To the Editor:
The MMAC1/PTEN gene is localized on chromosome 10q23.3 and encodes a putative tumor suppressor with structural homologies to known phosphatases and cytoskeletal proteins.1,2 Functional studies have shown that MMAC1/PTEN is a dual-specificity protein phosphatase that dephosphorylates tyrosine, serine, and threonine.3,4 Germline mutations of MMAC1/PTEN have been implicated as the predisposing factor in Cowden disease,5 and loss of function mutations have been found in a variety of sporadic solid cancers.1,2,6 Previous observations of 10q abnormalities in lymphoproliferative diseases7 8 suggest that MMAC1/PTEN may possibly be involved in lymphomagenesis.
We systematically studied 14 malignant and 4 benign lymphoid cell lines for deletions and mutations in all exons of the MMAC1/PTEN gene by combining polymerase chain reaction (PCR), denaturing gradient gel electrophoresis (DGGE) and direct sequence analysis.9Overall, alterations of MMAC1/PTEN were shown in 2 of the malignant cell lines and in none of the benign cell lines. The T-cell acute lymphoblastic leukemia cell line CCRF-CEM harbored homozygous deletion of a genomic region including exons 2 through 5. In the myeloma cell line HS-Sultan, we identified a C → T transition at the first base of codon 17, resulting in the substitution of glutamine with a premature termination signal. This cell line also showed loss of the wild-type allele.
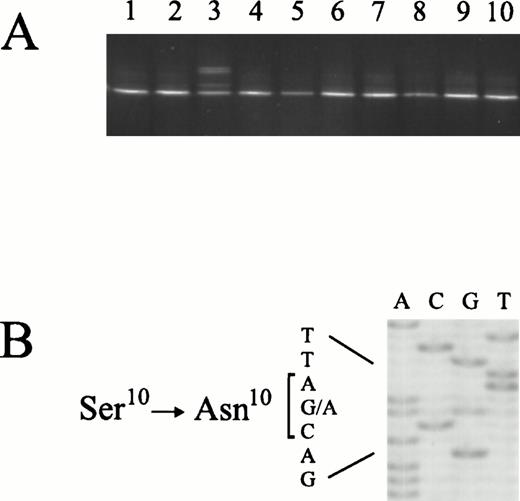
We next examined all exons of the MMAC1/PTEN gene by PCR/DGGE analysis in 170 primary lymphoid malignancies (Fig 1). This approach led to the detection of point mutations in 2 (5%) of 39 diffuse large B-cell lymphomas. One of the samples harbored a G → C transversion at the third base of codon 342, which is predicted to cause the substitution of lysine with asparagine. This mutation affects the last base of exon 8, which is considered to be part of the donor-splice site consensus sequence required for correct splicing of pre-mRNA.10 Several examples of mutations at position −1 of donor-splice sites that cause skipping of the preceding exon have been reported.11Whether the MMAC1/PTEN Lys342Asn mutation has functional consequences with respect to splicing remains to be investigated. Analysis of normal tissue from the patient showed only the normal sequence, documenting that the mutation occurred somatically. The second sample harbored a G → A transition at the second base of codon 10, causing substitution of serine with asparagine. This residue is conserved in auxilin and the hypothetical yeast PTPase, YNL128W.1 Semiquantitative DGGE analysis9showed unequal distribution of mutant and wild-type sequences, suggesting that the mutation was present in only a subpopulation of cells. None of the mutations we identified have been reported to occur in normal tissues.12
Detection and identification of MMAC1/PTENmutations in primary lymphomas. (A) PCR/DGGE analysis ofMMAC1/PTEN exon 1 in 10 tumor samples. (B) Direct sequence analysis of the sample displaying an aberrant band pattern in (A). The partial sequence ladder shows a G → A transition, resulting in the substitution of Ser10 with Asn10.
Detection and identification of MMAC1/PTENmutations in primary lymphomas. (A) PCR/DGGE analysis ofMMAC1/PTEN exon 1 in 10 tumor samples. (B) Direct sequence analysis of the sample displaying an aberrant band pattern in (A). The partial sequence ladder shows a G → A transition, resulting in the substitution of Ser10 with Asn10.
Although MMAC1/PTEN alterations were found in only a minority of lymphoma cell lines and tumor samples, our results, together with the recent demonstration of mutations in leukemia cell lines,6 indicate that MMAC1/PTEN may be a target in the pathogenesis or progression of some forms of hematological malignancy.
ACKNOWLEDGMENT
We are grateful to Dr H.E. Johnsen who generously provided cell lines, and to V. Ahrenkiel and L. Jensen for expert technical assistance. Supported by grants from the Danish Cancer Society, Kong Christian X's Foundation, the Hasselbalch Foundation, and Søeborg Ohlsens Foundation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal