Abstract
Rhesus D category VI (DVI) is the clinically most important partial D. DVI red blood cells were assumed to possess very low RhD antigen density and to be caused by twoRHD-CE-D hybrid alleles. Because there was no population-based work-up, we screened three populations in central Europe for DVI. Twenty-six DVI samples were detected and examined by exon-specific RHD polymerase chain reaction with sequence-specific primers (PCR-SSP). A new genotype, hereby designated D category VI type III, was characterized as a RHD-Ce(3-6)-D hybrid allele by sequencing of the cDNA, parts of intron 1, and by PCR-restriction fragment length polymorphism (PCR-RFLP) of intron 2. Rhesus introns 5 and 6 were sequenced and the 3′ breakpoints of all knownDVItypes shown to be distinct. We differentiated the 5′ breakpoints of DVItypeI andDVItype II by a newly devised RHD-PCR. Thus, the DVI phenotype originated in at least three independent molecular events. Each DVI type showed distinct immunohematologic features in flow cytometry. The number of RhD proteins accessible on the red blood cells' surface ofDVItype III was normal (about 12,000 antigens/cell; DVItypeI, 500;DVItype II, 2,400) based on the determination of an RhD epitope density profile. DVItype II and DVItype III occurred as CDe haplotypes, and DVItype I as a cDE haplotype.The distribution of the DVItypes varied significantly in three German-speaking populations. Genotyping strategies should take account of allelic variations in partial RhD. The reconsideration of previous serologic and clinical data for partial D in view of the underlying molecular structures may be worthwhile.
THE RHESUS BLOOD GROUP system is of great importance for transfusion medicine because of the high immunogenicity of its antigens. Rhesus antigens are carried by two highly homologous proteins, the RhD and RhCE proteins.1-6 The D antigen (ISBT 004.001; RH1) determined by the RhD protein is the most important Rhesus antigen and the leading cause for hemolytic disease of the newborn.7 About 17% of Caucasians lack the expression of the D antigen.8 The transfusion of a single unit of D positive red blood cells to a D negative patient is associated with an immunization rate of greater than 80%.9
The D antigen comprises several different antigenic epitopes. Rare individuals carry a partial D antigen10 and may produce alloantibodies directed against D epitopes that are lacking in their RhD protein. Initially, these individuals have been classified into six distinct categories (DII to DVII, DI being obsolete) based on the mutual reactivity with polyclonal anti-D sera from immunized partial D carriers.11Today, characterization of partial D is performed by differential reactivity with monoclonal anti-D antibodies.12,13 D category VI (DVI) is the clinically most important partial D. Severe cases of hemolytic disease of the newborn have occurred in RhD positive babies born to DVI mothers with anti-D.14 DVI is the most abundant serologically defined partial D occurring among weak D samples. DVI is reported to comprise about 6% to 10% of weak D samples8,15 and has a phenotype frequency of 1:6,200 in Germany (range, 0.02% to 0.05% in Caucasians).8,15,16 The majority of RhD positive individuals with allo-anti–D were DVI.17
DVI occurs in CDe, cDE, and cDe haplotypes.17The CDVIe haplotype is due to an RHD-RHCEhybrid molecule in which exons 4 to 6 of RHD were substituted by the respective exons of RHCE.18Samples with a cDVIE haplotype were initially assumed to carry a deletion of exons 4 to 6,18 but in fact, are due to an exon 4 to 5 hybrid.19,20 Most CDVIe haplotypes carry the low frequency Rhesus antigen BARC (ISBT 004.052; RH52).21 No other consistent serologic differences in DVI have been described.21
We recently completed a serologic random survey for partial D in southwestern Germany.8 22 Here we report the results of the molecular and immunohematologic work-up of DVI samples. We describe a novel molecular event that caused a DVIphenotype carrying a normal number of RhD proteins accessible on the red blood cells' surface. We show that the three DVItypes may be readily discriminated by flow cytometry based on distinct immunohematologic features. We demonstrate considerable differences in the distribution of DVI types within German-speaking populations showing the importance of a full molecular description for Rhesus genotyping purposes, eg, in prenatal testing.
MATERIALS AND METHODS
Random Survey and Blood Samples
EDTA- or citrate-anticoagulated blood samples came from southwestern Germany (DRK-Blutspendedienst Baden-Württemberg, Ulm, Germany), northern Germany (DRK-Blutspendedienst Niedersachsen, Oldenburg) and Tyrol, Austria (Zentralinstitut für Bluttransfusion und Immunologische Abteilung Innsbruck, Innsbruck, Austria). As described previously,8 blood samples in Ulm screened for differential reactivity with a monoclonal IgM anti-D (BS226; Biotest, Dreieich, Germany; not reactive with DVI) and with polyclonal anti-D in antiglobulin technique were checked for the DVIphenotype by a panel of monoclonal anti-D (D-Screen; Diagast, Lille, France). The DVI phenotype was further confirmed by reactivity with monoclonal anti-D BS221, H41 (Biotest), and BRAD-2 (International Blood Group Reference Laboratory, Bristol, UK), as well as absence of reactivity with BS227, BS229, BS231, BS232 (Biotest) and RUM-1 (Bio Products Laboratory, Elstree, UK). Similarly, blood donors in Oldenburg and Innsbruck with weak D phenotype were screened by theRHD exon-specific polymerase chain reaction with sequence-specific primers (PCR-SSP; see below).
Molecular Biology
DNA was prepared using a modified salting out procedure23or QIAAmp Blood DNA isolation kit (Qiagen, Hilden, Germany). RNA was isolated using RNeasy kit (Qiagen). Reverse transcription was performed with oligo-dT-priming and Moloney murine leukemia virus (MMLV) reverse transcriptase (Sigma, Deisenhofen, Germany). RHD exon-specific PCR-SSP was performed as previously described.24 cDNA was amplified in a nested PCR-reaction (High Fidelity PCR system, Boehringer Mannheim, Mannheim, Germany) with external primers RR1 and RR3 and internal primers Rh5 and Rh7. The 5′ part of intron 1 was amplified with primers RB13 and RB45. Restriction fragment length polymorphism-PCR (RFLP-PCR) of intron 2 was performed using the primers of Poulter et al.25 The 3′ region of intron 3 was amplified with primers RB46 and RB5, RB12 and RI4R2. The intron 3 length polymorphism data were based on seven RhD-negative and 20 RhD-positive samples. Intron 5 was amplified using primers RA9B and Rh2 and intron 6 using primers RB25, RB7, and RB27.
Nucleotide sequencing was performed with a DNA sequencing unit (Prism dye terminator cycle-sequencing kit with AmpliTaq FS DNA polymerase; ABI 373A, Applied Biosystems, Weiterstadt, Germany). We subcloned the PCR product into pMos-T-vectors (pMos-T-kit, United States Biochemicals, Cleveland, OH). Three independent cDNA clones were sequenced using T7 promoter primers, U19 reverse plasmid primers, and internal Rhesus primers. Genomic sequences were established from cloned PCR fragments using both primer walking and nested deletion strategies (Nested deletion kit, Pharmacia, Freiburg, Germany) and verified by sequencing of PCR products using internal Rhesusprimers. Intron 1 sequences were based on six RhD-negative (three ccee, two ccEE, one CCee), eleven RhD-positive samples and one sample of each DVI type, intron 3 sequences on six RHCE (two ce, Ce, cE each) and four RHD alleles, intron 5 and 6 sequences on at least two RHCE (ce) and RHD alleles. Intron DNA sequences were analyzed for the presence of repetitive sequences by the CENSOR program26 (censor@charon.lpi.org).
Primer Sequences
RB13, ctagagccaaacccacatctcctt (promoter,27 position -675 to -653 relative to the A of the start codon of the cDNA); RR1, tgttggagagaggggtgatg (5′ untranslated, -60 to -41); Rh5,28 gcacagagacggacacag (5′ untranslated, -19 to -2); Rh1,29 tatctagagacggacacaggATGAGC (5′ untranslated to exon 1, -17 to 6); RB45, acactgttgrctgaatttcggtgc (intron 1, antisense); RA21,25 gtgccacttgacttgggact (intron 2, sense); RA22,25 gtggacccaatgcctctg (intron 2, antisense); RB46, tggcaagaacctggaccttgacttt (intron 3, sense); RA9B, GGTGCCTGCCAAAGCCTCTACCC (exon 4, 554 to 576); RB5, GGCAGACAAACTGGGTATCGTTGC (exon 4, 627 to 604); RB12, tcctgaacctgctctgtgaagtgc (intron 4, antisense, RHD-specific); RI4R2, ttggctcactgcaacctccaccac (intron 4, antisense,RHCE-specific); RB25, agcagggaggatgttacag (intron 4, sense); Rh2,29 AGAAGGGATCAGGTGACACG (exon 5, 900 to 881); RB7, ATCTCTCCAAGCAGACCCAGCAAGC (exon 7, 1022 − 998); RB27, AGCCCAgtgacccacatg (exon7/intron 7); Rh7, acgtacaaatgcaggcaa (3′ untranslated, 1330 − 1313); RR3, cagtctgttgtttaccagatg (3′ untranslated, 1512 − 1431, RHD-specific).
Immunohematology
Monoclonal anti-D were provided by the Workshop on Monoclonal Antibodies against Human Red Blood Cells and Related Antigens.30 All monoclonal anti-D were tested for agglutination in a gel matrix test (LISS-Coombs 37°C, DiaMed-ID Micro Typing System, DiaMed, Cressier sur Morat, Switzerland). As detailed in the Results, positive reactivities were obtained with BIRMA-D6; BTSN6; BTSN10; HIRO-3; HIRO-4; HIRO-7; HIRO-8; H41; LHM76/55; LHM76/59; LHM-77/64; LOR11-2D9; LOR17-6C7; LOR29-F7; MCAD-6; MS26; NAU3-2E8; NAU6-4D5; P3G6; P3x21223B10; P3x290; 822; negative with AUB-2F7/Fiss; BIRMA-D56; BRAD-3; BRAD-5; BS229; BS231; BS232; B9A4B2; CAZ7-4C5; CLAS1-126; C205-29; D6D02; D10; D89/47; D90/12; D90/17; F5S; HeM-92; HG/92; HIRO-1; HIRO-2; HIRO-6; HM10; HS114; H2D5D2F5; ID6-H8; LHM50/2B; LHM50/3.5; LHM59/19; LHM59/20; LHM59/25; LHM70/45; LHM-76/58; LHM169/80; LHM 169/81; LHM174/102; LORA; LOR12-E2; LOR17-8D3; LOR28-7E6; LOR28-21D3; L87.1G7; MAR-1F8; MS201; NaTH28-3C11; NaTH53-2A7; NaTH87-4A5; NAU6-1G6; NOI; NOU; P3AF6; P3F17; P3F20; P3x35; P3x61; P3187; RAB.B15; RUM-1; SALSA-12; SAL17-4E8; SAL20-12D5; T3D2F7; VOL-3F6; ZIG-189; 17010C9; 175-2; 819; and weak positive or variable with BIRMA-DG3; BTSN4; D90/7; LORE. Furthermore, reactivity with two polyclonal anti-D produced by carriers of the DVI phenotype (CcDVIee and ccDVIEe), as well as with anti-BARC (ISBT 004.052; RH52) serum and eluate (kindly provided by Drs Geoff Daniels and Carole A. Green, Bristol, UK) was checked.
Determination of RhD antigen density was performed by indirect immune fluorescence as described previously.31 32 All 22 IgG monoclonal anti-D reactive to RhD epitopes present in DVI 30 were used (BIRMA-D6, BTSN6, BTSN10, HIRO-3, HIRO-4, HIRO-7, HIRO-8, H41, H41.11B7, LHM76/55, LHM76/59, LHM 77/64, LOR11-2D9, LOR17-6C7, LOR29-F7, MCAD-6, MS26, NAU3-2E8, NAU6-4D5, P3G6, P3x290, and 822). The secondary antibody was goat antihuman immunoglobulins, fluorescein isothiocyanate (FITC)-conjugated (Sigma).
All blood samples were stored on fluid nitrogen. The fluorescence was compared with that of a standard CDe/cde red blood cell (13,000 RhD antigens per cell). Background fluorescence was determined with RhD-negative samples. The number of RhD epitopes detected on the sample cells was calculated as [median fluorescence of sample—background fluorescence] / [median fluorescence of standard cell—background fluorescence] × RhD antigen density of standard cell.
Markers were set to count all red blood cells, even if a fraction of red blood cells appeared unstained. To account for a log-normal distribution, we based the parametric statistical analysis on the logarithms of the RhD antigen densities.
RESULTS
Detection of Three Independent Molecular Events Causing D Category VI
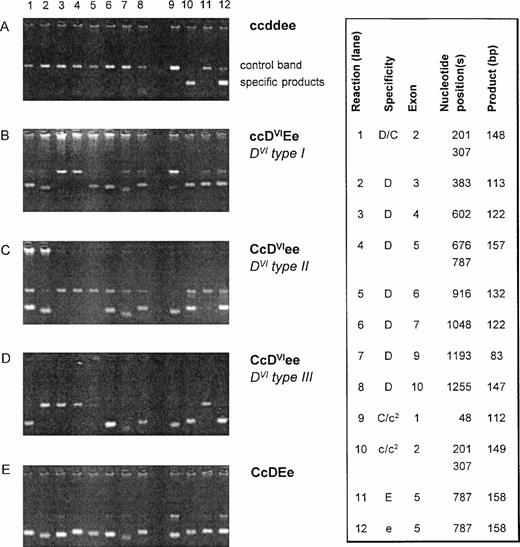
Twenty-six DVI samples were examined usingRHD-specific PCR-SSP for exons 2, 3, 4, 5, 6, 7, 9, and 10 (3′ untranslated).24 All DVI samples differed in their PCR-SSP pattern from the wild-type RHD allele and showed one of three PCR-SSP patterns (Fig 1). Two PCR-SSP patterns were compatible with the previously described genomic rearrangements associated with DVItype I (lack of RHDexons 4 and 5) and DVItype II (lack of RHDexons 4 to 6).18,19 33 A third pattern could not be explained by any known RHD/RHCE variation and is hereby calledDVItype III.
PCR-SSP of DVI samples. Agarose gels of representative DVI samples are shown along with negative and positive controls. The specificities of the 12 reactions for the RHD/RHCE exons are shown on the right side (box). The nucleotide position(s) detected by the PCR-SSP are given along with the expected sizes of the specific products. The control band represents a 434-bp product of the growth hormone gene. For DVI samples, three different reaction patterns are observed: pattern I (B) lacks specific signals for RHD exons 4 and 5 and is compatible with DVItype I.19 Pattern II (C) lacks those for RHD exons 4 to 6 being compatible with DVItype II.18 Pattern III (D) lacking specific products forRHD exons 3 to 6 is novel. c2 indicates the c(cyt48) allele.
PCR-SSP of DVI samples. Agarose gels of representative DVI samples are shown along with negative and positive controls. The specificities of the 12 reactions for the RHD/RHCE exons are shown on the right side (box). The nucleotide position(s) detected by the PCR-SSP are given along with the expected sizes of the specific products. The control band represents a 434-bp product of the growth hormone gene. For DVI samples, three different reaction patterns are observed: pattern I (B) lacks specific signals for RHD exons 4 and 5 and is compatible with DVItype I.19 Pattern II (C) lacks those for RHD exons 4 to 6 being compatible with DVItype II.18 Pattern III (D) lacking specific products forRHD exons 3 to 6 is novel. c2 indicates the c(cyt48) allele.
Molecular Characterization of DVI Type III
Coding sequence.
Because the PCR-SSP pattern of DVI type IIIwas novel, we determined the full-length coding sequence of its cDNA (EMBL/GenBank/DDBJ nucleotide sequence database accession numberZ97026). The DVI type III cDNA comprising all 10Rhesus gene exons represented a RHD-CE-D cDNA, in which the complete exons 3, 4, 5, and 6 of the RHD gene were replaced by the corresponding exons of the RHCE gene.
The exons 3 to 6 are derived from the RHCe allele.
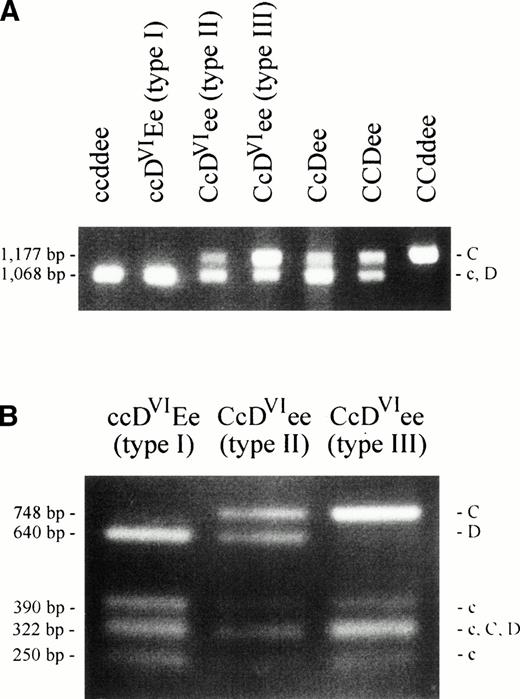
We applied a PCR-RFLP method for the characterization of theRhesus genes' intron 2.25 A length polymorphism discriminates between the RHC and RHc/RHD alleles of the two Rhesus genes (Fig 2A). An RFLP allows the further separation of the RHC, RHc andRHD alleles (Fig 2B). We excluded the presence ofRHD-specific sequences in DVI type III at the position of this polymorphism in intron 2 (Fig 2B). The discrimination between an RHC- or RHc-origin of theDVI type III intron 2 was achieved by the length polymorphism. The DVI type III sample showed an enhanced band of 1,177 bp size (RHC) compared with that of 1,068 bp size (RHc) (Fig 2A). This indicated that two copies ofRHC-like intron 2 sequences were present in theCDVIe/ce genotype, one from theDVI type III allele, the other from the Ceallele of the CDVIe haplotype. We concluded that the RHCE-derived genomic sequences of the DVItype III allele originated from the RHCe allele and extended 5′ of this polymorphism, which is located in the middle of intron 2.
PCR-RFLP of intron 2 of the Rhesus genes. An intron 2 polymorphism was analyzed by PCR amplification and digestion by PstI as previously described.25 Agarose gels are shown with fragment lengths25 and fragment specificities indicated. (A) The 1,177-bp product is specific for RHCalleles, the 1,068-bp product is representative for RHD orRHc or both. The CcDVIee type III sample shows a strong band at the RHC position and a weaker band at theRHD/RHc position. In contrast, the CcDVIee type II and the CcDee samples show a weak band at RHC position and a strong band at the RHD/RHc position. (B) The PCR products shown in (A) were digested with PstI to separate RHD fromRHc-specific products. The DVI type III sample lacks the RHD-specific fragment (640 bp), whereas all other RhD positive samples show this fragment.
PCR-RFLP of intron 2 of the Rhesus genes. An intron 2 polymorphism was analyzed by PCR amplification and digestion by PstI as previously described.25 Agarose gels are shown with fragment lengths25 and fragment specificities indicated. (A) The 1,177-bp product is specific for RHCalleles, the 1,068-bp product is representative for RHD orRHc or both. The CcDVIee type III sample shows a strong band at the RHC position and a weaker band at theRHD/RHc position. In contrast, the CcDVIee type II and the CcDee samples show a weak band at RHC position and a strong band at the RHD/RHc position. (B) The PCR products shown in (A) were digested with PstI to separate RHD fromRHc-specific products. The DVI type III sample lacks the RHD-specific fragment (640 bp), whereas all other RhD positive samples show this fragment.
Exon 1 is of RHD origin.
The guanosine at nucleotide position 48 relative to the A of the translation start codon in the DVI type III cDNA was compatible with both an RHD or an RHcorigin.3 28 To prove the RHD derivation of exon 1, we characterized the 5′ portion of intron 1 for bothRhesus genes (EMBL/GenBank/DDBJ nucleotide sequence database accession number Z97362 and Z97363). DVI type IIIpresented all three nucleotide substitutions and the insertion characteristic for the RHD allele (Fig 3). This observation indicated that the genomic sequences of the DVI type III allele 5′ of this part of intron 1 were derived from the RHDgene. The molecular characteristics of DVI type III were summarized and compared with other published alleles (Fig 4).
5′ portion of the Rhesus genes' intron 1. One hundred thirty nucleotides of intron 1 adjacent to exon 1 are shown for the DVI type III allele along with the commonRHCE and RHD genes. The RHCE and RHDgenes differ by three nucleotide substitutions and one insertion (boxed). The DVI type III allele is identical toRHD at these positions. As expected, DVI type I and DVI type II alleles are also identical toRHD (not shown). Nucleic acid sequence accession numbers wereZ97362 (RHCE) and Z97363 (RHD).
5′ portion of the Rhesus genes' intron 1. One hundred thirty nucleotides of intron 1 adjacent to exon 1 are shown for the DVI type III allele along with the commonRHCE and RHD genes. The RHCE and RHDgenes differ by three nucleotide substitutions and one insertion (boxed). The DVI type III allele is identical toRHD at these positions. As expected, DVI type I and DVI type II alleles are also identical toRHD (not shown). Nucleic acid sequence accession numbers wereZ97362 (RHCE) and Z97363 (RHD).
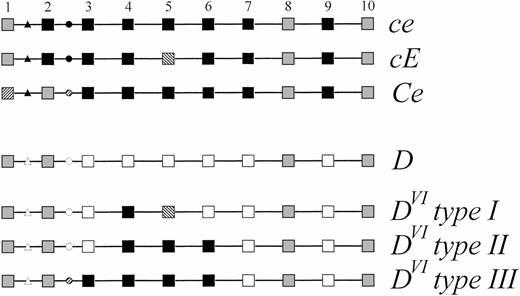
Schematic representation of the genomic structure ofDVI type III compared with other alleles of theRHD and RHCE genes. The 10 exons of the Rhesusgenes are symbolized by squares and numbered 1 to 10, the introns are represented by lines. The triangles denote the base substitutions and insertion occurring in intron 1 distinguishing RHD andRHCE. The circles denote the intron 2 polymorphism distinguishing RHD, RHC and RHc. Black symbols represent RHCE-specific sequences, open symbolsRHD-specific sequences. Sequences shared by at least oneRHCE and the wild-type RHD allele are indicated by gray symbols, sequences specific for RHC or for RHE are hatched. The nucleic acid and amino acid sequence accession number ofD category VI type III was Z97026.
Schematic representation of the genomic structure ofDVI type III compared with other alleles of theRHD and RHCE genes. The 10 exons of the Rhesusgenes are symbolized by squares and numbered 1 to 10, the introns are represented by lines. The triangles denote the base substitutions and insertion occurring in intron 1 distinguishing RHD andRHCE. The circles denote the intron 2 polymorphism distinguishing RHD, RHC and RHc. Black symbols represent RHCE-specific sequences, open symbolsRHD-specific sequences. Sequences shared by at least oneRHCE and the wild-type RHD allele are indicated by gray symbols, sequences specific for RHC or for RHE are hatched. The nucleic acid and amino acid sequence accession number ofD category VI type III was Z97026.
Demonstration of Distinct Breakpoints in the Three DVITypes
The 3′ breakpoints of DVItype II and DVItype III are different.
To define the 3′ limits of the conversion regions of the threeDVI types, we established the complete nucleotide sequence ranging from exon 5 to exon 7 including both introns 5 and 6. We found the breakpoint of DVI typeI to be located at the border of intron 5 and exon 6 in a nucleotide range of 215 bp between -100 bp and +115 bp relative to the first nucleotide of exon 6 (Fig 5). The breakpoint ofDVI type III was located in intron 6 between +360 bp and +963 bp (range of 603 bp) relative to the first nucleotide of intron 6. Finally, the breakpoint of DVI type II34 was also located in intron 6 between +1,781 and +1,821 bp (range of 40 bp) relative to the first nucleotide of intron 6.
Exon 6 of the Rhesus genes and parts of the adjacent introns. The sequence of the RHCE gene extending 202 bp 5′ of exon 6 to 1860 bp 3′ of exon 6 is shown. Numbers indicate the position relative to the first base of exon 5 in theRHCE gene. Exon 6 (bases 1902 to 2040) is demarcated by uppercase letters. Dashes denote nucleotides in the RHD gene that are identical, dots denote deletions. The breakpoint regions forDVI type I, DVI type II andDVI type III are indicated by asterisks. Repetitive DNA elements are marked by carets. The full intron 5 and intron 6 sequences of RHCE and RHD were deposited in EMBL/GenBank/DDBJ under accession numbers Z97333 (RHCE; 5,134 bp) and Z97364 (RHD; 5,146 bp).
Exon 6 of the Rhesus genes and parts of the adjacent introns. The sequence of the RHCE gene extending 202 bp 5′ of exon 6 to 1860 bp 3′ of exon 6 is shown. Numbers indicate the position relative to the first base of exon 5 in theRHCE gene. Exon 6 (bases 1902 to 2040) is demarcated by uppercase letters. Dashes denote nucleotides in the RHD gene that are identical, dots denote deletions. The breakpoint regions forDVI type I, DVI type II andDVI type III are indicated by asterisks. Repetitive DNA elements are marked by carets. The full intron 5 and intron 6 sequences of RHCE and RHD were deposited in EMBL/GenBank/DDBJ under accession numbers Z97333 (RHCE; 5,134 bp) and Z97364 (RHD; 5,146 bp).
An intron 3 length polymorphism differentiates the 5′ breakpoints of DVI type I and DVI type II.
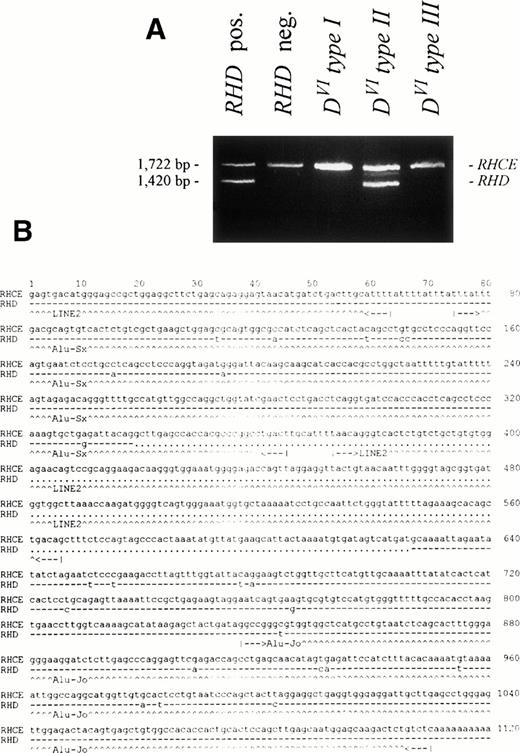
We found a 288-bp deletion in intron 3 of the RHD gene, when compared with the RHCE gene. Based on this deletion, a PCR typing method for RHD was devised. In this intron 3 PCR,DVI type I and DVI type IIIsamples reacted like RHD negative controls, whileDVI type II samples displayed the shorter,RHD-specific band (Fig 6A). This indicated that the conversion point of DVI type Ihad to be 5′ of the intron 3 deletion. We confirmed the conversion point of DVI type II adjacent to an Alu repeat34 (data not shown, see Z97030 and Z97031) and 5′ of the conversion point were two additional Alu repeats with inverse orientation, one of which was partly deleted in RHD(Fig 6B).
Intron 3 length polymorphism of the Rhesus genes. The 3′ region of intron 3 was amplified by PCR using primers RB46 and RB5. (A) The agarose gel shows a 1,722-bp product for theRHCE gene. The 1,420-bp product is representative of theRHD gene. In the DVI type II sample, aRHD-specific product is found. DVI type Iand DVI type III samples show noRHD-specific product. (B) The nucleotide sequence of the 3′ part of intron 3 of the RHCE gene starting 1,556 nucleotides 5′ from exon 4 and of the corresponding parts of theRHD gene comprising the diagnostic 288 bp deletion are shown. Dashes denote nucleotides in the RHD gene that are identical, dots denote deletions. Nucleic acid sequence accession numbers were Z97030(RHCE; 1,580 bp) and Z97031 (RHD; 1,278 bp).
Intron 3 length polymorphism of the Rhesus genes. The 3′ region of intron 3 was amplified by PCR using primers RB46 and RB5. (A) The agarose gel shows a 1,722-bp product for theRHCE gene. The 1,420-bp product is representative of theRHD gene. In the DVI type II sample, aRHD-specific product is found. DVI type Iand DVI type III samples show noRHD-specific product. (B) The nucleotide sequence of the 3′ part of intron 3 of the RHCE gene starting 1,556 nucleotides 5′ from exon 4 and of the corresponding parts of theRHD gene comprising the diagnostic 288 bp deletion are shown. Dashes denote nucleotides in the RHD gene that are identical, dots denote deletions. Nucleic acid sequence accession numbers were Z97030(RHCE; 1,580 bp) and Z97031 (RHD; 1,278 bp).
Linkage of the DVItypes to different Rhesus haplotypes.
We observed the three DVI types associated with specific Rhesus haplotypes: all DVI type Isamples (n = 14) were found in the cDVIE haplotype, all DVI type II (n = 9), and DVItype III (n = 3) in the CDVIe haplotype. Because the genomic structure of DVI type III isD-Ce(3-6)-D, a conversion event among the two Rhesusgenes in cis-position may be the cause of this hybrid allele.
Regional frequency variation of the DVI types.
The distribution of the different DVI types varied depending on the regional origin of the samples (Table 1). In Tyrol (Austria), all samples were DVI type I, while in southwestern Germany,DVI type I and DVI type II were observed about equally frequently. In northern Germany, the only DVI samples that we found so far were DVItype II.
Distribution of DVI Types in German-Speaking Populations
| Regional Origin . | D Category VI Samples Observed (n) . | Total* . | ||
|---|---|---|---|---|
| Type I . | Type II . | Type III . | ||
| Tyrol (Austria)-151 | 9 | 0 | 0 | 9 |
| Southwestern Germany-151 | 5 | 7 | 3 | 15 |
| Northern Germany | 0 | 2 | 0 | 2 |
| Total | 14 | 9 | 3 | 26 |
| Regional Origin . | D Category VI Samples Observed (n) . | Total* . | ||
|---|---|---|---|---|
| Type I . | Type II . | Type III . | ||
| Tyrol (Austria)-151 | 9 | 0 | 0 | 9 |
| Southwestern Germany-151 | 5 | 7 | 3 | 15 |
| Northern Germany | 0 | 2 | 0 | 2 |
| Total | 14 | 9 | 3 | 26 |
*The DVI samples were found by a serologic survey of RhD-positive samples including weak D (southwestern Germany) and by molecular screenings of weak D samples (northern Germany and Tyrol). All DVI samples of the serologic survey were found in weak D as previously published.8
The observed distributions of the various DVI types in Tyrol and Southwestern Germany were statistically significantly different (P < .01, Brandt-Snedecor-χ2-test for 2 × 3 contingency tables with correction for multiple testing (n = 3) according to Bonferroni-Holm).
Serology of DVI Samples
Polyclonal antibodies.
One sample of each DVI type was tested with two polyclonal anti-D and anti-BARC (Table 2).DVI type III qualified as a D category VI, because it was nonreactive with anti-D produced by probands ofDVI type I and DVI type II. Further, DVI type III carried the BARC antigen (ISBT 004.052; RH52). Anti-BARC did not differentiateDVI type II and DVI type III.
Reactivity of DVI Types With Polyclonal Anti-D and Anti-BARC
| Proband's Antiserum . | Proband's Genotype . | D Category VI Samples . | RhD Positive Controls (n) . | ||
|---|---|---|---|---|---|
| Type I (n) . | Type II (n) . | Type III (n) . | |||
| Anti-D | DVI type I | − (1) | − (1) | − (1) | ++++ (2) |
| Anti-D | DVItype II | − (1) | − (1) | − (1) | ++++ (2) |
| Anti-BARC* | — | ND | +++ (6) | ++++ (2) | − (3) |
| Proband's Antiserum . | Proband's Genotype . | D Category VI Samples . | RhD Positive Controls (n) . | ||
|---|---|---|---|---|---|
| Type I (n) . | Type II (n) . | Type III (n) . | |||
| Anti-D | DVI type I | − (1) | − (1) | − (1) | ++++ (2) |
| Anti-D | DVItype II | − (1) | − (1) | − (1) | ++++ (2) |
| Anti-BARC* | — | ND | +++ (6) | ++++ (2) | − (3) |
Abbreviation: ND, not determined.
Anti-BARC (ISBT 004.052; RH52) eluate kindly provided by Drs G. Daniels and C. A. Green.
Monoclonal anti-D.
One sample of each DVI type was tested with the full panel of monoclonal anti-D provided in the recent Workshop on Monoclonal Antibodies against Human Red Blood Cells and Related Antigens.35 The three DVI types did not differ in their reaction pattern (Table 3, upper panel). All positive and most negative reactivities reported by the Workshop coordinator30 were confirmed. Four anti-D (BIRMA-DG3; BTSN4; D90/7; LORE), reported to be nonreactive,30 showed discrepant results and were tested with additional DVI samples (Table 3, lower panel). We found variable, ie, negative or weak positive, reactivity. This reactivity would have been considered negative by the Workshop criteria30 and thus our observations were in full agreement with the Workshop results.
RhD Epitopes Expressed by the DVI Types as Detected by Monoclonal Anti-D
| RhD Epitopes . | D Category VI . | Monoclonal Anti-D* Tested (n) . | ||
|---|---|---|---|---|
| Type I (n = 1) . | Type II (n = 1) . | Type III (n = 1) . | ||
| epD1 | Negative | Negative | Negative | 2 |
| epD2 | Negative | Negative | Negative | 2 |
| epD3 | Negative | Negative | Negative | 1 |
| epD4 | Negative | Negative | Negative | 1 |
| epD7 | Negative | Negative | Negative | 2 |
| epD10 | Negative | Negative | Negative | 7 |
| epD11 | Negative | Negative | Negative | 1 |
| epD12 | Negative | Negative | Negative | 6 |
| epD13 | Negative | Negative | Negative | 6 |
| epD15 | Negative | Negative | Negative | 9 |
| epD17 | Negative | Negative | Negative | 9 |
| epD18 | Negative | Negative | Negative | 8 |
| epD21 | Negative | Negative | Negative | 5 |
| epD22 | Negative | Negative | Negative | 2 |
| epD31 | Negative | Negative | Negative | 2 |
| epD32 | Negative | Negative | Negative | 1 |
| epD33 | Negative | Negative | Negative | 1 |
| epD34 | Negative | Negative | Negative | 1 |
| epD35 | Negative | Negative | Negative | 1 |
| epD5 | Positive | Positive | Positive | 5 |
| epD6 | Positive | Positive | Positive | 1 |
| epD23 | Positive | Positive | Positive | 8 |
| epD36 | Positive | Positive | Positive | 4 |
| epD37 | Positive | Positive | Positive | 4 |
| Reactivity With D Category VI Samples | Monoclonal Anti-D | |||
| Type I (n = 4) | Type II (n = 5) | Type III (n = 3) | ||
| epD3 | Negative | Variable† | Variable | LORE |
| epD15 | Negative | Variable | Variable | BIRMA-DG3 |
| epD15 | Negative | Variable | Variable | BTSN4 |
| epD15 | Variable | Variable | Variable | D90/7 |
| RhD Epitopes . | D Category VI . | Monoclonal Anti-D* Tested (n) . | ||
|---|---|---|---|---|
| Type I (n = 1) . | Type II (n = 1) . | Type III (n = 1) . | ||
| epD1 | Negative | Negative | Negative | 2 |
| epD2 | Negative | Negative | Negative | 2 |
| epD3 | Negative | Negative | Negative | 1 |
| epD4 | Negative | Negative | Negative | 1 |
| epD7 | Negative | Negative | Negative | 2 |
| epD10 | Negative | Negative | Negative | 7 |
| epD11 | Negative | Negative | Negative | 1 |
| epD12 | Negative | Negative | Negative | 6 |
| epD13 | Negative | Negative | Negative | 6 |
| epD15 | Negative | Negative | Negative | 9 |
| epD17 | Negative | Negative | Negative | 9 |
| epD18 | Negative | Negative | Negative | 8 |
| epD21 | Negative | Negative | Negative | 5 |
| epD22 | Negative | Negative | Negative | 2 |
| epD31 | Negative | Negative | Negative | 2 |
| epD32 | Negative | Negative | Negative | 1 |
| epD33 | Negative | Negative | Negative | 1 |
| epD34 | Negative | Negative | Negative | 1 |
| epD35 | Negative | Negative | Negative | 1 |
| epD5 | Positive | Positive | Positive | 5 |
| epD6 | Positive | Positive | Positive | 1 |
| epD23 | Positive | Positive | Positive | 8 |
| epD36 | Positive | Positive | Positive | 4 |
| epD37 | Positive | Positive | Positive | 4 |
| Reactivity With D Category VI Samples | Monoclonal Anti-D | |||
| Type I (n = 4) | Type II (n = 5) | Type III (n = 3) | ||
| epD3 | Negative | Variable† | Variable | LORE |
| epD15 | Negative | Variable | Variable | BIRMA-DG3 |
| epD15 | Negative | Variable | Variable | BTSN4 |
| epD15 | Variable | Variable | Variable | D90/7 |
*Clone identifications were listed in Materials and Methods as provided by Nantes Workshop.35 All monoclonal anti-D were tested with an identical random sample of each DVI type.
Reactivity with the monoclonal anti-Ds indicated on the left side was either weak positive or negative.
Flow Cytometric Analysis of the DVI Types
Epitope density profiles.
Fifteen DVI samples and three control samples were tested with the 22 IgG monoclonal anti-D of the Workshop30 that bind the RhD epitopes of D category VI. In contrast to the control samples, the number of RhD epitopes per cell detected on theDVIsamples varied considerably depending on the monoclonal antibody used (Fig 7). This variation in the number of epitopes detected did not correlate with the epitope specificity30 of the anti-D (data not shown,P = .23 in the analysis of variance). DVItypeI and DVItype II presented consistently low numbers of RhD epitopes per cell with all anti-D. Interestingly, many monoclonal anti-D detected normal, if not enhanced, numbers of RhD epitopes per cell in DVItype III.
Epitope density profiles of samples with the three DVI types and with normal RhD. On the abscissa, ranges of epitope densities (sites/cell) as detected by various anti-D are given. On the ordinate, the number of anti-D representing the particular ranges of sites/cell are shown. One representative sample is shown for each DVI type. Epitope density profiles obtained with four additional DVI type I, six additional DVI type II, and two additional DVI type III samples were similar.
Epitope density profiles of samples with the three DVI types and with normal RhD. On the abscissa, ranges of epitope densities (sites/cell) as detected by various anti-D are given. On the ordinate, the number of anti-D representing the particular ranges of sites/cell are shown. One representative sample is shown for each DVI type. Epitope density profiles obtained with four additional DVI type I, six additional DVI type II, and two additional DVI type III samples were similar.
RhD antigen density (antigens/cell).
Using the results of all 22 anti-D, we calculated the RhD antigen densities as correlates of the number of RhD proteins accessible on the red blood cells' surface (Table 4). The RhD antigen density of DVI type III was similar to the CcDee control and several fold higher than that ofDVI typeI and DVI type II. Still, the RhD antigen densities of DVItypeI and DVI type II differed significantly.
RhD Antigen Density of DVI Types
| D Category VI . | RhD Antigen Densities (antigens per cell)* . | % of Reference3-152 . | DVI Samples Tested (n)3-153 . | ||
|---|---|---|---|---|---|
| Median . | Mean3-151 . | Range . | |||
| Type I | 502 | 489 | 204-1,169 | 3 | 5 |
| Type II | 2,458 | 2,049 | 634-3,941 | 20 | 7 |
| Type III | 13,294 | 12,699 | 11,018-13,981 | 106 | 3 |
| D Category VI . | RhD Antigen Densities (antigens per cell)* . | % of Reference3-152 . | DVI Samples Tested (n)3-153 . | ||
|---|---|---|---|---|---|
| Median . | Mean3-151 . | Range . | |||
| Type I | 502 | 489 | 204-1,169 | 3 | 5 |
| Type II | 2,458 | 2,049 | 634-3,941 | 20 | 7 |
| Type III | 13,294 | 12,699 | 11,018-13,981 | 106 | 3 |
*The RhD antigen density of a sample was calculated as median of the epitopes per cell detected by the 22 IgG monoclonal anti-D.
Geometric mean of the RhD antigen densities.
Median RhD antigen density as percentage of control cells with comparable Rhesus phenotypes (CcDee 12,532 RhD antigens/cell; ccDEe 19,062).
The RhD antigen densities of all three DVItypes were significantly different from one another (P < .007, t-test with correction for multiple testing (n = 3) according to Bonferroni-Holm).
Distinct immunohematologic features of the DVI types.
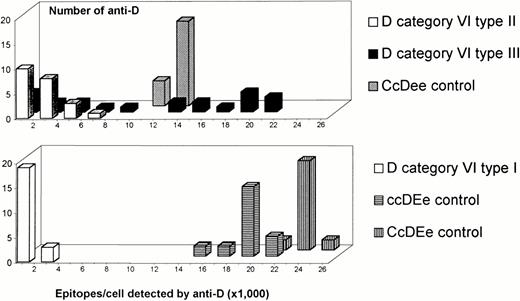
Two of four monoclonal anti-D that are binding to epD3730detected fair numbers of RhD epitopes per cell for DVItype I, but rather low numbers for DVItype IIand DVItype III. This deviation from the actual RhD antigen densities (Table 4) was neither observed with the two other monoclonal anti-D binding to epD37 nor any other anti-D binding to the remaining RhD epitopes present in DVIsamples. This heterogeneity of anti-D's binding to epD37 may represent a flow cytometric split: epD37a (BTSN10 and HIRO-3) was detected equally well in all DVI types, epD37b (MCAD-6 and 822) was reduced in DVI type II and DVI type III. The binding characteristic of MCAD-6 was used to discriminate the threeDVItypes by immunohematologic methods, which also allowed separation from normal controls (Fig 8).
Distinct immunohematologic features of the three DVI types. The RhD antigen density is plotted on the ordinate. On the abscissa, the relative epitope detection by MCAD-6 is shown. This parameter was calculated as follows: [epitopes per cell detected by MCAD–6] ÷ [RhD antigen density] × 100%. Data of 15 DVI samples and three controls are shown. ○, DVI type I, n = 5; ▵, DVI type II, n = 7; □, DVI type III, n = 3; •, controls, n = 3.
Distinct immunohematologic features of the three DVI types. The RhD antigen density is plotted on the ordinate. On the abscissa, the relative epitope detection by MCAD-6 is shown. This parameter was calculated as follows: [epitopes per cell detected by MCAD–6] ÷ [RhD antigen density] × 100%. Data of 15 DVI samples and three controls are shown. ○, DVI type I, n = 5; ▵, DVI type II, n = 7; □, DVI type III, n = 3; •, controls, n = 3.
DISCUSSION
The population-based study showed that the variability of the DVI phenotype is greater than previously reported for the underlying molecular structures18-20 and the RhD antigen densities.15,36-39 We characterized a D-Ce(3-6)-Dhybrid allele of the RHD gene. In accordance with the previous nomenclature,18 this new allele was dubbedDVItype III. Its DVI type III phenotype is associated with an almost normal number of RhD proteins accessible on the red blood cells' surface. All three DVItypes and RhD controls showed distinct immunohematologic features in flow cytometry. The distribution of the DVItypesvaried significantly even within German-speaking populations.
The observation of a D-Ce(3-6)-D hybrid allele, which represented a DVI phenotype, contributed considerably to the understanding of the immunoreactivity in partial D. The DVI phenotype is caused by several different genotypes that are strictly associated with specific Rhesus haplotypes. As a common feature, all known DVIgenotypes sharedRHCE exons 4 and 5 and RHD exon 7. Substitutions ofRHD exon 4 or exon 5 alone by the corresponding exon ofRHCE result in different partial D (exon 4: DFR, exon 5: DVa),40 the additional substitution of exon 7 results in the loss of most41 or all42 RhD immunoreactivity. Our report of a D-Ce(3-6)-D allele proved that in contrast to exon 7, RHD exon 3 is not necessary for a DVI phenotype. This observation supported the current RhD loop model.43,44 All polymorphic sites of exon 3 and exon 6 are believed to occur in the transmembrane and intracellular portions, and hence, may not be expected to influence RhD immunoreactivity very much. In contrast, the polymorphic amino acids of the extracellular loops 3, 4, and 6 are determined by exons 4, 5, and 7, respectively. In concordance with several recent reports,19,20,45 we were unable to find the “deletion type”18 that has been proposed for the cDE haplotype of DVI. However, the ccDVIee phenotype observed in one individual37likely represented a fourth D category VI genotype (proband lost to follow-up; J.W. Jones, personal communication, 1996). Interestingly, the D-Ce(3-6)-D hybrid protein (DVItype III) is complementary to the Ce-D(2-6)-Ce hybrid protein. The latter hybrid protein causes some Evans (D··) phenotypes,46 47 encodes several RhD epitopes, and lacks all CcEe antigens.
An unexpected feature of DVI type III was its almost normal number of RhD proteins per cell. The determination of epitope density profiles in DVI samples gave unequivocal evidence that the lack of certain RhD epitopes need not correlate with the loss of RhD proteins per cell. The observation of the DVI type III phenotype provided a formal proof that the limited RhD immunoreactivity detected with polyclonal anti-Ds in DVI type I and DVI type II37 cannot be explained by the lack of these RhD epitopes only, but must be due to a reduced number of RhD proteins accessible on the red blood cells' surface. The Rhesus protein conformation is likely to influence its red blood cell membrane integration. However, there is currently no conclusive model to predict the effect of any substitution on RhD protein expression. This is exemplified by the different DVItypes showing a paradoxical, inverse correlation between the size of the substituted protein segments and the RhD antigen density. Substitution of exon 348 increases RhD antigen density37 in DIIIc, also. Furthermore, single residue substitutions as occurring in DVII49,50 and DNU51 may have considerable effects on RhD antigen density.32 37
It is intriguing to note that all three DVI typesmay be explained by gene conversion events occurring among bothRhesus genes in cis position. The molecular structures of mostRhesus hybrids (DIIIb,52DVa,40 hybrid-VS,42DBT53) are also compatible with this proposed mechanism. Only one RHD hybrid characterized so far (Rh D-E variant ISBT4954) seemed to be caused by a gene conversion in trans position. The impression that conversions in trans position were predominant in RHCE hybrids (RN,55R0Har,56 and rG57) is likely due to an observation bias because RHCE hybrids will almost exclusively be detected in RhD negative samples.
We referred to DVI type III as aD-Ce(3-6)-D hybrid, but a D-Ce(2-6)-D hybrid could not formally be excluded. The approximately 4,500-bp region encompassing exon 2 was reported to be identical between the RHD andRHCe alleles and to contain many repetitive elements.58 The 5′ conversion point ofDVI type III resided in the region between the polymorphisms in intron 1 and intron 2 (Figs 2 and 3). A further characterization did not seem worthwhile because of the long stretch of identical sequences and repetitive elements in that region. The 5′ conversion points of several independent gene conversion events with substitutions in the RHCe allele by RHDsequences in D−− probands were shown by Kemp et al58 to occur also in this stretch of identical sequences. It is tempting to speculate that the sequence identity over more than 4,000 bp including many repetitive elements facilitated conversion events. A similar accumulation of repetitive Alu and LINE elements (Fig 3) occurred adjacent to the breakpoint region of DVI type II in intron 3, which hosted the conversion points of fourRHD/RHCE hybrids.34
Characterization of the 3′ breakpoint regions of the threeDVI types (Fig 5) showed that their breakpoints were not clustered. The breakpoint of DVI typeI occurred in a stretch of 195 bp covering parts of intron 5 and exon 6, that of DVI type III in a stretch of identity between RHD and RHCE over 605 bp including an Alu repeat. The extent of the whole gene conversion sequence thus varied between about 4,800 bp (DVI type II) and > 19,500 bp (DVI type III). The breakpoint region ofDVI type II in intron 6 was identical to that recently described by Matassi et al.34 These findings are compatible with a common origin (identity by descent) of allDVI type II samples described so far in France, the Netherlands, and Germany.
Our quantitative RhD epitope analysis of molecularly characterized samples clarified several previously controversial issues of DVI immunohematology. First, the use of epitope density profiles addressed the problem of variable antibody affinities in partial D. Studies based on single or few monoclonal antibodies15,36 were likely to underestimate the true number of RhD proteins accessible on the red blood cells' surface, in particular, if the anti-D used36 happened to lack affinity for DVI.12 Even with conditions believed to be saturating, variable epitope densities were obtained with different anti-D.37 We established epitope density profiles using a panel of monoclonal IgG directed to different RhD epitopes present in the partial D tested.32 Such epitope density profiles in DVI showed the variability of anti-D affinities. In difference to other partial D like DVII and DNU,32 there was no narrow antigen density peak (Fig 7), and therefore, the median of the results of all antibodies was used. This robust approach may slightly underestimate the RhD antigen density as correlate of the true number of RhD proteins accessible on the red blood cells' surface because antibodies of marginal affinities to DVI were not excluded. However, our results forDVItypeI and DVItype II were in good agreement with previous reports.37
Second, the immunohematologic features were correlated with molecular structures instead of serologic haplotypes. Previously, the influence of the molecular structures were not checked. Controversial results indicating low36-38 or variable15,39 RhD antigen densities may simply reflect the absence or presence ofDVI type III samples in the CcDVIee group tested. The close linkage of Rhesus haplotype and molecular structure also explains the observation37 that the presence of C suppresses RhD antigen density in normal RhD samples,59,60 but not in DVIsamples.37 In DVI, the slight suppressive effect of antigen C was overwhelmed by the effects of the molecular structures as the principal determinants of RhD protein expression.
Third, the quantitative analysis by flow cytometry separated overall RhD antigen density caused by variations in the number of RhD proteins accessible on the red blood cells' surface from variable expression of certain RhD epitopes. Flow cytometry allowed differentiation of theDVI types from one another and from normal RhD. Furthermore, we could demonstrate a flow cytometric split of RhD epitope epD37. However, the only qualitative serologic difference that we could correlate with the molecular structures was a paucity, but not lack, of epD37b on DVI type II andDVI type III. We suspect that some previously reported serologic splits8,16,18,21,61,62 that were mainly observed with weak overall antibody reactivity8,16,18,62may be due to quantitative differences in RhD epitope expression rather than lack of certain RhD epitopes. We propose that a meaningful report of a serologic split in partial D should exclude the confounding effect of low antigen densities. This exclusion may be achieved by inverse reaction patterns of different monoclonal antibodies,12 by quantitative methods like flow cytometry,31,32 and enzyme-linked immunosorbent assay (ELISA)37 or by the demonstration of different underlying molecular structures.
Our findings have several practical implications for RhD phenotyping and RHD genotyping. Comprehensive RHD genotyping is a complex task24 because of many Rhesus hybrid genes18,19,33,40,42,52,53,55-57,63 and of RhD-negative phenotypes still harboring RHD-specific sequences.6,42,64,65,DVI type III adds to this complexity. It would type RHD negative in a standard intron 2-based PCR method25 previously believed to type DVI samples reliably as RHD positive. The population study (Table 1) provided further evidence for the allelic variation between closely related populations, which influences the specificity and sensitivity of Rhesus genotyping. An absolute match of phenotype and genotype is unlikely to be achieved by current technology because sporadic nonfunctional alleles occur rather frequently in genes66 includingRhesus.67 Hence, the expense of a genotyping system must be weighed against its residual failure rate in phenotype prediction. DVI is the clinically most importantRHD variant and it might be advantageous to dissociate this variant both from RHD positive and RHD negative. To this end, a simple system testing intron 4 and exon 7 may suffice because any D category VI genotype is likely to lack bothRHD exon 4 and 5 and to retain RHD exon 7.
DVI recipients should be transfused with RhD negative blood to limit anti-D immunization,17 a rationale that prompted RhD negative transfusion in patients carrying weak D. This essentially RhD antigen density-based transfusion strategy is today considered wasteful, as it became apparent that most weak D patients may be safely transfused RhD positive. The wastage might be reduced by lowering of the weak D threshold for RhD negative transfusion. However, this measure would trigger RhD positive transfusion in partial D likeDVItype III, while still many RhD negative units would be transfused to weak D patients not requiring RhD negative transfusion. In this context, a strategy based on two monoclonal anti-D that do not react with DVI is advantageous.8,17This RhD epitope-based transfusion strategy abandons RhD antigen density as the trigger for RhD negative transfusions and became mandatory in Germany in 1996.68 It should be advocated in all regions where DVI is the single clinically important partial D. For donor typing, weak D is considered Rhesus positive.69 DVItype III proved that DVI erythrocytes may carry rather high RhD antigen densities. The threshold of RhD antigen density and the RhD epitopes that most likely cause anti-D immunization are not fully established. We think the transfusion of DVI red blood cells should be restricted to RhD positive individuals, until further evidence for lack of immunogenicity is established.
ACKNOWLEDGMENT
We thank Drs Hans-Hermann Sonneborn and Manfred Ernst, Dreieich, Germany, for generously supplying us with their monoclonal anti-Ds; Drs Geoff Daniels and Carole A. Green, Bristol, England, for providing anti-BARC serum and eluate; Dr Jeff W. Jones, Liverpool, England, for the determination of absolute RhD antigen density on red blood cell samples that were used as standards; Elisabeth Hörner, Olga Zarupski, and Esther Rainer for expert technical assistance; and Bryan Hillesheim for preparing red blood cell and DNA samples.
Supported by the University of Ulm (Forschungsförderungsprojekt P. 239) and the DRK-Blutspendedienst Baden-Württemberg, Stuttgart, Germany.
Address reprint requests to Willy A. Flegel, MD, Abteilung Transfusionsmedizin, Universität Ulm, and DRK-Blutspendezentrale Ulm, Helmholtzstrasse 10, D-89081 Ulm, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.



![Fig. 8. Distinct immunohematologic features of the three DVI types. The RhD antigen density is plotted on the ordinate. On the abscissa, the relative epitope detection by MCAD-6 is shown. This parameter was calculated as follows: [epitopes per cell detected by MCAD–6] ÷ [RhD antigen density] × 100%. Data of 15 DVI samples and three controls are shown. ○, DVI type I, n = 5; ▵, DVI type II, n = 7; □, DVI type III, n = 3; •, controls, n = 3.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/6/10.1182_blood.v91.6.2157/2/m_blod4060708.jpeg?Expires=1763503141&Signature=FbxqA59WKDWnyoHd5XemqLyA08Ae1QEuLCEtq3tP3uugc7eBYnsfjJu0xzMfQxndFEGUnlYAULoTf0I1MLJV1Q7HWE60XXcozi2a-3m0FFwZDNFO8A30TfzBvsu8mmjlp-FN3Ihrm2Olc0Sk-i7u2eb0J5cRV25cvkQLavSaYVJ3fbHBvtuaH~e06v6T6WeHAq3JqdqtroIoOzmJpAddwpNFbuMSf9dPfMX8EZXxXEV5RsONieJ~lL-FsdQ31mIAJrDo9wfp2ZwP3hFCN0cdeHNnMYAhHn6bvPcDEyNaE6AF6XacPtTTtVW8i88F6GGkhytSSijBZOMZ58AttPfEpw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal