Abstract
Glanzmann thrombasthenia is an inherited bleeding disorder due to a functional reduction or absence of platelet GPIIb/IIIa (αIIbβ3) integrin receptors. Based on a prolonged bleeding time and absence of platelet aggregation in response to physiologic agonists, a 55-year-old white man was diagnosed as having Glanzmann thrombasthenia. The patient's platelet fibrinogen level was ≈5% of normal. As judged by complex-dependent monoclonal antibody (MoAb) binding, surface expression of platelet GPIIb/IIIa receptors was less than 5.5% of normal, whereas the binding of an anti-GPIIIa specific MoAb (7H2) was ≈12% of normal. Immunoblot analysis of the patient's platelet lysates showed ≈35% of normal levels of GPIIIa, ≈30% of normal levels of GPIIb, and an abnormally migrating fragment of GPIIb. Biotinylation of the surface proteins on the patient's platelets followed by immunoprecipitation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed only GPIIb and GPIIIa subunits of normal size. Surface expression of platelet αvβ3 receptors was 192% of normal, suggesting that the patient's' defect was in GPIIb. Sequence analysis of the patient's GPIIb cDNA identified a T to C transition at nucleotide 643, predicting a Leu214Pro substitution. Direct sequencing of GPIIb exon 6 indicated that the patient is homozygous for the mutation. The nature of the Leu214Pro mutation was analyzed by expression in Chinese hamster ovary (CHO) cells. As judged by subunit-specific MoAb binding, surface expression of mutant receptors was ≈60% of normal, but these receptors were not recognized by the complex-dependent monoclonal antibodies, 10E5 and 7E3. In addition, mutant receptors pretreated with the ligand-induced binding site MoAb AP5 were not recognized by the activation-dependent MoAb PAC-1 and mutant expressing CHO cells did not adhere to immobilized fibrinogen. These data suggest that the Leu214Pro mutation in GPIIb disrupts the structural conformation, and either directly or indirectly, the ligand binding properties of the heterodimeric complex. This is in accord with studies from other integrins that have implicated a β-turn in a homologous region as important in ligand binding. Thus, the Leu214Pro mutation appears to produce the Glanzmann thrombasthenia phenotype by both qualitative and quantitative abnormalities. In addition, the mutation appears to confer susceptibility of the GPIIb subunit to proteolysis.
THE PLATELET GPIIb/IIIa receptor has served as a prototype for the integrin family of adhesion receptors because of its important role in platelet physiology and the early recognition of its capacity to undergo activation. The receptor mediates the interaction of activated platelets with ligands, including fibrinogen, von Willebrand factor, vitronectin, and fibronectin.1 The α subunit of the receptor (GPIIb or αIIb) is platelet-specific and the β subunit (GPIIIa or β3) is more widely expressed.1,2 Association of β3 with αv forms the αvβ3 vitronectin receptor, which is present on endothelial cells, osteoclasts, megakaryocytes, smooth muscle cells, and many cultured cells.3 The GPIIb/IIIa receptor is abundantly expressed on the platelet surface (≈80,000 molecules per platelet),4 whereas the αvβ3receptors are expressed in very low numbers (50 to 100 molecules per platelet).5
A number of key steps have been identified in the normal biosynthesis of integrin subunits with many of the studies performed on the platelet GPIIb/IIIa receptor complex.6-9 In megakaryocytes, proGPIIb is synthesized as a single chain precursor (Mr ≈140,000) that associates with GPIIIa (Mr ≈90,000) in the endoplasmic reticulum (ER). The expression and proper folding of both subunits are required for normal complex formation, maturation in the ER, and additional processing of proGPIIb into heavy (GPIIbα) (Mr ≈120,000) and light (GPIIbβ) (Mr ≈20,000) chains, which remain associated by a single disulfide bond.10 Cleavage of the proGPIIb subunit may be mediated by the Golgi-associated serine proteinase furin or a furin-like proteinase11 within a conserved arginine containing consensus sequence12 that is found in other α-chain subunits including human and rodent GPIIb.13 The mature GPIIb/IIIa receptor complex is transported to the cell surface and is expressed in an “unactivated” or “low-affinity” form that requires an activation event to attain high-affinity binding for soluble fibrinogen and other adhesive glycoproteins.14Fibrinogen mediates platelet aggregation by binding to GPIIb/IIIa receptors on adjacent platelets via a γ-chain carboxyl-terminal dodecapeptide sequence, HHLGGAKQAGDV, that is found only in fibrinogen.15 Even the “unactivated” form of the GPIIb/IIIa receptor can, however, mediate adhesion to immobilized fibrinogen16-18 and uptake of fibrinogen into α-granules.19
Amino acid residues and structural domains that are crucial for biogenesis and function of platelet GPIIb/IIIa receptors have been identified by characterizing naturally occurring inherited mutations in patients with Glanzmann thrombasthenia.20 This disease is a rare, inherited recessive bleeding disorder characterized by quantitative or qualitative abnormalities of GPIIb or GPIIIa, resulting in a life-long bleeding diathesis characterized by mucocutaneous hemorrhage. The platelets of patients with Glanzmann thrombasthenia do not aggregate in response to physiologic agonists such as adenine diphosphate (ADP), thrombin, or epinephrine as a result of the GPIIb/IIIa abnormality. A total of 27 GPIIb and 23 GPIIIa genetic defects responsible for Glanzmann thrombasthenia have been identified.21-23
This study characterizes the mutational defect in a Glanzmann thrombasthenia patient with a T to C transition in GPIIb exon 6 resulting in a Leu214Pro amino acid substitution. Decreased levels of the mutant receptor were expressed on the patient's platelets, but this quantitative abnormality was not as severe as it is in many other patients with Glanzmann thrombasthenia, suggesting the possibility of an additional qualitative abnormality. To assess this possibility, the function of the mutant receptor was analyzed by expression in Chinese hamster ovary (CHO) cells. The proline substitution in GPIIb resulted in a disruption of the ligand-binding conformation of the receptor complex as shown by (1) the inability of GPIIb/IIIa complex-dependent monoclonal antibodies to bind to the receptor, (2) the inability of the mutant receptor to bind PAC-1, and (3) the inability of transfected CHO cells to adhere to immobilized fibrinogen.
MATERIALS AND METHODS
Subject.
The patient (L.W.) is a 55-year-old product of a nonconsanguineous marriage who has been the subject of previous reports.24 25He suffered from repeated bouts of epistaxis, excessive bleeding after dental extractions and lacerations, and episodes of pharyngeal and gastric bleeding. Several bleeding episodes required platelet transfusions. At age 39, he developed painful swelling of his ankles and x-rays obtained 2 years later showed destructive changes in both ankle joints with total loss of the entire tibial talar joint, ebernution of the cartilaginous tissues, and cystic changes. Bilateral ankle fusion was performed and histologic examination of the synovial tissue showed evidence of remote hemorrhage and no active inflammation. Studies for rheumatoid factor have been consistently negative.
Laboratory tests have demonstrated absent platelet aggregation in response to ADP (50 μmol/L), but a normal initial response to ristocetin. Clot retraction, observed at 37°C in citrated platelet-rich plasma (PRP) that had been clotted with thrombin and calcium, was absent at 1 hour and only partial at 24 hours.
Surface expression of platelet GPIb, GPIIb/IIIa, and αvβ3 receptors.
The preparation of PRP and the assessment of surface expression of platelet receptors based on the binding of radiolabeled monoclonal antibodies was performed as previously described.26Platelet surface GPIb expression was assessed by the binding of murine monoclonal antibody (MoAb) 6D127; GPIIb/IIIa expression was assessed by the binding of the complex-dependent murine MoAb 10E526 and the Fab fragment of the mouse/human chimeric antibody 7E328 (which also reacts with αvβ3); and GPIIIa expression was assessed by the binding of the murine MoAb 7H2.29 Surface expression of the αvβ3 vitronectin receptor was assessed by the binding of murine monoclonal antibodies LM142,30 specific for human αv, and LM609,30 specific for the αvβ3complex (generously provided by Dr David Cheresh, Scripps Clinic, La Jolla, CA).
Platelet fibrinogen levels and immunoblot analyses.
The preparation of platelets and the analysis of sodium dodecyl sulfate (SDS)-solubilized platelets for fibrinogen, GPIIb, and GPIIIa levels by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot were performed as previously described.31 Platelet fibrinogen was quantified relative to myosin heavy chain by scanning densitometry as previously described.19 For immunoblot analyses, samples of SDS-solubilized untreated or surface-biotinylated platelet proteins (see below) were first separated by SDS-PAGE and then electrophoresed onto polyvinylidene difluoride (PVDF) membranes (Millipore, Burlington, MA). The membranes were analyzed with a murine MoAb specific for GPIIIa, 7H2,29 and a murine MoAb specific for the heavy chain of GPIIb, PMI-1 (a generous gift of Dr Mark Ginsberg, Scripps Clinic, La Jolla, CA)8,32 33 followed by an horseradish peroxidase (HRP)-conjugated rabbit antimouse kappa light chain specific antibody. For identification of bands in immunoprecipitates from lysates of surface-biotinylated platelets, HRP-streptavidin was used followed by detection using the avidin-ECL chemiluminescence detection system (Amersham, Arlington Heights, IL). To obtain a semiquantitative assessment of patient platelet GPIIb and GPIIIa content, immunoblot band intensities of multiple dilutions of normal and patient samples were compared visually after normalizing for myosin heavy chain.
Platelet surface biotinylation and immunoprecipitation analysis.
PRP from 30 mL of whole blood anticoagulated with EDTA (10 mmol/L) yielded a total of ≈4 × 109 platelets. Platelets were washed three times in phosphate-buffered saline (PBS) (137 mmol/L NaCl; 2.7 mmol/L KCl; 4.3 mmol/L Na2HPO4, 1.4 mmol/L KH2PO4; pH 7.4) containing EDTA (10 mmol/L), and the platelet pellet was put on ice for 5 minutes. A fresh solution of Sulfo-NHS-LC-biotin (5 mmol/L) (Pierce, Rockford, IL) in PBS was added to each pellet to yield a final platelet concentration of ≈2 × 109/mL. The pellets were quickly resuspended and incubated on ice for 30 minutes with occasional mixing. The biotinylated platelets were added to tubes prechilled on ice containing 5 mmol/L glycine in 5 mL Tris-buffered saline (TBS) (10 mmol/L Tris-Cl, pH 7.4; 150 mmol/L NaCl; 0.05% NaN3) and EDTA (10 mmol/L) (TSE) and incubated on ice for 10 minutes. Tubes were centrifuged (900g) for 10 minutes and platelets were washed two times with TSE containing glycine (5.0 mL each). Platelet pellets were solubilized in lysis buffer (TBS containing 0.5% NP-40 and 2 mmol/L phenylmethylsulfonyl fluoride) at a concentration of ≈4 × 108/mL, incubated on ice for 30 minutes with occasional mixing, and centrifuged (12,000g) for 30 minutes at 4°C. Supernatants were added to fresh tubes containing 1% deoxycholate and 0.1% SDS, centrifuged as in the previous step, and transferred to fresh tubes for immunoprecipitation analysis.
Platelet lysates (1.0 mL each) were precleared by adding 100 μL of a 1:1 protein G sepharose (Pharmacia, Piscataway, NJ) slurry that was washed and equilibrated in immunoprecipitation (IP) buffer (100 mmol/L Tris-Cl, pH 7.4, 150 mmol/L NaCl, 0.5% NP-40, 1% deoxycholate, 0.1% SDS). This step was repeated one time. Immunoprecipitation of platelet GPIIb/IIIa was accomplished with antibody 10E5, GPIIb with Tab34 (generously provided by Dr Roger McEver, Oklahoma City, OK), and GPIIIa with 7H2. Antibody (5 μg/mL) was added to 20 μL (normal control) or 300 μL (patient L.W.) aliquots, tubes were rotated for 1 hour at room temperature followed by addition of protein G Sepharose slurry (50-μL/tube) and rotation of tubes for an additional 45 minutes at room temperature. After centrifugation (12,000g for 10 seconds), beads were washed five times in IP buffer containing 600 mmol/L NaCl, 1% deoxycholate, 0.1% SDS (1.0 mL/tube), and then proteins were eluted from the beads by heating to 95°C in sample buffer. Samples were then analyzed by SDS-PAGE in 6.5% gels under reduced and nonreduced conditions and transferred onto PVDF membranes (Millipore). Biotinylated proteins were identified using the avidin-ECL chemiluminescence detection system (Amersham), according to the manufacturer's instructions.
Identification of mutation by polymerase chain reaction (PCR) and sequencing.
Primers were synthesized (Operon Technologies, Inc, Alameda, CA), resuspended in ddH2O (1.0 mmol/L), and stored at −80°C. The sequences of the GPIIb specific primers that were used for reverse transcriptase (RT)-PCR were kindly provided by Dr Peter Newman (The Blood Center of SE Wisconsin, Milwaukee) and are listed in Table 1. The sequences of the GPIIb-specific sense 5′AGGCGAGTAGGGAGCAAAAG3′and antisense 5′GAAAATATCCGCAACTGGAG3′ primers used for the amplification of exon 6 were previously described.35 The protocols for RT-PCR and PCR amplification reactions and for the sequence determinations of PCR amplified fragments, including cloned fragments, were performed as previously described.19 The identification of the mutant sequence was obtained from amplified PCR fragments that were subcloned into the PCR II vector according to the manufacturer's protocol (InVitrogen Corp, San Diego, CA). Confirmation of the mutation was obtained from the patient's DNA by direct sequence of amplified fragments of GPIIb exon 6.
GPIIb cDNA Oligonucleotide Primers
| Primer . | Sequence . | Nt No.* . |
|---|---|---|
| A-sense | GGCCAGAGCTTTGTGTCCACTGC | 4-26 |
| C-antisense | AAAGCAGCTACCTACGGGCGTC | 487-466 |
| D-sense | CAAACTTTACAAACCTTCAAGGC | 338-360 |
| B-antisense | CTGGGTTGCTGGAGTCAAAGG | 779-759 |
| E-sense | CGTGGTCACTCAGGCCGGAGAGC | 613-635 |
| F-antisense | TCTCCATATACAGTGGAGCGCCC | 1037-1015 |
| G-sense | GGATTCCTACTACCAGAGGCTGC | 904-1026 |
| I-antisense | CTGTCCAGGACCTGGGAGGGACG | 1320-1298 |
| J-sense | GGCTACAATGACATTGCAGTGGC | 1202-1224 |
| K-antisense | TGCAGCTCGGCATTTAGGGATAG | 1620-1598 |
| L-sense | CTAAATGCCGAGCTGCAGCTG | 1604-1624 |
| H-antisense | GGCCCCCTCGCCCTCGTTGGC | 2026-2006 |
| M-sense | GAGCTGCAGATGGACGCAGCC | 1988-2008 |
| O-antisense | CCCCAGCTGTCCAAGCTGTTCTG | 2424-2401 |
| P-sense | GAATCGCGATGTTGGTGAGCG | 2193-2013 |
| Q-antisense | CAGTCCACCTTGAGAGGGTTG | 2610-2590 |
| R-sense | GTGAATGGTCTTCACCTCAGC | 2474-2494 |
| S-antisense | CAGGAAGGCCAGCACCGTGAC | 2821-2801 |
| T-sense | GTACTGTGGTGCAGTGTGACC | 2748-2768 |
| N-antisense | CCAACCCAAAGCTTGGAGGC | 3210-3191 |
| Primer . | Sequence . | Nt No.* . |
|---|---|---|
| A-sense | GGCCAGAGCTTTGTGTCCACTGC | 4-26 |
| C-antisense | AAAGCAGCTACCTACGGGCGTC | 487-466 |
| D-sense | CAAACTTTACAAACCTTCAAGGC | 338-360 |
| B-antisense | CTGGGTTGCTGGAGTCAAAGG | 779-759 |
| E-sense | CGTGGTCACTCAGGCCGGAGAGC | 613-635 |
| F-antisense | TCTCCATATACAGTGGAGCGCCC | 1037-1015 |
| G-sense | GGATTCCTACTACCAGAGGCTGC | 904-1026 |
| I-antisense | CTGTCCAGGACCTGGGAGGGACG | 1320-1298 |
| J-sense | GGCTACAATGACATTGCAGTGGC | 1202-1224 |
| K-antisense | TGCAGCTCGGCATTTAGGGATAG | 1620-1598 |
| L-sense | CTAAATGCCGAGCTGCAGCTG | 1604-1624 |
| H-antisense | GGCCCCCTCGCCCTCGTTGGC | 2026-2006 |
| M-sense | GAGCTGCAGATGGACGCAGCC | 1988-2008 |
| O-antisense | CCCCAGCTGTCCAAGCTGTTCTG | 2424-2401 |
| P-sense | GAATCGCGATGTTGGTGAGCG | 2193-2013 |
| Q-antisense | CAGTCCACCTTGAGAGGGTTG | 2610-2590 |
| R-sense | GTGAATGGTCTTCACCTCAGC | 2474-2494 |
| S-antisense | CAGGAAGGCCAGCACCGTGAC | 2821-2801 |
| T-sense | GTACTGTGGTGCAGTGTGACC | 2748-2768 |
| N-antisense | CCAACCCAAAGCTTGGAGGC | 3210-3191 |
*Nucleotide (numbering according to Poncz et al36).
Generation of GPIIb Leu214Pro mutant cDNA construct.
The GPIIb and GPIIIa cDNA constructs in the pcDNA3 mammalian cell expression vector (Invitrogen Corp) were kindly provided by Dr Peter Newman. Using primers A and B (Table 1), an RT-PCR amplified fragment with the T to C mutation at nucleotide 64336 was synthesized from the patient's RNA. First-strand cDNA synthesis was performed using Superscript II (GIBCO-BRL Life Technologies, Grand Island, NY), according to the manufacturer's instructions. PCR amplification was performed by preheating the following reaction mixture (50 μL total volume) to 96°C for 5 minutes: 0.4 μmol/L primer A, 0.4 μmol/L primer B, 0.2 mmol/L deoxynucleotides (dNTP), 1.5 mmol/L MgCl2 , 20 μL cDNA, 2.5 U AmpliTaq (Perkin Elmer, Norwalk, CT) in buffer (20 mmol/L Tris-Cl, pH 8.3, 50 mmol/L KCl). After preheating, 30 cycles of 92°C for 45 seconds, 55°C for 30 seconds, and 72°C for 30 seconds with a final extension at 72°C for 10 minutes were performed. The 776-bp fragment was digested withKsp I and BbrPI (Boehringer Mannheim, Indianapolis, IN) and gel-purified by electroelution for ligation into a gel-purified preparation of pcDNA3/GPIIb vector in which the normal 524-bpKsp I-BbrPI fragment had been removed. XL-1 BlueEscherichia coli cells (Stratagene, La Jolla, CA) were transformed by electroporation (Bio-Rad Laboratories, Melville, NY) according to the maufacturer's instructions. Single colonies were picked for restriction enzyme analysis and the cloned insert was sequenced to confirm the presence of the mutation and to eliminate any PCR and cloning artifacts.
Flow cytometic analysis of transfected CHO cells.
CHO cells were transiently transfected with normal pcDNA3/GPIIb and pcDNA3/GPIIIa or mutant pcDNA3/GPIIb and normal pcDNA3/GPIIIa cDNA constructs using LipofectAmine reagent (Life Technologies, Gaithersburg, MD) according to a published protocol.37Briefly, GPIIb and GPIIIa cDNA constructs (2 μg each) or pcDNA3 alone (4 μg) were mixed with LipofectAmine (20 μL) in Dulbecco's modified Eagle's medium (DMEM) (200 μL) for 10 minutes. This mixture was added to CHO cells (plated 24 hours before transfection at 2 × 106 cells/100-mm tissue culture dish) and incubated at 37°C for 6 hours. The transfection efficiency of the GPIIb and GPIIIa expressing cDNA constructs in CHO cells was determined by cotransfection with another plasmid, pXGH5, that expresses human growth hormone (hGH) (Nichols Institute Diagnostics, San Juan Capistrano, CA). The secreted hGH was measured in the medium from each dish using the HGH-TGES kit (Nichols Institute Diagnostics, San Juan Capistrano, CA). Briefly, medium (100 μL from each dish was incubated with 125I-labeled and biotin-labeled hGH antibodies and an avidin-coated bead for 90 minutes at room temperature. The samples were washed and counted in a Packard Autogamma 5650 (Packard Instrument Co, Downers Grove, IL).
GPIIb/IIIa surface expression was measured by flow cytometry approximately 48 hours after transfection. Cells (≈3 to 5 × 106) were incubated with 10 μg/mL of monoclonal antibodies 10E526 (anti-GPIIb/IIIa complex), 7E328 (anti-GPIIb/IIIa and αvβ3), and Tab34 (anti-GPIIb) in 100 μL of DMEM on ice for 60 minutes, washed with PBS, and incubated with fluorescein isothiocyanate (FITC)-labeled donkey antimouse F(ab')2 fragments (Jackson ImmunoResearch, West Grove, PA). To assess the ability of the receptors to bind the PAC-138 MoAb, transfected CHO cells (≈3 to 5 × 106) were resuspended in TSBG buffer (50 mmol/L Tris, pH 7.4; 150 mmol/L NaCl; 5 mmol/L glucose) containing MgCl2(1.4 mmol/L) and 100 μg/mL of the ligand-induced binding site (LIBS) MoAb AP539 (generously provided by Dr Thomas Kunicki, Scripps Clinic, La Jolla, CA) or the GPIIIa-specific MoAb AP340 (generously provided by Dr Peter Newman) and FITC-labeled PAC-1 (20 μL of 0.1 mg/mL) (Becton Dickinson Immunocytometry Systems, San Jose, CA) in a total volume of 100 μL. The samples were analyzed with a FACScan flow cytometer (Becton Dickinson, Mountain View, CA) using LYSYS II software.
Adhesion of transfected CHO cells to immobilized fibrinogen.
Adhesion of transfected CHO cells to immobilized fibrinogen was performed as previously described.19 Briefly, CHO cells were transfected by electroporation and analyzed 48 hours later. The cells (1.5 × 108 cells/mL in 2 mL) were51Cr-labeled for 1 hour at 37°C and incubated with TBS alone and complex-dependent antibodies (50 μg/mL) 10E526(anti-GPIIb/IIIa), 7E328 (anti-GPIIb/IIIa + αvβ3), and LM60930(anti-αvβ3) for 30 minutes at room temperature. Cells (50 μL/well) were added to the fibrinogen-coated plates and incubated for 1 hour at room temperature, washed, and adherent cells were lysed in 2% SDS (100 μL/well) for 30 minutes at room temperature. Lysates were counted in a Packard Autogamma 5650 (Packard Instrument Company, Downers Grove, IL). Cells bound per well were calculated from the specific activity of a control aliquot (50 μL) of 51Cr-labeled cells.
RESULTS
Expression of platelet surface receptors.
The binding of radiolabeled 6D1 antibody (anti-GPIba) to the patient's platelets was 108% of the control value demonstrating that the patient's GPIb receptor was expressed at normal levels (Table 2). The binding of antibody 10E5 (anti-GPIIb/IIIa) was 2.6% of normal and the binding of c7E3 Fab (anti-GPIIb/IIIa + αvβ3) was 5.4% of normal. In contrast, the patient's platelets bound ≈12% of the normal amount of antibody 7H2 (anti-GPIIIa), which does not require GPIIb/IIIa complex formation. The discordance between the binding values for 7H2 versus those of 10E5 and 7E3, raised the possibility that there may be GPIIb/IIIa complexes on the platelet surface that are not recognized by 10E5 and 7E3 because of abnormalities in complex formation. Antibody LM609 (anti-αvβ3) was tested at two different concentrations, and in both cases, the patient's platelets bound more antibody molecules than did the normal platelets (≈192% of normal). The increased level of surface αvβ3 receptors indicates that GPIIIa (β3) is probably normal, making it likely that the patient's abnormality is in GPIIb.
Surface Expression of Platelet Receptors
| Receptor/ Subunit . | 125I-Antibody (concentration) . | Molecules of Antibody/Platelet . | Surface Expression (% of normal) . | |
|---|---|---|---|---|
| Patient . | Control . | |||
| GPIbα | 6D1 (20 μg/mL) | 19,100 18,900 | 17,100 18,200 | 108.0 |
| GPIIb/IIIa | 10E5 (20 μg/mL) | 700 1,400 | 38,500 40,900 | 2.6 |
| GPIIb/IIIa + αvβ3 | c7E3 Fab (18 μg/mL) | 2,800 2,800 | 49,200 54,900 | 5.4 |
| GPIIIa | 7H2 (20 μg/mL) | 6,200 | 53,000 | 12.0 |
| αvβ3 | LM609 (38 ng/mL) (162 ng/mL) | 25 53 | 15 25 | 172.0 212.0 |
| Receptor/ Subunit . | 125I-Antibody (concentration) . | Molecules of Antibody/Platelet . | Surface Expression (% of normal) . | |
|---|---|---|---|---|
| Patient . | Control . | |||
| GPIbα | 6D1 (20 μg/mL) | 19,100 18,900 | 17,100 18,200 | 108.0 |
| GPIIb/IIIa | 10E5 (20 μg/mL) | 700 1,400 | 38,500 40,900 | 2.6 |
| GPIIb/IIIa + αvβ3 | c7E3 Fab (18 μg/mL) | 2,800 2,800 | 49,200 54,900 | 5.4 |
| GPIIIa | 7H2 (20 μg/mL) | 6,200 | 53,000 | 12.0 |
| αvβ3 | LM609 (38 ng/mL) (162 ng/mL) | 25 53 | 15 25 | 172.0 212.0 |
Platelet fibrinogen level and immunoblot analyses.
Platelet α-granule fibrinogen levels depend on the expression of functional GPIIb/IIIa receptors41 and so we analyzed the patient's platelet fibrinogen content. Based on scanning densitometry of SDS-PAGE gels, the patient's platelet fibrinogen level was ≈5% of normal. This value is similar to the levels of fibrinogen (3% to 11% of normal) identified in the platelets of patients with Glanzmann thrombasthenia having no GPIIb/IIIa or only trace amounts of GPIIb/IIIa.42,43 This value is much less than the 36% of normal levels we found in a patient with only ≈10% of normal surface GPIIb/IIIa whose mutant GPIIb/IIIa receptors were able to support cell adhesion to fibrinogen.19
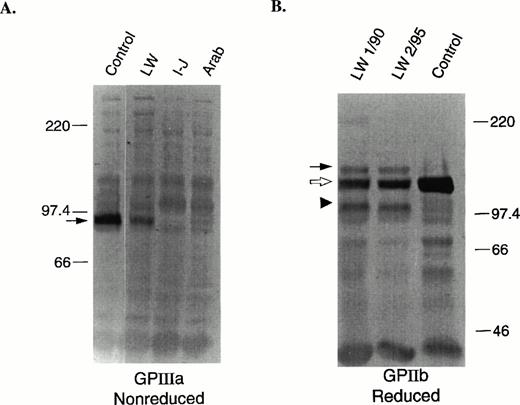
To determine the levels of total GPIIb and GPIIIa in the patient's platelets, immunoblot analyses were performed. Semiquantitative estimates of GPIIb and GPIIIa levels were determined by analysis of multiple dilutions of patient and control solubilized platelets using the antibodies 7H2 (anti-GPIIIa) and PMI-1 (anti-GPIIb). Under nonreduced conditions, GPIIIa migrates at an Mr of ≈95,000 (Fig 1A, arrow). The protein level of GPIIIa in the patient's platelets was determined to be ≈35% of normal. This level is considerably higher than the low and undetectable levels of GPIIIa, respectively, in the platelets of two other patients with Glanzmann thrombasthenia who have mutations in GPIIb (Arab) and GPIIIa (Iraqi-Jewish).44 This level is also higher than the estimate of surface GPIIIa (Table 2), suggesting that a disproportionate amount of GPIIIa is intracellular. The GPIIb in the patient's platelets was analyzed under nonreduced (data not shown) and reduced (Fig 1B) conditions to determine the levels of proGPIIb, which migrates at Mr 140,000 (Fig 1B, closed arrow), and mature processed GPIIb heavy chain, which migrates at Mr120,000 (Fig 1B, open arrow). A semiquantitative estimate of mature GPIIb heavy chain in the patient's platelets was ≈25% of normal. The total of proGPIIb and mature GPIIb was estimated to be ≈30% of normal, which is similar to the estimate of GPIIIa content (≈35%), but much higher than the <1% of normal GPIIb content we found in other patients with Glanzmann thrombasthenia.45 Of interest was the identification of an abnormally migrating immunoreactive GPIIb fragment of Mr ≈100,000 (Fig 1B, arrow head) in both reduced samples of the patient's platelets obtained more than 5 years apart. Nonreduced samples of the patient's platelets also showed an abnormal GPIIb-immunoreactive fragment, but the Mr was 120,000 (data not shown). Because the epitope for the PMI-1 antibody is located near the carboxy-terminal end of the GPIIb heavy chain,33 these data suggest that the abnormal band may be due to proteolytic cleavage of a fragment from the amino-terminus of GPIIb.
Immunoblot analyses of GPIIIa (nonreduced) and GPIIb (reduced). SDS-solubilized platelets (1 μL of 5 × 107platelets/mL) were electrophoresed into a 7.5% polyacrylamide gel, electrotransferred onto PVDF membranes, and developed as previously described.31 (A) Control, patient, and two other GT patient samples with mutations in GPIIIa (I-J: Iraqi-Jewish) and GPIIb (Arab)44 were run under nonreduced conditions. The membranes were incubated with the anti-GPIIIa specific murine MoAb, 7H2.29 The arrow indicates the position of GPIIIa. (B) Two patient samples prepared on different dates (January 1990 and February 1995) and a control sample were run under reduced conditions. The membranes were incubated with the anti-GPIIb heavy chain specific murine MoAb, PMI-1.32 33 The solid arrow indicates the position of proGPIIb, the open arrow, mature processed GPIIb, and the arrow head an abnormally migrating fragment.
Immunoblot analyses of GPIIIa (nonreduced) and GPIIb (reduced). SDS-solubilized platelets (1 μL of 5 × 107platelets/mL) were electrophoresed into a 7.5% polyacrylamide gel, electrotransferred onto PVDF membranes, and developed as previously described.31 (A) Control, patient, and two other GT patient samples with mutations in GPIIIa (I-J: Iraqi-Jewish) and GPIIb (Arab)44 were run under nonreduced conditions. The membranes were incubated with the anti-GPIIIa specific murine MoAb, 7H2.29 The arrow indicates the position of GPIIIa. (B) Two patient samples prepared on different dates (January 1990 and February 1995) and a control sample were run under reduced conditions. The membranes were incubated with the anti-GPIIb heavy chain specific murine MoAb, PMI-1.32 33 The solid arrow indicates the position of proGPIIb, the open arrow, mature processed GPIIb, and the arrow head an abnormally migrating fragment.
Surface biotinylation and immunoprecipitation of platelet GPIIb/IIIa receptors.
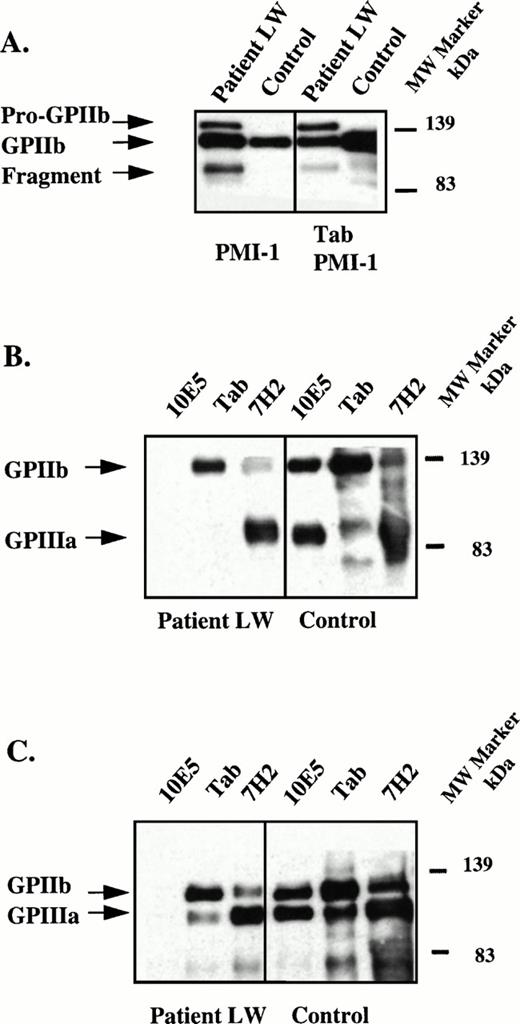
Because an abnormal fragment of GPIIb was identified in the patient's platelet lysate, the molecular weights of the GPIIb and GPIIIa subunits expressed on the surface of the patient's platelets were determined. Platelet surface proteins were biotinylated and the receptor was immunoprecipitated from platelet lysates using the GPIIIa specific MoAb, 7H2, the GPIIb specific MoAb, Tab, and the complex-dependent MoAb, 10E5. To insure that the anti-GPIIb MoAb Tab recognized the abnormal fragment of GPIIb, immunoblots of the biotinylated platelet lysates using PMI-1 were performed with (Fig 2A, right) and without (Fig 2A, left) immunoprecipitation using Tab. As already shown in Fig 1B, the abnormally migrating fragment was demonstrated in the immunoblot using PMI-1 (Fig 2A, left) and this fragment was also identified in the sample after immunoprecipitation with Tab (Fig 2A, right). These data show that the GPIIb-specific MoAb, Tab, recognized the abnormally migrating GPIIb fragment in the patient's platelets.
Immunoblot and immunoprecipitation of solubilized, surface-biotinylated platelets. (A) Left panel, immunoblot of patient and control platelet lysates using the GPIIb-specific MoAb, PMI-1.32,33 This pattern is essentially identical to that in Fig 1 except that because the control sample was diluted 20-fold, the bands in the patient's sample are more intense than the bands in the control sample. Right panel, immunoprecipitation of lysates using the GPIIb-specific MoAb, Tab34 and immunoblotting with PMI-1. Samples were electrophoresed under reduced conditions and arrows mark the positions of pro-GPIIb, mature GPIIb, and the GPIIb fragment. These data indicate that Tab recognizes all forms of the mutant GPIIb. (B) Patient and normal control surface-biotinylated platelet lysates were immunoprecipitated with the GPIIb/IIIa complex-specific MoAb, 10E5,26 the GPIIb-specific MoAb, Tab, and the GPIIIa-specific MoAb, 7H2.29 Immunoprecipitates were electrophoresed under nonreduced conditions and blotted onto PVDF membranes. The membranes were treated with HRP-streptavidin and the bands developed. Arrows mark the positions of GPIIb and GPIIIa. Because only surface-labeled molecules are detected by the avidin reagent, the failure to identify the patient's abnormal GPIIb bands indicates that these were not present on the surface of the patient's platelets. (C) Experiment conducted as in (B) except immunoprecipitates were reduced before electrophoresis.
Immunoblot and immunoprecipitation of solubilized, surface-biotinylated platelets. (A) Left panel, immunoblot of patient and control platelet lysates using the GPIIb-specific MoAb, PMI-1.32,33 This pattern is essentially identical to that in Fig 1 except that because the control sample was diluted 20-fold, the bands in the patient's sample are more intense than the bands in the control sample. Right panel, immunoprecipitation of lysates using the GPIIb-specific MoAb, Tab34 and immunoblotting with PMI-1. Samples were electrophoresed under reduced conditions and arrows mark the positions of pro-GPIIb, mature GPIIb, and the GPIIb fragment. These data indicate that Tab recognizes all forms of the mutant GPIIb. (B) Patient and normal control surface-biotinylated platelet lysates were immunoprecipitated with the GPIIb/IIIa complex-specific MoAb, 10E5,26 the GPIIb-specific MoAb, Tab, and the GPIIIa-specific MoAb, 7H2.29 Immunoprecipitates were electrophoresed under nonreduced conditions and blotted onto PVDF membranes. The membranes were treated with HRP-streptavidin and the bands developed. Arrows mark the positions of GPIIb and GPIIIa. Because only surface-labeled molecules are detected by the avidin reagent, the failure to identify the patient's abnormal GPIIb bands indicates that these were not present on the surface of the patient's platelets. (C) Experiment conducted as in (B) except immunoprecipitates were reduced before electrophoresis.
We next assessed which glycoproteins were expressed on the platelet surface by first immunoprecipitating all of the immunoreactive proteins, but only detecting the ones that were surface-biotinylated by using an avidin-HRP conjugated reagent. In the nonreduced (Fig 2B) and reduced (Fig 2C) samples immunoprecipitated with 10E5, Tab, and 7H2; only GPIIb and GPIIIa subunits of normal Mr were detected by this technique. The surface GPIIb immunoprecipitated by both Tab and 7H2 only contained the band of normal mobility, indicating that the abnormal GPIIb molecules do not become expressed on the platelet surface. The 10E5 MoAb failed to immunoprecipitate the patient's surface exposed GPIIb/IIIa receptors presumably because the abnormality affects the 10E5 epitope. These data are consistent with the surface expression studies in which 125I-10E5 antibody binding to the patient's platelets was only 2.6% of normal (Table 2).
Identification of a GPIIb mutation.
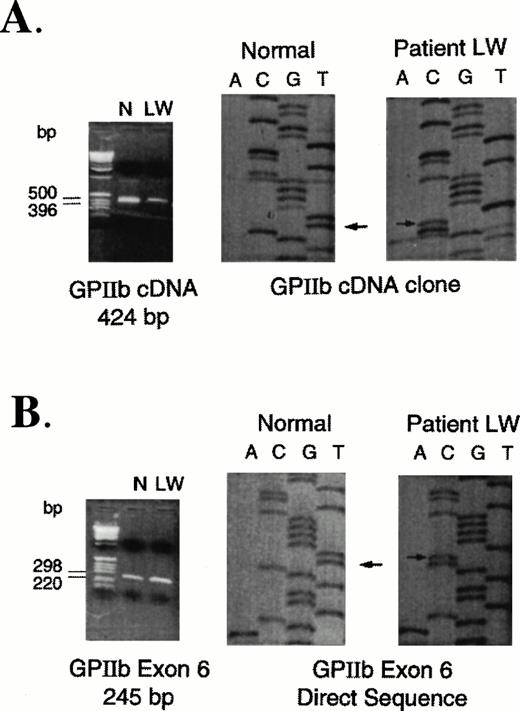
Based on increased expression of platelet surface αvβ3 receptors and the presence of an abnormal fragment of GPIIb in the patient's platelet lysates (Fig 2), sequence analysis of GPIIb RNA was performed. Using 10 pairs of primers (Table 1) that hybridize to sequences specific for GPIIb cDNA, PCR products were generated by RT-PCR from total RNA of the patient and a normal control. The amplified products from patient and control samples were cloned and sequenced. The only abnormality that was identified was a T to C mutation (Fig 3A) at nucleotide 64336 that corresponds to a Leu214Pro substitution. The mutation was confirmed by performing another RT-PCR reaction using primers E and F (Table 1) and sequencing the cloned product. Because Leu 214 is encoded within exon 6 of the GPIIb gene,46specific primers were also used to amplify this exon from high molecular weight DNA isolated from patient and control peripheral blood mononuclear cells. Direct sequence analysis of exon 6 PCR fragments showed only the T to C mutation in the patient's sample indicating that the patient did not have any normal DNA (Fig 3).
PCR-amplified fragments and sequence analyses of control and patient RNA and DNA samples. (A) RT-PCR amplification of RNA extracted from normal control (N) and patient (LW) platelets. Using primers A and C (Table 1), a 424-bp fragment was cloned and sequenced. Arrows indicate the T (normal) to C (patient) nucleotide base change. (B) PCR amplification of DNA extracted from peripheral mononuclear cells isolated from a normal control (N) and the patient (LW). Using primers specific for amplification of GPIIb exon 6, the 245-bp fragments were directly sequenced as described previously. Arrows indicate the T (normal) to C (patient) nucleotide base change. No normal sequence was identified in the patient's DNA.
PCR-amplified fragments and sequence analyses of control and patient RNA and DNA samples. (A) RT-PCR amplification of RNA extracted from normal control (N) and patient (LW) platelets. Using primers A and C (Table 1), a 424-bp fragment was cloned and sequenced. Arrows indicate the T (normal) to C (patient) nucleotide base change. (B) PCR amplification of DNA extracted from peripheral mononuclear cells isolated from a normal control (N) and the patient (LW). Using primers specific for amplification of GPIIb exon 6, the 245-bp fragments were directly sequenced as described previously. Arrows indicate the T (normal) to C (patient) nucleotide base change. No normal sequence was identified in the patient's DNA.
Surface expression of GPIIb/IIIa on CHO cells.
CHO cell expression studies were performed to assess the effect of the Leu214Pro mutation in GPIIb/IIIa receptor expression and function. A GPIIb cDNA construct containing the T to C transition at position 64347 was generated by RT-PCR amplification of a fragment including the mutation from the patient's RNA and subsequently cloned into the cDNA construct. The mutant construct was coexpressed with a normal GPIIIa cDNA construct in CHO cells, as was normal GPIIb with normal GPIIIa. To determine the surface expression of normal and mutant GPIIb/IIIa receptors, flow cytometry was performed using the GPIIb-specific MoAb, Tab (which previously was shown to immunoprecipitate the patient's GPIIb; Fig 2) and the GPIIb/IIIa complex-dependent antibodies 10E5 and 7E3 (Table 3). Because both GPIIb and GPIIIa subunits are required for cell surface expression, the GPIIb surface expression levels are indicative of GPIIb/IIIa receptor expression. In both experiments, transfection efficiencies were similar for the normal and mutant constructs assessed by the secretion of hGH (Table 3, legend). CHO cells cotransfected with mutant GPIIb and unmutated GPIIIa cDNA constructs bound ≈60% of the amount of Tab antibody that bound to CHO cells expressing normal GPIIb/IIIa receptors. Despite the high level of GPIIb and GPIIIa expression, the binding of the 10E5 and 7E3 complex-dependent antibodies was ≈10% of normal and <1% of normal, respectively. Thus, consistent with the data from the patient's platelets, the mutant GPIIb/IIIa receptor adopts a conformation that disrupts the 10E5 and 7E3 epitopes.
Flow Cytometric Analyses of Transfected CHO Cells
| No. . | Antibody . | Subunit/Receptor . | MFI* . | |
|---|---|---|---|---|
| WT . | Patient LW . | |||
| 1 | Tab | GPIIb | 42.6† | 25.8 |
| 10E5 | GPIIb/IIIa | 41.3 | 4.2 | |
| 2 | Tab | GPIIb | 56.8 | 34.6 |
| 7E3 | GPIIb/IIIa + αvβ3 | 50.1 | <1.0 | |
| No. . | Antibody . | Subunit/Receptor . | MFI* . | |
|---|---|---|---|---|
| WT . | Patient LW . | |||
| 1 | Tab | GPIIb | 42.6† | 25.8 |
| 10E5 | GPIIb/IIIa | 41.3 | 4.2 | |
| 2 | Tab | GPIIb | 56.8 | 34.6 |
| 7E3 | GPIIb/IIIa + αvβ3 | 50.1 | <1.0 | |
*Mean fluorescence intensity in arbitrary fluorescence units.
Each value represents the average MFI, minus background values from mock transfected cells, from 2 separate experiments. Values for mock transfected cells: experiment 1 Tab = 54.4 and 10E5 = 52.2, experiment 2 Tab = 120.5 and 7E3 = 115.6. Transfection efficiencies were determined by secreted hGH levels using 125I-anti-hGH antibody (expressed in cpm): experiment 1 WT = 4797 and LW = 5539, experiment 2 WT = 1913 and LW = 2148.
PAC-1 binding to LIBS-activated normal and mutant GPIIb/IIIa receptors.
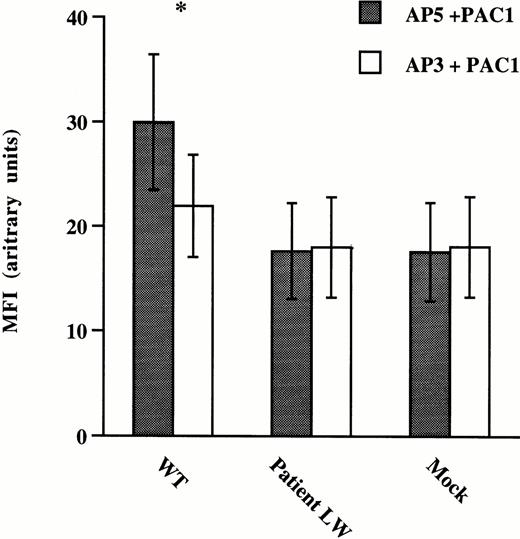
To assess ligand-binding function of mutant GPIIb/IIIa receptors expressed on CHO cells, antibody binding studies using the activation-dependent MoAb, PAC-1, were performed on CHO cells incubated with the LIBS antibody AP5 (Fig 4). The anti-GPIIIa specific antibody, AP3, was used as a negative control for AP5 because it does not induce activation of the GPIIb/IIIa receptor. FITC–PAC-1 binding to normal and mutant GPIIb/IIIa expressing CHO cells was measured using flow cytometry. The binding of PAC-1 to CHO cells expressing wild-type GPIIb/IIIa receptors activated with AP5 was significantly increased over cells incubated with the nonactivating AP3 antibody. In contrast, despite the high levels of mutant GPIIb/IIIa receptors expressed on the CHO cell surface (Table 3), the binding of PAC-1 to CHO cells expressing mutant GPIIb/IIIa receptors activated with AP5 was the same as the binding to mutant cells incubated with AP3 and the same as PAC-1 binding to mock transfected cells incubated with either AP3 or AP5. These data suggest that the mutant receptor cannot be induced to make the transition to a high-affinity ligand binding conformation.
Binding of the PAC-1 MoAb to CHO cells expressing normal and mutant GPIIb/IIIa receptors. CHO cells were transfected with vector alone (Mock), mutant GPIIb and normal GPIIIa (Patient LW), and normal or wild-type GPIIb and GPIIIa (WT) cDNA constructs. The cells were incubated with FITC–PAC-1 in the presence of the GPIIIa-specific LIBS antibody AP539 or a control anti-GPIIIa-specific antibody AP3.40 Flow cytometric analyses were performed and data are presented as mean fluorescence intensity (MFI) ± standard deviation (SD) from five separate experiments. *A two-tailed Student's t-test was performed showing that the binding of PAC-1 to WT cells in the presence of AP5 was significantly greater than the binding in the presence of AP3 (P < .004), while the binding of PAC-1 to AP5-activated Patient LW (P = .35) transfected cells was not greater than the binding in the presence of AP3.
Binding of the PAC-1 MoAb to CHO cells expressing normal and mutant GPIIb/IIIa receptors. CHO cells were transfected with vector alone (Mock), mutant GPIIb and normal GPIIIa (Patient LW), and normal or wild-type GPIIb and GPIIIa (WT) cDNA constructs. The cells were incubated with FITC–PAC-1 in the presence of the GPIIIa-specific LIBS antibody AP539 or a control anti-GPIIIa-specific antibody AP3.40 Flow cytometric analyses were performed and data are presented as mean fluorescence intensity (MFI) ± standard deviation (SD) from five separate experiments. *A two-tailed Student's t-test was performed showing that the binding of PAC-1 to WT cells in the presence of AP5 was significantly greater than the binding in the presence of AP3 (P < .004), while the binding of PAC-1 to AP5-activated Patient LW (P = .35) transfected cells was not greater than the binding in the presence of AP3.
Adhesion of normal and mutant GPIIb/IIIa-expressing CHO cells to immobilized fibrinogen.
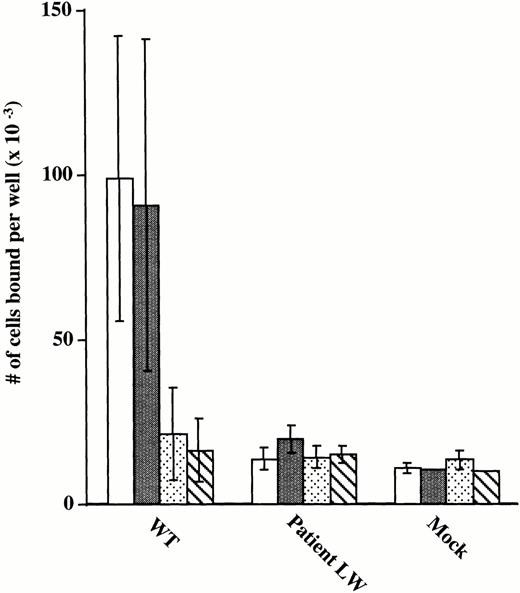
To further assess ligand-binding function of the mutant GPIIb/IIIa receptors, the adhesion of CHO cells expressing wild-type and mutant receptors to microtiter wells precoated with immobilized fibrinogen was tested (Fig 5). Cells expressing normal GPIIb/IIIa receptors adhered to the fibrinogen-coated wells and the adhesion was nearly completely inhibited by antibodies 7E3 (anti-GPIIb/IIIa + αvβ3) and 10E5 (anti-GPIIb/IIIa). Inhibition with antibody LM609 (anti-αvβ3) was minimal because adhesion of αvβ3 receptors to immobilized fibrinogen requires the presence of MnCl2,48 which was not added in the experiment. These data provide evidence that adhesion of the transfected CHO cells to immobilized fibrinogen is mediated via the GPIIb/IIIa receptor. In contrast, the cells expressing the mutant receptor bound to the fibrinogen-coated wells no better than the background binding of mock transfected cells. To assess the possibility that adhesion by wild-type cells is mediated exclusively by cells expressing the highest level of receptors, flow cytometry using the GPIIb-specific MoAb, Tab, was performed on wild-type cells after adhesion to immobilized fibrinogen. The profile of adherent wild-type–expressing cells was broad, encompassing cells with quite variable expression of GPIIb/IIIa receptors. This expression profile for adherent wild-type cells overlapped the profile of Tab binding to the entire population of mutant-expressing cells, showing that the mutant cells expressed sufficient levels of receptors for adhesion to immobilized fibrinogen (data not shown). In conclusion, these data indicate that the Leu214Pro mutation in GPIIb disrupts the ligand-binding properties of the GPIIb/IIIa receptor complex.
Adhesion of transiently transfected CHO cells to immobilized fibrinogen. 51Cr-labeled CHO cells transfected with vector alone (Mock), mutant GPIIb and normal GPIIIa cDNA constructs (Patient LW), and wild-type GPIIb and GPIIIa cDNA constructs (WT) were added to microtiter wells precoated with fibrinogen (100 μL of 20 μg/mL). Cells were incubated with buffer (□) or MoAbs specific for αvβ3 (LM609) (▪),30 GPIIb/IIIa + αvβ3(c7E3) (□),28 and GPIIb/IIIa (10E5) (▧).26The number of cells per well was calculated according to the specific activity of a 50-μL aliquot of cells sampled at the beginning of the experiment, as previously described.19 Data are presented as mean ± SD of 4 data points obtained from two separate experiments.
Adhesion of transiently transfected CHO cells to immobilized fibrinogen. 51Cr-labeled CHO cells transfected with vector alone (Mock), mutant GPIIb and normal GPIIIa cDNA constructs (Patient LW), and wild-type GPIIb and GPIIIa cDNA constructs (WT) were added to microtiter wells precoated with fibrinogen (100 μL of 20 μg/mL). Cells were incubated with buffer (□) or MoAbs specific for αvβ3 (LM609) (▪),30 GPIIb/IIIa + αvβ3(c7E3) (□),28 and GPIIb/IIIa (10E5) (▧).26The number of cells per well was calculated according to the specific activity of a 50-μL aliquot of cells sampled at the beginning of the experiment, as previously described.19 Data are presented as mean ± SD of 4 data points obtained from two separate experiments.
DISCUSSION
We have identified a patient with typical clinical and laboratory features of Glanzmann thrombasthenia who has a Leu214Pro mutation in the GPIIb subunit of the platelet GPIIb/IIIa receptor. One unusual aspect of the patient's history is long-standing seronegative arthritis. Although histology of his joint lining indicated evidence of distant hemorrhage, it is unlikely that his platelet disorder was causal because he did not give a history suggestive of repeated hemarthroses and spontaneous hemarthroses are rare in Glanzmann thrombasthenia.20
The leucine residue at position 214 in human GPIIb is conserved in rodent GPIIb13 and a number of human α-chain subunits including αv, α1, α2, and α513,49 suggesting that this residue may be located in a site important for the conformational integrity of the receptor. Within the normal architecture of a protein molecule, proline residues can usually be found in regions of direction change; thus the probability of a proline residue being located in a β-turn structure is high.50 A predicted secondary structure of the amino terminal domain of integrin α-chain subunits has been determined51 and the Leu214 residue in GPIIb immediately precedes a predicted β-turn structure comprising amino acids 215-224 (GAPGGYYFLG) that is highly conserved in non-I domain α-chain subunits.13,49 Within the β-turn structure is a proline residue at position 217 and this residue is highly conserved in rodent GPIIb and in all non-I domain containing α-chain subunits.13,49 Importantly, this β-turn structure has been implicated in ligand-binding function of the GPIIb/IIIa receptor52 and the α4β1 and α5β1integrin receptors.53 Due to significant homology among integrin α-subunits, this region has been proposed as a site involved in the ligand-binding function of non-I domain containing integrins.53 The Leu214Pro mutation identified in this study results in the creation of a proline-glycine-alanine-proline (PGAP) sequence at the amino-terminal end of the β-turn structure. The rigidity of the two proline residues linked by flexible glycine and alanine residues may create a kink in the secondary structure affecting the conformation of the β-turn. The inability of mutant GPIIb/IIIa receptors to be recognized by conformation-dependent antibodies, to be activated into a high-affinity ligand binding conformation, and to adhere to immobilized fibrinogen suggests that this structural alteration affects ligand-binding and that the Leu214Pro mutation has either an indirect or direct effect on this site.
In addition to disrupting the ligand-binding conformation of the GPIIb/IIIa receptor, the Leu214Pro mutation resulted in decreased surface expression of the receptor on the patient's platelets (Table2). Platelet surface receptor levels were determined to be ≈12% of normal using an anti-GPIIIa specific antibody. A total of 27 mutations have been identified in the gene encoding GPIIb (including the mutation in this study).21,22 The majority of patients have surface expression levels of less than 10% of normal and one patient was reported to have expression levels greater than 20% of normal.54 Three patients have been reported to have receptor surface expression levels similar to that of patient LW55-57 and transfection studies showed comparable receptor expression levels to those identified on the patient's platelets. In characterizing the Leu214Pro mutation in CHO cells, an interesting finding was that surface expression levels of the receptor were ≈60% of normal compared with the ≈12% of normal levels on the surface of the patient's platelets. The discrepancy between the receptor expression levels on the surface of platelets and CHO cells is likely due to differences in proteolysis of the mutant GPIIb subunit between the cell types. Thus, when both intracellular and surface GPIIb/IIIa were measured by immunoblot analysis, the patient's total platelet GPIIb/IIIa content was actually ≈30% to 35% of normal, with the proteolytic fragment of GPIIb a significant component. In contrast, this fragment was not present in lysates of CHO cells expressing the mutant GPIIb/IIIa receptor (data not shown). There are several possible reasons for the mutant GPIIb fragment being detectable in the patient's platelets, but not in transfected CHO cells: (1) proteolysis may occur over a period of time during platelet formation and survival—the CHO cell expression studies were performed over 48 hours using cells that were transiently transfected with normal and mutant cDNA constructs, whereas platelets are formed over 8 to 10 days and circulate for another 10 days; (2) the enzyme(s) in platelets responsible for the cleavage of the GPIIb subunit may not be expressed or activated in CHO cells; and (3) the conformation of the GPIIb subunit that may result in susceptibility to proteolytic cleavage may not be formed during biogenesis of the receptor complex in CHO cells. It is interesting to speculate that the absence of mutant GPIIb cleavage in CHO cells contributed to the higher surface expression than was found in the patient's platelets.
By immunoblot with an antibody to the carboxy-terminus of GPIIb (residues 875-891), the patient's GPIIb fragment was Mr120,000 with nonreduced platelets (which is ≈20,000 less that proGPIIb or nonreduced GPIIb). On reduction, a fragment of Mr ≈100,000 was identified, which presumably is the same fragment identified under nonreducing conditions, but without the GPIIb light chain (Mr ≈20,000). These data suggest that the GPIIb fragment is formed by proteolysis of the amino-terminal end of the GPIIb heavy chain. If the cleavage interferes with transport of GPIIb to the platelet surface, which seems plausible, as the signal sequence is likely to be included, this might explain the relatively large amount of intraplatelet GPIIb/IIIa compared with platelet surface-expressed receptors, as well as the failure to find the cleaved GPIIb fragment on the surface of the patient's platelets. The Mr of the fragment suggests that cleavage occurs near amino acid 200, which is near to the mutation site, suggesting that the additional proline may expose a nearby site that is susceptible to proteolysis.
We could not detect any normal GPIIb sequence in our analyses of the patient's RNA or DNA, and this raises interesting questions because both the patient and his mother deny that he is the product of a consanguineous relationship. We excluded the presence of a large deletion affecting a possible second GPIIb allele by Southern blot and fluorescent in situ hybridization (FISH) analyses (V. Najfeld, unpublished data, November 1996). Another Glanzmann thrombasthenia patient was recently reported in whom only a mutant GPIIIa could be identified even though consanguinity was denied.58 Haplotype analyses of chromosome 17 suggested that homozygosity was due to uniparental disomy from the patient's mother. We would like to perform similar studies, but the patient's father is dead and his mother has not yet been available to study; the patient denies having any children.
In conclusion, we have identified a new GPIIb mutation, Leu214Pro, that produces a Glanzmann thrombasthenia phenotype due to both qualitative and quantitative abnormalities. This mutation alters the conformation of the GPIIb/IIIa receptor and disrupts ligand-binding. It is located within the first 334 residues of GPIIb, a region that has been identified as contributing to the ligand-binding pocket of the GPIIb/IIIa receptor,59 and amino-terminal to a putative binding site for the fibrinogen γ-chain dodecapeptide sequence (residues 294-314).60,61 The presumed defect in ligand binding capacity of the patient's GPIIb/IIIa receptor is supported by the finding that the patient's platelets contained only ≈5% of normal levels of plasma fibrinogen even though they contained ≈12% of the normal number of receptors on the platelet surface. The platelet fibrinogen level in this patient is much lower than the ≈36% of normal that we identified in another patient with Glanzmann thrombasthenia whose platelets contained only ≈10% of the normal numbers of surface receptor.19 This latter patient has a Cys374Tyr mutation in GPIIIa that affects surface expression, but not the ability of the receptor to mediate adhesion to immobilized fibrinogen. Thus, even though these two patients had mutations resulting in comparable levels of surface GPIIb/IIIa receptors, the characterization of parameters such as platelet fibrinogen levels and receptor function in transfected cells provided important structure/function information on biosynthesis, ligand-binding, and protein trafficking functions of the receptor. Finally, our studies highlight the importance of rigidly classifying Glanzmann thrombasthenia mutations into quantitative versus qualitative abnormalities, as a single mutation may produce both effects.
ACKNOWLEDGMENT
We thank Dr Peter Newman for generously providing us with GPIIb and GPIIIa cDNA constructs, oligonucleotide primer sequences for cDNA analyses, the AP3 MoAb, and very helpful technical advice; to Dr Mark Ginsberg for kindly supplying the PMI-1 MoAb, Dr David Cheresh for kindly supplying LM609 and LM142 MoAbs, Dr Rodger McEver for kindly supplying the Tab MoAb, and Dr Thomas Kunicki for kindly supplying the AP5 MoAb. We are grateful to Dr Vesna Najfeld, Director Tumor Cytogenetics Laboratory at Mount Sinai Medical Center, for performing FISH analyses and to Lesley Scudder for her expert technical assistance.
Supported by Grant No. 19278 from the National Heart, Lung and Blood Institute (to B.S.C.) and Grant No. 91014650 from the National American Heart Association (to D.L.F.).
Address reprint requests to Deborah L. French, PhD, Division of Hematology, Department of Medicine, Box 1079, Mount Sinai School of Medicine, One Gustave L. Levy Place, New York, NY 10029.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal