors to human leukemogenesis.
A gene frequently deleted in acute leukemias (mainly T-cell–derived leukemias) is p16INK4A, with or without codeletion of p15INK4B, and both are located on the short arm of chromosome 9. The incidence of homozygous deletions ofp16INK4A has been as high as 80% in some series.97,109,110 115
Of the tumor suppressor genes, the retinoblastoma (RB1) and thep53 genes have been the most thoroughly investigated. TheRB1 gene, on chromosome 13q14, was isolated and cloned by virtue of its deletion in retinoblastoma.258 It binds to and inhibits several transcription factors (among them, E2F) and inhibits cell-cycle progression at the G1-S phase. It was shown to be deleted in 12% of T-ALL patients and about 20% of patients with CD10+ early pre-B-ALL in a study by Ahuja et al,259 which included both children and adults.
The p53 gene is located on the short arm of chromosome 17, band p13.1.260 Loss of wild-type function of p53 appears to facilitate growth and transformation of malignant cells and, in some cases, defer entry of cells into apoptotic pathways.261,262Several studies have analyzed p53 mutations in lymphoid malignancies, including ALL and non-Hodgkin's lymphomas, by SSCP, PCR, and nucleotide sequencing. The incidence of p53 alterations was low, ranging from 2% to 15%, and included both pediatric and adult cases.263-265 There is a correlation between p53mutation and progressive or relapsing disease, especially in T-cell ALL. In B-lineage disease, p53 mutations are frequently observed with translocation t(1;19) and with t(8;14) and FAB L3 morphology.266-270 In L3 ALL, mutations of p53 do not correlate with extensive tumor load or poor response to intensified treatment, at least as long as a normal p53 allele persists.271 Most data are based on results in children with ALL and very few have included adults.
How do tumor suppressor gene alterations influence each other and affect outcome?
Tsai et al272 analyzed RB1 and p53 gene expression in adult ALL (89 de novo, 26 relapsed or refractory). Lack of RB1 expression was not different between de novo and relapsing disease (64% v 58%), but overexpression ofp53 was significantly higher in cases of relapsed patients (42% v 21%). In about 10% of cases p53 andRB1 expression were both abnormal. Although the presence of the double abnormality did not distinguish between de novo and relapsing disease, it was associated with increased early mortality and suggested a poor response to therapy. There was no definite relationship between abnormalities of the tumor suppressor genes alone and disease outcome. However, loss of RB1 expression and abnormal p53 were more frequent in adult than in pediatric ALL populations.
Hangaishi et al273 examined inactivations of several tumor suppressor genes in lymphoid malignancies, includingp16INK4A, p15INK4B, p53, andRB1, by Southern blot analysis, immunoblotting, PCR, SSCP, and nucleotide sequencing of the p53 gene. Among 117 ALL cases,p16 deletions were found in 41% (homozygous deletions in 27%), deletions of p15 in 43% (homozygous deletions in 36%), deletions of the RB1 gene in 2% (but loss of the retinoblastoma protein in 33%), and aberrations of the p53gene in 10% (with loss of the second allele in 5%). In 45% of the samples, one or more aberrations of the four tumor suppressor genes were reported. Noteworthy is the high degree of loss of the retinoblastoma protein, which was also found in other lymphoid malignancies and was also observed by Tsai et al.272Inactivations of the tumor suppressor genes occurred independently of each other.
CONCLUSIONS AND FUTURE DIRECTIONS
Most if not all cases of ALL harbor cytogenetic or submicroscopic molecular abnormalities. Abundant evidence now exists that these aberrations are not “innocent bystanders,” but that they represent decisive early steps in a process that leads to the disruption of hematopoiesis, the development of leukemia, and then continues in the leukemic clone as part of a “clonal evolution.” Associations exist between some chromosomal rearrangements and morphologic and immunophenotypic characteristics of the leukemia cell (Table 3). Examples are translocation t(8;14), which occurs in mature B-cell ALL with almost exclusively L3 morphology. Others include TCR rearrangements in T-cell ALL, and the Ph translocation or abnormalities of 19p13, both of which are closely linked to a pre-B immunophenotype. More importantly, karyotypic changes provide important prognostic information independent of other variables such as age or initial leukocyte count, allowing “low-risk” and “high-risk” patients to be distinguished at the time of diagnosis.10,25 274
This has led to the concept of risk-adapted therapy in which treatment is tailored according to risk groups, and intensive therapy approaches, including BMT, can be reserved for patients who, by virtue of their cytogenetic profile, will otherwise do poorly with chemotherapy. Examples include the Ph translocation, 11q23 abnormalities, and translocation t(1;19).7,8,10,18,25,37,84,90 On the other hand, patients at low risk can be spared the morbidity or mortality of high-dose chemotherapy, when it is not needed, as for t(12;21) or trisomies 4 and 10.30,231,233,234,236 This strategy is beginning to be applied successfully in cases of childhood ALL, where a large body of data guides clinical decision making. Outcomes often improve significantly once poor-prognosis groups are identified and adequately treated, as was seen in cases of t(1;19) or mature B-cell ALL (Burkitt's leukemia).148,151,171 208
These developments are in sharp contrast to the situation in adult ALL where an insufficient database and concomitant lack of experience have delayed the adoption of risk-oriented therapeutic concepts in most cases. Expansion of our knowledge of cytogenetic abnormalities in adult ALL, and investigation of how these changes pertain to clinical behavior and prognosis are of paramount importance. To achieve this goal, cytogenetic and molecular abnormalities at the time of diagnosis should be defined. Traditional cytogenetic studies are not sufficient, and thorough molecular analyses are needed. Examples for which evaluations by routine cytogenetics underestimate the frequency of rearrangements include Ph− butBCR-ABL+ leukemias,71-76 TCR gene rearrangements involving the TAL1gene,157,172,176,177 cryptic t(12;21) with rearrangements of the TEL gene,229,234,235 and deletions or mutations of the p16 gene in abnormalities of the short arm of chromosome 9.109-113
The analysis of cytogenetic abnormalities is more than a prognostic tool and should provide better understanding of the molecular biology of leukemias. Many genes have been implicated in leukemogenesis (Table3). These include oncogenes, putative or proven tumor suppressor genes, genes that act as transcription-regulating factors, and genes that are involved in the regulation of apoptosis and differentiation. The study of these genes, as well as their transcriptional and translational products, will further increase our understanding of crucial pathways of leukemic transformation.
ACKNOWLEDGMENT
We are grateful to Arlene Hoffman for her excellent assistance in editing this manuscript and many helpful suggestions. Due to the vast amount of literature and space limitations, we could not include many reports that also made important contributions to the topic discussed, and we apologize to their authors.
Address reprint requests to Hagop M. Kantarjian, MD, Department of Leukemia, Box 61, M.D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030.