Ship is a recently identified SH2-containing inositol polyphosphate 5-phosphatase that has been implicated as an important signaling molecule in cell-culture systems. To understand the physiologic function of Ship in vivo, we performed expression studies of Ship during mouse development. Results of this study demonstrate the expression of ship to be in late primitive-streak stage embryos (7.5 days postcoitus [dpc]), when hematopoiesis is thought to begin, and the expression is restricted to the hematopoietic lineage in mouse embryo. In adult mice, Ship expression continues to be in the majority of cells from hematopoietic origin, including granulocytes, monocytes, and lymphocytes, and is also found in the spermatids of the testis. Furthermore, the level of Ship expression is developmentally regulated during T-cell maturation. These results suggest a possible role for Ship in the differentiation and maintenance of the hematopoietic lineages and in spermatogenesis.
A HALLMARK OF EARLY hematopoiesis in the mouse embryo is the appearance of primitive nucleated erythroid cells in the extraembryonic yolk sac mesoderm during late primitive-streak stage (7.5 to 8.5 days postcoitus [dpc]). This event occurs in blood islands, which are aggregations of mesenchymal cells in the yolk sac. Within each blood island, the central cells separate from those at the periphery to form two distinctive populations of cells: hematopoietic stem cells and endothelial cells. It is believed that both arise from a common mesoderm precursor known as the hemangioblast.1,2 In an attempt to gain insight into the molecular events of early hematopoiesis, we have isolated Ship, a gene expressed in 5-fluorouracil (5FU)-treated bone marrow.3
ship encodes an inositol polyphosphate 5-phosphatase that hydrolyzes phosphotidylinositol(3,4,5) polyphosphate [PtdIns(3,4,5)P3] and inositol(1,3,4,5)polyphosphate (IP4).4,5 PtdIns(3,4,5)P3 is the major product of PI3 kinase and is a common second messenger in delivering signals from multiple surface receptors. Recently, Ship has been shown to reduce the level of PtdIns(3,4,5)P3 and inhibit biologic effects induced by PI 3 kinase activation inXenopus oocytes.6 IP 4 is another in vitro substrate identified for Ship. It has been proposed that IP 4 activates calcium influx through the cytoplasmic membrane7 and that recruitment of Ship to the membrane could account for the reported inhibition of calcium entry during negative signaling.8 9
In addition to the enzymatic inositol 5-phosphatase domain in the central portion of the protein, Ship contains an SH2 domain at the N-terminal end, three putative SH3-interacting motifs, and two potential PTB domain-binding sites (NPXY) at the C-terminal to the inositol phosphatase region. The SH2 domain was shown to be essential for the tyrosine phosphorylation of Ship.10 It is likely that this phosphorylation occurs when Ship binds to a tyrosine kinase11 or to proteins that can bring it to a tyrosine kinase. The NPXY motifs of Ship were shown to be essential for the association with Shc during T-cell receptor signaling.12Indeed, Ship was first identified as a Shc-associated tyrosine-phosphorylated protein in cells of hematopoietic origin, following cytokine stimulation,13,14 and Shc association with a 140-kD protein, most likely Ship, was often detected in leukemic cell lines.15,16 Since Shc is a mediator of signal transduction from growth factor receptors to the Ras pathway,17 Ship's involvement in Ras signaling has been a target for investigation. Taken together, the data indicate Ship may be an important signaling molecule that can be engaged in diversified pathways.
To gain insight into the possible function of Ship, we investigated its expression during mouse development. Here, we report the expression ofship to be within the hematopoietic lineage and spermatids. Furthermore, we show that this expression is first detected at the onset of hematopoietic differentiation during mouse development.
MATERIALS AND METHODS
Reagents.
Rabbit antihuman von Willebrand factor antibody was from DAKO (Carpinteria, CA) and was shown to react with mouse von Willebrand factor specifically. The immunohistochemical staining, peroxidase substrates, and biotin/avidin blocking kit were from Vector Laboratories (Burlingame, CA). Poly(A+) RNA blots were from Clontech (Palo Alto, CA). All other reagents were purchased from Fisher Scientific (Pittsburgh, PA) unless otherwise indicated.
The following antibodies were used in flow cytometry: FcBlock (2.4G2), phycoerythrin (PE)-conjugated anti-αβTCR (H57-597), PE-conjugated anti-B220 (RA3-6B2), PE-conjugated anti-CD4 (L3T4), biotinylated anti-IgD (11-26C-2a), biotinylated anti-IgM (R6-60.2), and biotinylated anti-CD8α (53-6.7) (all from PharMingen, San Diego, CA). Biotinylated antibodies were visualized using Streptavidin-Red670 (Gibco/BRL). Fluorescein isothiocyanate (FITC)-conjugated goat antirabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) was used to label rabbit anti-Ship antibody in all of the flow cytometry assays.
Northern, RNA in situ hybridization, and reverse-transcriptase polymerase chain reaction analysis.
A 1.5-kb DNA fragment corresponding to the 3′-end and a 1.3-kb fragment from the 5′-end of the ship cDNA were independently random prime-labeled and hybridized to a poly(A) RNA blot (Clontech) in 50% formamide solution that contained 4X SSPE, 1% sodium dodecyl sulfate (SDS), 0.5% blotto, and 10% dextran sulfate at 42°C overnight. The blot was washed twice with 0.1× SSC and 0.1% SDS at 55°C for 15 minutes each and autoradiographed.
Embryos at different stages of mouse development were isolated, fixed overnight in 4% paraformaldehyde, dehydrated with ethanol and xylene, processed for paraffin embedding, sectioned at 6 μm, and mounted on 3-amino-propyltriethoxysilane–treated slides (Sigma, St Louis, MO). After removal of paraffin, the sections were predigested with protease K (Boehringer Mannheim, Mannheim, Germany), acetylated with acetic anhydride, dehydrated, and hybridized as described by Frohman et al,18 except that base hydrolysis of the probe was omitted. Antisense 35S-labeled riboprobe was synthesized following Xho I digestion of a partial Ship cDNA clone, pE41, which contains sequences 1 to 1342 of the gene. Sense probe was made from pE41 plasmid digested with Not I.
For reverse-transcriptase polymerase chain reaction (RT-PCR), Trizol (Life Technologies Inc, Gaithersburg, MD) was used to extract total RNA from embryos. These embryos were genotyped by Southern blot analysis of DNA prepared from cone and trophoblast cells cultured in embryonic stem (ES) cell medium without leukemia inhibitory factor (LIF) for 10 to 14 days to ensure the loss of maternal cells.19 Five micrograms of RNA was reverse-transcribed using Superscript reverse transcriptase (BRL) in 20-μL reactions. PCR was then performed using 0.5 μL of the cDNA and the following primers: 5′-CAGAATCTACCAACAGGCGTT-3′ of Shipsequence 987 to 1007 and 5′-GAGAAACCAGGACGTGATCTT-3′ of complementaryShip sequence 1408 to 1388 to yield a product of 421 bp. Primers for Flt1 selected were Flt1A—TGTGGAGAAACTTGGTGACCT, and Flt1B—TGGAGAACAGCAGGACTCCTT, to yield a fragment of 504 bp.
Antibody production and immunohistochemistry.
DNA sequences coding for the region between the SH2 domain and the inositol phosphatase homologous sequences of Ship (nucleotide 827 to 1350) were released from plasmid pE41 by SacI andNotI digestion and subcloned into a glutathione S-transferase (GST) expression vector. GST fusion protein was expressed inEscherichia coli and recovered from cell lysate with glutathione-agarose beads.20 Antiserum against mouse Ship was generated by immunizing rabbits with purified GST fusion protein. Specific antibody from rabbit serum was affinity-purified by passing through a GST-conjugated Sepharose-4B column followed by adsorption to the antigen-conjugated Sepharose. Antibody was eluted from the column using 100 mmol/L glycine pH 2.5 and dialyzed against 1× phosphate-buffered saline (PBS) at 4°C overnight. The antibody was aliquotted and stored at −70°C in 1% bovine serum albumin (BSA) and 0.1% NaN3.
Mouse embryos of different stages were isolated and fixed in Histochoice MB solution (Amresco, Solar, OH) for 4 hours, soaked in 30% sucrose overnight, embedded in O.C.T. compount (tissue talc; Miles, Elkhart, IN), and sectioned at 8 μm with a Jung CM3000 cryostat (Leica, Heerbrugg, Switzerland). The frozen sections were mounted on superfrost/plus microscope slides, air-dried, and fix-permeabilized in 50% methanol/acetone at −20°C for 10 minutes. All slides were stored at −70°C until use.
The sections were brought to room temperature in a sealed box, rinsed twice in PBS, and immunostained with Vectastain Elite ABC kit (Vector Laboratories) as per the manufacturer's protocol. After immunostaining, the slides were rinsed twice with tap water and counterstained with Harris' hematoxylin (BDH, Dorset, UK) for 1 minute, rinsed with running water for 30 seconds, dehydrated, and mounted with cover glass.
Flow cytometry.
Spleen, thymus, and bone marrow cells were prepared according to standard procedures.21 A total of 106 cells were incubated in staining buffer (PBS, 1% fetal bovine serum, and 0.1% NaN3) with saturating amounts of antibodies against lineage-specific surface antigens at 4°C for 30 minutes. Cells were then fixed and permeabilized with FIX and PERM solutions as per the manufacturer's instructions (CALTAG, South San Francisco, CA). To detect Ship expression, 0.2 μg of anti-Ship antibody was included in the permeabilization step, followed by incubation with 0.5 μg FITC-conjugated antirabbit IgG in 100 μL PBS at room temperature for 15 minutes. The cells were thoroughly washed with PBS and analyzed using a FACSCalibur flow cytometer and CELLQuest software (Becton Dickinson, Mountain View, CA). Windows for the analysis of lymphocytes, granulocytes, and monocytes were set using forward and side scatter on the CELLQuest software (Becton Dickinson).
RESULTS
Ship is expressed in the hematopoietic lineages and in the adult testis.
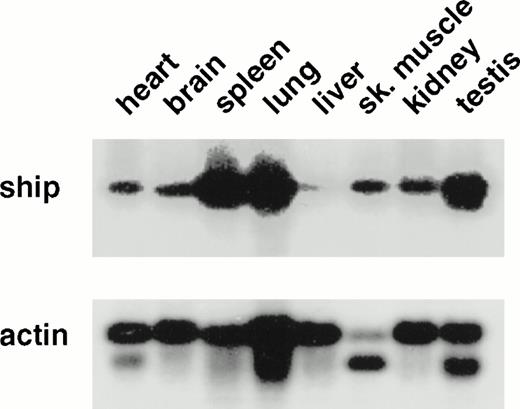
To determine the pattern of ship expression, we first performed Northern analysis on poly(A+) RNA from various adult mouse tissues. A single 5.0-kb transcript was detected in all the tissues analyzed. However, the level of ship expression was different among the tissues. The highest levels were found in spleen, lung, and testis, and the lowest in the liver (Fig1). This variation suggested cell-type specificity of ship expression. To localize shipexpression to specific cell types during development, RNA in situ hybridization was performed on embryo sections from different stages of mouse development. No hybridization was detected in any organs in the sagittal sections of 12.5- to 15.5-dpc embryos, except for punctated signals within the developing liver (data not shown). Since definitive hematopoiesis begins in the embryonic mouse liver at approximately 12 dpc,22 and these early hematopoietic cells appear intermingled with hepatocytes, the RNA in situ hybridization results raised the possibility that ship is expressed in hematopoietic cells in the developing liver.
Northern analysis: 2 μg poly(A+) RNA from various adult mouse tissues was loaded in each lane. A 1.3-kb fragment from the 5′ end of ship cDNA was used as probe. The membrane was reprobed with mouse actin probe to measure the amount of RNA in each lane. The same result was obtained with a 1.5-kb probe from the 3′-end of the ship cDNA clone.
Northern analysis: 2 μg poly(A+) RNA from various adult mouse tissues was loaded in each lane. A 1.3-kb fragment from the 5′ end of ship cDNA was used as probe. The membrane was reprobed with mouse actin probe to measure the amount of RNA in each lane. The same result was obtained with a 1.5-kb probe from the 3′-end of the ship cDNA clone.
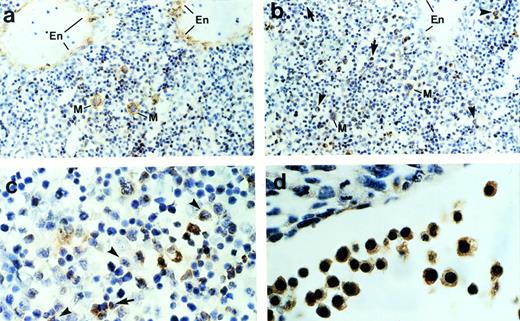
To determine further the cell types that express ship,immunohistochemical staining was performed on sagittal sections of mouse embryos. Consistent with the RNA in situ hybridization result, immunohistochemical staining detected ship expression only in the liver and occasional single cells under the skin. Normally, hematopoietic cells can be distinguished from other mesenchymal cells in the liver by the distinct shape of their nuclei and their irregular sizes. Close examination of the ship-expressing cells in the liver indicate that they are hematopoietic cells. Megakaryocytes can undergo as many as seven duplications of the nuclei and cytoplasmic constituents without cell division. As a result, they appear as distinctively giant cells with large, irregular, multilobular nuclei (Fig 2a and b, cells labeled “M”). When compared with the section stained with antibody against von Willebrand factor (Fig 2a), a protein synthesized by megakaryocytes and endothelial cells, it was evident that ship was expressed in megakaryocytes and not in endothelial cells (Fig 2b). Furthermore, Ship-positive staining could be found in monocyte-macrophages, identified by their intermediate cell size and cashew nut-shaped nuclei (Fig 2b and c, arrowheads), and in granulocytes, specified by their small sizes and horseshoe or sometimes ring-shaped nuclei (Fig 2b and c, arrows); the ship-expressing cells found scattered under the skin are mast cells identified by their extensive cytoplasm packed with large granules (data not shown). Staining on 10.5-dpc embryo sections showed ship expression in the large nucleated erythroblasts (Fig 2d). The hematopoietic identity of theship-expressing cells was substantiated by coexpression ofship with CD43, a surface marker expressed by most hematopoietic stem cells,23 through fluorescence coimmunostaining (data not shown).
Immunohistochemical staining of sections across the liver from a 15.5-dpc mouse embryo. (a) Section stained with anti–von Willebrand factor antibody. Cells expressing von Willebrand factor are visualized by a brown color. En, endothelial cells; M, megakaryocyte. (b) Sections stained with anti-Ship antibody. Representative Ship-expressing cells with distinctive morphology of granulocyte or monocyte/macrophage are denoted by arrows and arrowheads, respectively. (c) Section as in panel b under higher magnification. (d) Section across yolk sac of a 10.5-dpc mouse embryo stained with anti-Ship antibody. The large round cells with big nuclei are nucleated erythroblasts. Original magnifications: ×200 for a and b; ×400 for c and d.
Immunohistochemical staining of sections across the liver from a 15.5-dpc mouse embryo. (a) Section stained with anti–von Willebrand factor antibody. Cells expressing von Willebrand factor are visualized by a brown color. En, endothelial cells; M, megakaryocyte. (b) Sections stained with anti-Ship antibody. Representative Ship-expressing cells with distinctive morphology of granulocyte or monocyte/macrophage are denoted by arrows and arrowheads, respectively. (c) Section as in panel b under higher magnification. (d) Section across yolk sac of a 10.5-dpc mouse embryo stained with anti-Ship antibody. The large round cells with big nuclei are nucleated erythroblasts. Original magnifications: ×200 for a and b; ×400 for c and d.
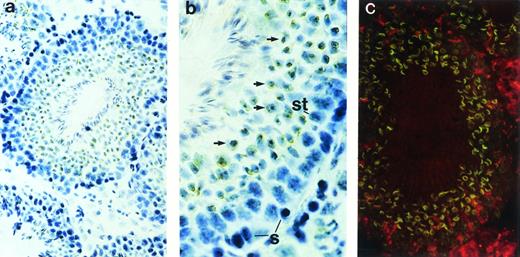
Although restriction of ship expression to hematopoietic cells could explain the abundance of ship message in adult spleen—a lymphoid organ in adult life—the high levels ofship transcript in the lung and testis needed to be addressed. Immunohistochemical staining on sections from carefully washed adult lung showed no significant positive staining (data not shown). However, positive signals were detected in the seminiferous tubules of testes (Fig 3). In the active seminiferous tubules, spermatogonia (Fig 3b, labeled “S”), the germ cells characterized by large round nuclei with condensed chromatin, are found in the basal layer of the seminiferous epithelium. Also near the basal layer are Sertoli cells (Fig 3b, labeled “St”), which are identified by their triangular nuclei, dispersed chromatin, and extensive cytoplasm. As spermatogonia go through meiosis, newly derived spermatids (Fig 3b, arrows) settle next to spermatogonia, away from the basal layer. Mature spermatozoa develop from spermatids and locate themselves close to the lumen of seminiferous tubules, with their tails floating inside the lumen.22 Immunohistochemical and fluorescence staining with anti-Ship antibody clearly demonstrated ship expression in spermatids, but not in spermatogonia or Sertoli cells (Fig 3). Moreover, Ship is predominantly localized to the membrane of spermatids (Fig 3c).
Immunohistochemical staining with anti-Ship antibody on sections across adult testis. (a) Cross section of a seminiferous tubule shows positive staining within the tubule. (b) Cross section of a seminiferous tubule photographed under higher magnification with basal layer of the tubule on the right, lumen on the left. S, spermatogonia; St, Sertoli cells; arrows point to spermatids. (c) Indirect fluorescence staining with anti-Ship antibody on adjacent section of panel a. ship expression is visualized by the green fluorescence in spermatids. Original magnifications: ×200 for a and c; ×400 for b.
Immunohistochemical staining with anti-Ship antibody on sections across adult testis. (a) Cross section of a seminiferous tubule shows positive staining within the tubule. (b) Cross section of a seminiferous tubule photographed under higher magnification with basal layer of the tubule on the right, lumen on the left. S, spermatogonia; St, Sertoli cells; arrows point to spermatids. (c) Indirect fluorescence staining with anti-Ship antibody on adjacent section of panel a. ship expression is visualized by the green fluorescence in spermatids. Original magnifications: ×200 for a and c; ×400 for b.
Ship is expressed at the beginning of hematopoiesis.
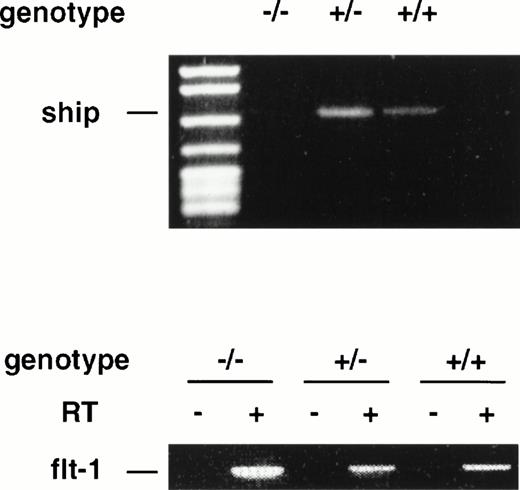
During late embryonic development, the fetal liver is the center of definitive hematopoiesis,22 and primitive hematopoiesis initiates from blood islands in the yolk sac at a much earlier stage (at ∼7.5 dpc) during development.1 To determine ifship expression coincides with the onset of hematopoietic development, we performed RT-PCR on RNAs isolated from 7.5-dpc and 8.5-dpc embryos of wild-type and flk-1 mutant mice.flk-1 codes for a tyrosine kinase receptor that is specifically expressed in early hematopoietic and endothelial cells. Mice that carry a null mutation of the flk-1 gene die at 8.5-dpc due to the lack of hematopoietic and endothelial cells.19 RT-PCR analysis on RNA isolated from 7.5-dpc and 8.5-dpc flk-1–deficient embryos showed no expression ofship, whereas ship was expressed in wild-type and heterozygous embryo littermates at both stages (Fig4 and data not shown). These results demonstrate that ship is expressed at the earliest time of hematopoietic development and that its expression is restricted to hematopoietic and/or endothelial cells at the stage of development.
RT-PCR analysis on RNA isolated from 7.5-dpc embryos of wild-type (+/+), heterozygote (+/−), and homozygote (−/−)flk-1 mutant mice. RT-PCR on flt-1 was included as an internal control for the quality of RNA preparation. RT+ and RT−, with or without reverse transcriptase in reverse transcription.
RT-PCR analysis on RNA isolated from 7.5-dpc embryos of wild-type (+/+), heterozygote (+/−), and homozygote (−/−)flk-1 mutant mice. RT-PCR on flt-1 was included as an internal control for the quality of RNA preparation. RT+ and RT−, with or without reverse transcriptase in reverse transcription.
The level of ship expression is developmentally regulated during T-cell maturation.
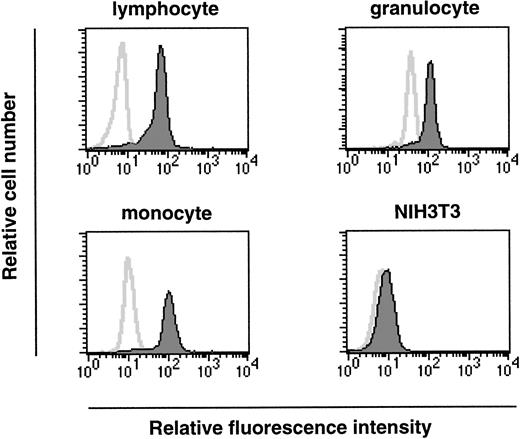
As we have shown, ship is expressed at the onset of hematopoiesis (7.5 dpc) and this expression is maintained within a variety of hematopoietic lineages in fetal liver. To investigate a possible role for Ship during hematopoietic cell development in adults, permeabilized bone marrow cells were labeled with FITC-conjugated antirabbit antibody in the presence or absence of rabbit anti-Ship antibody, and the expression in various subpopulations of hematopoietic cells was determined by the intensity of fluorescence staining in the gated cell population. Essentially all cells in the lymphocyte lineage, cells in the myeloid lineage (mostly granulocytes, as in Fig5), and cells in the size group of early hematopoietic progenitor cells and monocytes (monocyte in Fig 5) showed significantly increased intensity of fluorescence labeling in the presence of anti-Ship antibody, which indicates that ship is expressed in these cells.
Flow-cytometric analysis on single-cell populations from bone marrow of 6-week-old female mice. Cell populations were gated according to cell-size distribution and are indicated on the top of each plot. Histograms in each plot represent the fluorescence intensity of cells treated with (filled area) or without (solid line) anti-Ship antibody. Each plot is representative of 3 independent experiments.
Flow-cytometric analysis on single-cell populations from bone marrow of 6-week-old female mice. Cell populations were gated according to cell-size distribution and are indicated on the top of each plot. Histograms in each plot represent the fluorescence intensity of cells treated with (filled area) or without (solid line) anti-Ship antibody. Each plot is representative of 3 independent experiments.
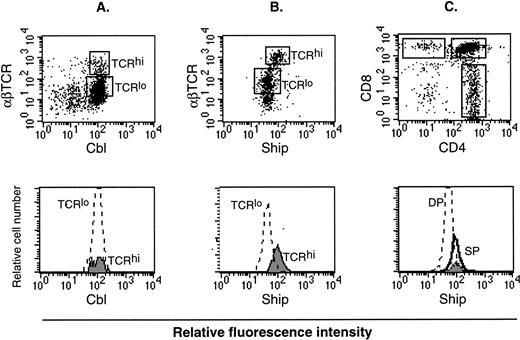
Lymphocytes consist of B-cell and T-cell populations. It is known that αβ T cells go through three distinct stages during thymic development, defined by expression of the CD4 and CD8 surface markers. These are, in order of increasing maturity, CD4−CD8− double-negative (DN), CD4+CD8+ double-positive (DP), and CD4+ or CD8+ single-positive (SP). During this process, the level of αβ T-cell receptor (αβ TCR) expression also increases.24 Therefore, mature T cells can be labeled as SP cells or αβ TCRhi cells. On the other hand, the process of B-cell maturation is defined by expression of surface molecules to be B220+IgM− premature, IgMhiIgD− immature, and IgMloIgD+ mature cells.25 Flow cytometry analysis on thymocytes labeled with different colored anti-Ship and anti-αβ TCR antibodies showed higher Ship-labeled fluorescence intensity in TCRhi mature T cells than in TCRlo immature T cells (Fig6B). No difference in the fluorescence intensity was detected among T cells of these two gated populations when thymocytes were stained for Cbl (Fig 6A) or Shc (data not shown), proteins known to be expressed constitutively in these cells. With the use of three-color labeling, we were able to gate the CD4+, CD8+ and the DP (CD4+CD8+) cells and compare the relative Ship-labeled fluorescence intensity among these three subpopulations of the thymocytes. As shown in Fig 6C, the peak of Ship fluorescence intensity for CD4+ cells overlaps with that of CD8+ cells and is at a value higher than that for DP cells. This result substantiated our previous observation on the two-color staining, to show that the level of shipexpression in mature T cells is higher than in immature T cells. No obvious difference in the level of Ship expression was detected in B220+IgM−, IgMhiIgD−, and IgMloIgD+ cells in the bone marrow (data not shown).
Flow cytometric analysis on adult lymphocytes. (A) PE-anti-αβTCR and FITC-anti-Cbl double staining on lymphocyte. (B) PE-anti-αβTCR and FITC-anti-Ship double staining on lymphocytes. The αβTCRhi and αβTCRlo populations are gated (indicated in the upper panel as TCRhi and TCRlo), and FITC fluorescence intensity among the gated cell populations are compared by overlay histogram (lower panel). (C) FITC-anti-Ship, PE-anti-CD4, Texas red-anti-CD8 triple-color staining on lymphocytes. CD4+, CD8+, and DP cells are gated as indicated in the upper panel. FITC fluorescence intensity in the gated cell populations, which correspond to the levels ofship expression, is compared in an overlay histogram (lower panel). SP in the lower histogram of panel C represents both CD4+ (solid line) and CD8+ (filled area) cells. Each plot is representative of 5 independent experiments.
Flow cytometric analysis on adult lymphocytes. (A) PE-anti-αβTCR and FITC-anti-Cbl double staining on lymphocyte. (B) PE-anti-αβTCR and FITC-anti-Ship double staining on lymphocytes. The αβTCRhi and αβTCRlo populations are gated (indicated in the upper panel as TCRhi and TCRlo), and FITC fluorescence intensity among the gated cell populations are compared by overlay histogram (lower panel). (C) FITC-anti-Ship, PE-anti-CD4, Texas red-anti-CD8 triple-color staining on lymphocytes. CD4+, CD8+, and DP cells are gated as indicated in the upper panel. FITC fluorescence intensity in the gated cell populations, which correspond to the levels ofship expression, is compared in an overlay histogram (lower panel). SP in the lower histogram of panel C represents both CD4+ (solid line) and CD8+ (filled area) cells. Each plot is representative of 5 independent experiments.
DISCUSSION
In this study, we have investigated the expression of shipduring mouse development by Northern analysis, RNA in situ hybridization, immunohistochemistry, and flow cytometry. We have shown the following: (1) ship expression during early development is only detected at sites of hematopoiesis, such as the yolk sac and the liver of mouse embryos; (2) ship expression is detected in hematopoietic cells, such as megakaryocytes, myeloid cells, monocyte/macrophages, tissue basophils (mast cells), and nucleated erythroblasts; (3) through the use of flk-1 null mutant mouse embryos, we have demonstrated that ship is not expressed in embryos that do not form hematopoietic and endothelial cells, which suggests ship's expression in one or both of these lineages; and (4) the expression of ship is not detected in endothelial cells by immunohistochemical staining (Fig 2b and d), nor is it detected in endothelial cell lines (data not shown). Taken together, these results demonstrate Ship expression to be exclusively in the hematopoietic lineages during embryonic mouse development. Furthermore, the expression of Ship in 7.5-dpc embryos suggests thatship is expressed in the presumed common precursor of endothelial and hematopoietic cells, the hemangioblast.1 2Whether Ship plays a role in hematopoietic commitment and differentiation from the hemangioblast remains to be investigated.
Although Ship is only detected in cells of the hematopoietic origin in prenatal animals, Northern analysis on RNA prepared from adult tissues detected ship message in a broad range of tissues4,26 (Fig 1). Could ship expression be turned on at a later stage in these adult organs? Results from immunohistochemical staining on sections of the lung, liver, and kidney (data not shown) showed no positive staining. Therefore, theship messages detected by Northern could be transcripts produced by (1) various amount of hematopoietic cells existing in each tissue; or (2) other cell types in the organ, and if so, they are not translated. However, ship is expressed in spermatids of the adult testis and the expression appears to be predominantly in the membrane (Fig 3c). Membrane localization was demonstrated to be necessary and sufficient for Ship's function,27 which suggests that the Ship in spermatids is probably in an active form. The role of Ship during spermatogenesis awaits further investigation.
Our studies also provided evidence that ship is broadly expressed in hematopoietic lineage. In fact, expression ofship was detected in nucleated erythroblasts in the yolk sac and in megakaryocytes, monocytes, neutrophils, and tissue basophils during definitive hematopoiesis. Consistent with a previous report,28 we also found that virtually all of the lymphocytes, granulocytes, and monocyte/progenitor cells in the bone marrow of adult mice express ship. Since these cells in various hematopoietic lineages have different sets of surface receptors for signal transduction, the wide expression pattern of shipsuggests that it may play a role in several distinct signaling pathways.
In B lymphocytes, ship was expressed at a relatively constant level throughout development. However, the level ofship expression in T lymphocytes was upregulated as T cells go through positive and negative selection from DP cells to become SP cells. It is known that the DP to SP transition is a checkpoint for selection of useful TCR specificities and is mediated by the αβ TCR complex.24 The regulated expression of shipduring this transition suggests that Ship plays an important role either during the selection process or in T-cell immune responses. The ability of Ship to interact with TCR complex ζ chain26implies that Ship can do so through mediating αβTCR-initiated signaling.
ACKNOWLEDGMENT
We thank Drs Josef Penninger and Jane McGlade for helpful discussions and critical reading of the manuscript. We are also grateful to Renu Sarao and Marissa Luchico for their help in preparation of the manuscript.
Address reprint requests to Daniel J. Dumont, PhD, Amgen Institute, 620 University Ave, Suite 706, Toronto, Ontario, Canada M5G 2C1.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal