The molecular mechanisms underlying the pathogenesis of aggressive lymphomas and the histological transformation of indolent variants are not well known. To determine the role of p16INK4a gene alterations in the pathogenesis of non-Hodgkin's lymphomas (NHLs) and the histological progression of indolent variants, we have analyzed the expression, deletions, and mutations of this gene in a series of 112 NHLs. Hypermethylation of the gene was also examined in a subset of tumors with lack of protein expression but without mutations or deletions of the gene. p16INK4a gene alterations were detected in 3 out of 64 (5%) indolent lymphomas but in 16 out of 48 (33%) primary or transformed aggressive variants. In the low-grade tumors, p16INK4a alterations were detected in 1 (4%) chronic lymphocytic leukemia (hemizygous missense mutation), 1 (6%) follicular lymphoma (homozygous deletion), and 1 (5%) typical mantle cell lymphoma (homozygous deletion). The two later cases followed an aggressive clinical evolution. In the aggressive tumors, p16INK4a gene alterations were observed in 2 (29%) Richter's syndromes (2 homozygous deletions), 3 (33%) transformed follicular lymphomas (1 homozygous deletion and 2 nonsense mutations), 3 (43%) blastoid mantle cell lymphomas (2 homozygous and 1 hemizygous deletions), 5 (28%) de novo large-cell lymphomas (1 homozygous deletion and 4 hypermethylations), 2 lymphoblastic lymphomas (2 homozygous deletions), and 1 of 2 anaplastic large cell lymphomas (hypermethylation). Protein expression was lost in all tumors with p16INK4a alterations except in the typical chronic lymphocytic leukemia (CLL) with hemizygous point mutation. Sequential samples of the indolent and transformed phase of three cases showed the presence of p16INK4a deletions in the Richter's syndrome but not in the CLL component of two cases, whereas in a follicular lymphoma the deletion was present in both the follicular tumor and in the diffuse large-cell lymphoma. In conclusion, these findings indicate that p16INK4a gene alterations are a relatively infrequent phenomenon in NHLs. However, deletions, mutations, and hypermethylation of the gene with loss of protein expression are associated with aggressive tumors and they may also participate in the histological progression of indolent lymphomas.
LYMPHOID MALIGNANCIES are a heterogeneous group of disease entities characterized by distinctive clinical, morphological, immunophenotypical, and genetic features.1These neoplasms can be generally divided into indolent and aggressive tumors on the basis of the clinical presentation, histology, clinical course, and response to therapy. The majority of aggressive non-Hodgkin's lymphomas (NHLs) are primary tumors recognized at diagnosis. In addition, indolent lymphomas are characterized by a relatively frequent transformation to more aggressive variants. The frequency of this transformation varies in different entities and, thus, it may occur in 1% to 10% of chronic lymphocytic leukemias (CLLs) small lymphocytic lymphomas2 but in 25% to 70% of low grade follicular lymphomas.3 Aggressive variants of mantle cell lymphomas (MCLs) are generally diagnosed at presentation. However, progression of typical MCLs into more aggressive variants can also occur in 24% to 39% of cases.4 5 Morphological transformation of indolent lymphomas is associated with a rapidly progressive clinical course and short survival of the patients.
Cytogenetic and molecular studies of NHLs have identified a series of gene alterations usually associated with specific disease entities. However, the molecular mechanisms responsible for the pathogenesis of most primary high-grade tumors and the progression of indolent lymphomas are not well known. p53 inactivation and c-myc rearrangements have been implicated in the transformation of a number of indolent lymphomas. Particularly, p53 mutations are relatively rare in low-grade tumors, but they are found in 20% to 50% of high-grade B-cell lymphomas.6 In addition, p53 mutations have also been associated with progression in 25% to 40% of indolent lymphomas including transformed CLLs,7 transformed follicular lymphomas (FCLs),8,9 aggressive variants of MCLs,4,10 and progressed mucosa-associated lymphoid tissue lymphomas.11 These findings indicate that p53 inactivation is an important pathway in the pathogenesis of a subset of primary high-grade and transformed lymphomas. However, they also suggest that other molecular mechanisms must be implicated in the development of these aggressive variants of tumors.
Cyclin-dependent kinase inhibitors (CDKIs) represent a class of negative regulatory elements of cell growth that suppress the kinase activity of the cyclin/CDK complexes. Among all these molecules, p16INK4a has been implied as a tumor-suppressor gene. Inactivation of this gene by homozygous deletions, mutations, and hypermethylation occurs in a wide array of human tumors.12-15 In hematologic disorders, homozygous deletions are frequently found in acute lymphoblastic leukemias (ALL), mainly of T-cell origin. In contrast, p16INK4a alterations in NHLs seem to be rare, and its possible implication in the pathogenesis and progression of these neoplasms is not well known. Some studies have detected p16INK4a deletions in primary large-cell lymphomas and sporadic transformed tumors.16-18 However, a clear association between p16INK4a alterations and aggressive variants of NHLs has not been observed in other series.19 20 In addition, the role of these alterations in the progression of indolent lymphomas has not been specifically addressed in previous studies.
To determine the possible implication of p16INK4a gene alterations in the development and progression of aggressive variants of NHLs, we have analyzed its gene structure and protein expression in a large series of NHLs, including a number of histologically transformed cases. Our results indicated that p16INK4a gene alterations are rare in low-grade lymphomas, but the inactivation of the gene by homozygous deletions, point mutations, or hypermethylation with loss of protein expression mainly occurred in primary aggressive and transformed lymphomas.
MATERIALS AND METHODS
Case selection.
Tumor specimens from 112 NHLs were selected based on the availability of frozen tissue for molecular analysis and classified according to the Revised European-American Classification of Lymphoid Neoplasms.1 The tumors were grouped into indolent and aggressive categories. Among indolent NHLs, we studied 24 CLLs, one hairy cell leukemia (HCL), 18 FCLs, and 21 typical MCLs. Aggressive NHLs comprised 7 large-cell lymphomas evolved from CLLs (Richter's syndrome), 9 diffuse large-cell lymphomas (LCLs) transformed from FCLs, 7 blastoid variants of MCLs, 18 de novo B-cell diffuse LCLs, 4 lymphoblastic lymphomas (LBLs), 3 of B-cell and 1 of T-cell phenotype, 1 Burkitt's lymphoma (BL), and 2 anaplastic large cell lymphomas (ALCLs). Data from 24 MCLs (18 typical and 6 blastoid variants) have been reported elsewhere.21 Frozen material from sequential samples were available in 2 CLLs and 1 FCL and their subsequent transformed large-cell lymphoma.
Southern blot analysis.
Genomic DNA was extracted from frozen material in 105 cases (24 CLLs, 7 LCLs transformed from CLLs, 1 HCL, 16 FCLs, 9 LCLs evolved from FCLs, 21 MCLs, 7 blastoid MCLs, 14 de novo LCLs, 1 BL, 3 LBLs, and 2 ALCLs) using proteinase K/RNAse treatment and phenol-chloroform extraction. Southern blot analysis could be performed in 95 of these cases (Table1). DNA from each case (15 μg) was digested with EcoRI, HindIII, and BamHI and analyzed as previously described.21,22 The p16INK4a probe used was a 0.8-kb EcoRI-XhoI fragment of the p16INK4a cDNA clone.23 The β-actin probe was also used as a loading control. The probes were radiolabeled using a random primer DNA labeling kit (Amersham Life Science, Buckinghamshire, UK) with [α-32P]dCTP. The intensity of the autoradiographic signals were quantified using a UVP-5000 video densitometer (UVP, San Gabriel, CA).
p16INK4a Gene Alterations and Protein Expression in a Series of 112 NHLs
| Diagnosis (Total Cases) . | Deletions . | Mutational Analysis . | Loss of Expression . | Total* . |
|---|---|---|---|---|
| Chronic lymphocytic leukemia | ||||
| Typical (24) | 0/18 | 1/24 | 0/21 | 1/24 (4%) |
| Richter's syndrome (7) | 2/7 | 0/5 | 1/6 | 2/7 (29%) |
| Hairy cell leukemia (1) | 0/1 | 0/1 | 0/1 | 0/1 |
| Follicular lymphoma | ||||
| Typical (18) | 1/15 | 0/16 | 1/18 | 1/18 (6%) |
| Transformed (9) | 1/9 | 2/8 | 3/8 | 3/9 (33%) |
| Mantle cell lymphoma | ||||
| Typical (21) | 1/21 | 0/20 | 1/19 | 1/21 (5%) |
| Blastoid (7) | 3/7 | 0/4 | 2/6 | 3/7 (43%) |
| Large cell lymphoma (18) | 1/11 | 0/14 | 5/18-151 | 5/18 (28%) |
| Burkitt's lymphoma (1) | 0/1 | 0/1 | 0/1 | 0/1 |
| Lymphoblastic lymphoma (4) | 2/3 | 0/1 | 2/4 | 2/4 (50%) |
| ALCL (2) | 0/2 | 0/2 | 1/2-152 | 1/2 (50%) |
| Total | 11/95 (12%) | 3/96 (3%) | 16/104 (15%) | 19/112 (17%) |
| Diagnosis (Total Cases) . | Deletions . | Mutational Analysis . | Loss of Expression . | Total* . |
|---|---|---|---|---|
| Chronic lymphocytic leukemia | ||||
| Typical (24) | 0/18 | 1/24 | 0/21 | 1/24 (4%) |
| Richter's syndrome (7) | 2/7 | 0/5 | 1/6 | 2/7 (29%) |
| Hairy cell leukemia (1) | 0/1 | 0/1 | 0/1 | 0/1 |
| Follicular lymphoma | ||||
| Typical (18) | 1/15 | 0/16 | 1/18 | 1/18 (6%) |
| Transformed (9) | 1/9 | 2/8 | 3/8 | 3/9 (33%) |
| Mantle cell lymphoma | ||||
| Typical (21) | 1/21 | 0/20 | 1/19 | 1/21 (5%) |
| Blastoid (7) | 3/7 | 0/4 | 2/6 | 3/7 (43%) |
| Large cell lymphoma (18) | 1/11 | 0/14 | 5/18-151 | 5/18 (28%) |
| Burkitt's lymphoma (1) | 0/1 | 0/1 | 0/1 | 0/1 |
| Lymphoblastic lymphoma (4) | 2/3 | 0/1 | 2/4 | 2/4 (50%) |
| ALCL (2) | 0/2 | 0/2 | 1/2-152 | 1/2 (50%) |
| Total | 11/95 (12%) | 3/96 (3%) | 16/104 (15%) | 19/112 (17%) |
*Number of cases with p16 alterations (p16 gene deletions, mutations and/or loss of protein expression)/Number of cases tested.
Four cases with loss of protein expression and germ line configuration of the gene showed hypermethylation of exon 1 (see Results).
This case showed hypermethylation of the gene (see Results).
Single-stranded conformation polymorphism (SSCP) analysis and DNA sequencing.
SSCP analysis was used to screen for p16INK4a gene mutations according to a previously described method.21,24Exons 1 and 2 of the p16INK4a gene were amplified by polymerase chain reaction (PCR) by using a simple set of flanking intronic primers. Primers for exon 1 were 5′-GAAGAAAGAGGAGGGGCTG-3′ and 5′-GCGCTACCTGATTCCAATTC-3′, and primers for exon 2 were 5′-CTCTACACAAGCTTCCTTTCC-3′ and 5′-GGGCTGAACTTTCTGTGCTGG-3′. We used a “touch-down” PCR strategy for the amplification of both exons as previously described.21 For the SSCP analysis, the PCR products of both exons were digested with Sma I, diluted in formamide-dye loading buffer, and electrophoresed on a 15% nondenaturing polyacrylamide gel with or without 10% glycerol at 150 V for 14 hours at room temperature. The gels were developed using a silver staining procedure as previously described.21
Samples with an altered mobility were sequenced using a commercial cycle sequencing kit (Perkin Elmer, Branchburg, NY) and α-33P dATP as previously described.21 A total of 0.5 μL of the p16INK4a gene PCR products were used as template for sequencing. The primers described previously and two internal primers for exon 2, 5′-ACTCTCACCCGACCCGTGCA-3′ and 5′-AGCTCCTCAGCCAGGTCCA-3′ were used for the sequencing reaction at a final concentration of 0.5 μmol/L. The reaction was performed according to the instructions supplied by the manufacturer. The presence of a mutation was confirmed by sequencing the other DNA strand.
Western blot analysis.
Protein extraction was obtained from additional frozen tissue available in 104 cases (Table 1). Protein extracts from the HeLa cell line were used as positive control. In each case, 10 frozen sections of 30 μm were incubated in 300 μL of ice-cold lysis buffer (50 mmol/L Tris-Cl, pH 8, 150 mmol/L NaCl, 0.4 mmol/L EDTA, 10 mmol/L NaF, 0.02% sodium azide, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, and 0.5% sodium deoxycholate) containing 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 1 μg/mL α-1-antitrypsin for 20 minutes at 4°C. The cell debris was sedimented by centrifugation at 14,000 revolutions per minute at 4°C for 25 minutes. The clarified supernatants were collected, and the protein content of the lysate was determined by the Lowry protein assay (Bio-Rad, Hercules, CA). Fifty micrograms of total cellular protein were run per lane on a 15% SDS-polyacrylamide gel and electroblotted to a nitrocellulose membrane (Amersham). The blocked membrane was incubated with the monoclonal antibody anti-p16INK4a, clone G175-405 (Pharmingen, San Diego, CA) at a final concentration of 1 μg/mL for 1 hour and 30 minutes, washed with phosphate-buffered saline 0.1% Tween-20, and exposed to sheep antimouse conjugated to horseradish peroxidase (Amersham) at a 1:1000 dilution for 1 hour and 30 minutes. After washing, antibody binding was detected by chemiluminescence detection procedures according to the manufacturer's recommendations (ECL; Amersham).
Methylation analysis.
A PCR assay was performed to analyze the methylation status of the first exon of the p16INK4a gene. Thirty units of methylation-sensitive restriction enzymes Sac II (Bio-Labs, Beverly, MA) and Sma I (Promega, Madison, WI) were used to digest 0.2 μg of DNA. Digested DNA was then used as a template in a multiplex PCR reaction for exon 1 of p16INK4aand β-globin. β-globin was amplified as internal control of the PCR reaction because the amplified fragment of this gene does not containSma I or Sac II sites. The primers used for the methylation analysis of p16INK4a exon 1 were described previously. Primers for the β-globin gene were 5′-ACACAACTGTGTTCACTAGC-3′ and 5′-CAACTTCATCCACGTTCACC-3′. We used a touch-down PCR strategy for the amplification. Conditions were one cycle at 95°C for 5 minutes; four cycles at 94°C for 45 seconds, at 68°C for 1 minute, and at 72°C for 1 minute; four cycles with annealing temperature at 67°C; 35 cycles with annealing temperature at 66°C; and a final step for 5 minutes at 72°C. PCR products were resolved on 2% agarose gel. Only cases with methylated DNA were expected to show amplified product of p16INK4a exon 1, whereas no amplification of this exon was obtained in nonmethylated cases. HeLa and Raji cell lines were used as negative and positive controls of methylation, respectively.
RESULTS
Analysis of p16INK4a gene deletions.
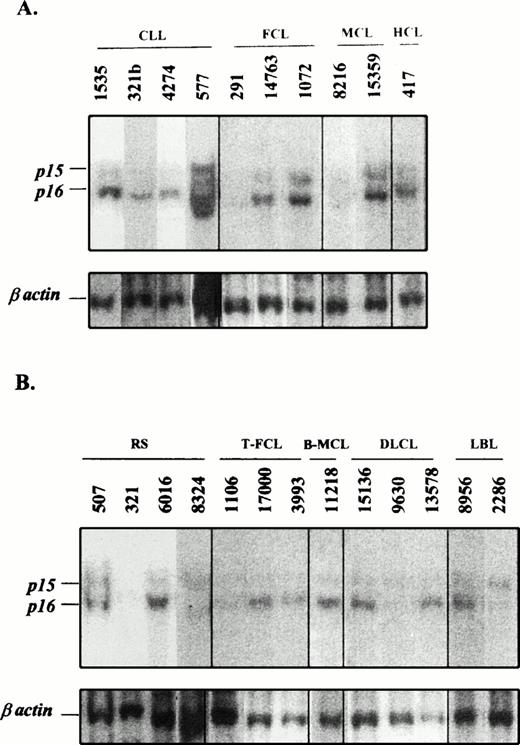
The p16INK4a gene was examined by Southern blot in 95 cases including 55 indolent and 40 aggressive lymphomas in which genomic DNA was available (Table 1 and Fig 1).p16INK4a homozygous deletions were detected in 10 tumors. In one additional blastoid MCL, in which no DNA was available, the cytogenetic analysis identified a hemizygous deletion affecting the 9p21 locus.21 Therefore, p16INK4a deletions were present in 9 (22.5%) aggressive lymphomas but only in 2 (3.6%) indolent tumors (Table 1). The two indolent lymphomas with p16INK4a deletions were a grade II FCL (case 291) and a typical MCL (case 8216; Fig 1A). In both cases, the p16INK4a deletion was considered to be homozygous and it was also associated with a homozygous deletion of the p15INK4b gene. Interestingly, these two cases showed a very aggressive clinical behavior. In fact, the FCL underwent a progression to a diffuse LCL in 12 months and the patient died 14 months after transformation. This transformed phase also showed a homozygous deletion of the p16INK4a gene (Fig 1B, case 1106). The MCL showed a relatively high proliferative index (2.6 mitosis × high power field) and the patient died 19 months after the initial diagnosis with no response to the therapy. The survival of these two patients was much shorter than the median survival of other patients with FCLs and typical MCLs in our institution, which were 102 and 48 months, respectively.
Southern blot analysis of 10 indolent (A) and 13 aggressive (B) NHLs. The indolent lymphoid neoplasms 321b (CLL), 577 (CLL), and 291 (FCL) in A have their subsequent sample of the progressed lymphoma in B, cases 321 (Richter's syndrome), 8324 (Richter's syndrome), and 1106 (transformed FCL), respectively. Genomic DNA was digested with HindIII restriction enzyme and hybridized with the exon 2 of p16INK4a and β-actin probes. (A) Indolent lymphomas showed germline configuration except cases 291 (FCL) and 8216 (typical MCL), which had a homozygous deletion of the p16INK4a gene. (B) Analyses of transformed and aggressive variants of NHLs showed homozygous deletions of both p15INK4b and p16INK4a genes in cases 321 and 8324 (Richter's syndrome), 1106 (transformed FCL), and 9630 (diffuse LCL). Case 2286 (lymphoblastic lymphoma) showed a homozygous deletion of p16INK4a but no p15INK4b. The identification number of these cases is the same as that in Figs 2, 3, and 4.
Southern blot analysis of 10 indolent (A) and 13 aggressive (B) NHLs. The indolent lymphoid neoplasms 321b (CLL), 577 (CLL), and 291 (FCL) in A have their subsequent sample of the progressed lymphoma in B, cases 321 (Richter's syndrome), 8324 (Richter's syndrome), and 1106 (transformed FCL), respectively. Genomic DNA was digested with HindIII restriction enzyme and hybridized with the exon 2 of p16INK4a and β-actin probes. (A) Indolent lymphomas showed germline configuration except cases 291 (FCL) and 8216 (typical MCL), which had a homozygous deletion of the p16INK4a gene. (B) Analyses of transformed and aggressive variants of NHLs showed homozygous deletions of both p15INK4b and p16INK4a genes in cases 321 and 8324 (Richter's syndrome), 1106 (transformed FCL), and 9630 (diffuse LCL). Case 2286 (lymphoblastic lymphoma) showed a homozygous deletion of p16INK4a but no p15INK4b. The identification number of these cases is the same as that in Figs 2, 3, and 4.
The nine aggressive lymphomas with p16INK4a deletions were two (29%) LCLs transformed from CLLs (Richter's syndrome), one (11%) LCL transformed from FCL, three (43%) blastoid MCLs, one (9%) de novo diffuse B-cell LCL, and two (67%) LBLs, one of B- and one of T-cell phenotype (Table 1). All p16INK4a deletions were associated with p15INK4b deletions except in one LBL in which p15INK4b was in germline configuration. No isolated p15INK4b deletions were detected in any case. DNA from sequential samples of the indolent tumor and the subsequent transformed LCL were analyzed in the two CLLs and the FCL with p16INK4ahomozygous deletions. In the two CLLs, the p16INK4ahomozygous deletions were detected in the LCL but not in the CLL component, whereas in the FCL (case 291 previously commented) it was present in both the follicular tumor and in the diffuse LCL (Fig 1).
Mutational analysis of p16INK4a gene.
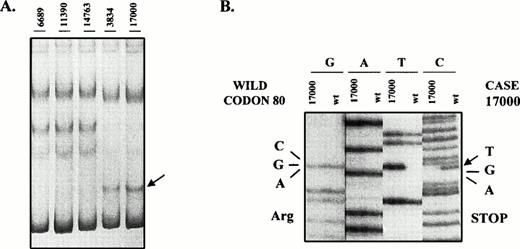
To determine whether mutations of the p16INK4a gene were present in these lymphomas, we analyzed exon 1 and 2 of the p16INK4a gene by PCR-SSCP. Cases with anomalous migrating bands were sequenced (Table 1). Mutations were only found in one indolent lymphoma (1.6%) and 2 aggressive tumors (5.5%). The mutated indolent lymphoma was a typical CLL. This case showed a mutation at codon 143 (GCC → ACC), resulting in a change of alanine by threonine. This case showed a p16INK4a germline by Southern blot analysis and normal residual bands in the SSCP and sequencing analysis suggesting that this mutation was hemizygous. No differences in the clinical course were observed in this case when compared with the nonmutated CLLs. The other two mutated cases were two diffuse LCLs progressed from FCLs. The two cases showed the same nonsense mutation at codon 80 with the change CGA (Arginine) → TGA (Stop) (Fig2). Codon 80 is considered a mutational hot spot within p16INK4a and it is frequently mutated in cell lines and neoplasms.12 25 No signal of the wild allele was observed in the SSCP and sequencing analysis and no protein expression was detected in any of these two cases by Western blot (see later). Five additional tumors, two FCLs, one typical MCL, and two LCLs showed an abnormal migrating band in exon 2. This altered mobility was the result of the known polymorphism at codon 148 with the change GCG (Alanine) to ACG (Threonine).
SSCP analysis of p16INK4a exon 2. The abnormal mobility observed in two transformed FCLs (case 17000 and 3834) (A) is the result of the mutation in codon 80 with the change CGA (Arg) → TGA (Stop) (B). The western blot analysis of these two cases is shown in Fig 3.
SSCP analysis of p16INK4a exon 2. The abnormal mobility observed in two transformed FCLs (case 17000 and 3834) (A) is the result of the mutation in codon 80 with the change CGA (Arg) → TGA (Stop) (B). The western blot analysis of these two cases is shown in Fig 3.
P16INK4a gene expression.
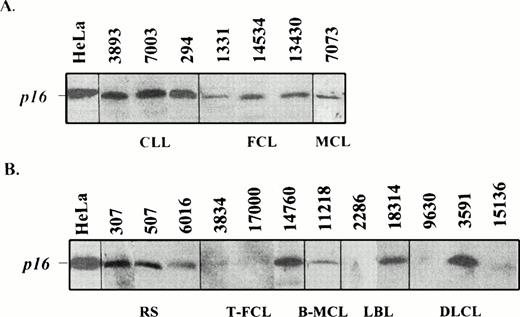
To assess the possible alterations of p16INK4a gene expression in these lymphomas, we examined the protein levels by Western blot analysis in 104 cases. Some MCLs had also been studied previously by Northern blot.21 Complete loss or very weak p16 protein expression was observed only in 2 of the 59 (3.4%) indolent lymphomas but in 14 of the 45 (32%) aggressive lymphomas (Table 1 and Fig 3). Protein expression was observed in all other tumors. The two indolent lymphomas with no protein expression were the FCL (case 291) and the typical MCL (case 8216) with homozygous deletion of the gene. The CLL with a mutated allele showed protein expression at similar levels than other nonmutated CLLs. Nine of the 14 aggressive lymphomas with loss of protein expression had shown genetic abnormalities in the Southern blot or mutational analysis including biallelic p16INK4a gene deletions in 7 cases and 2 homozygous nonsense mutations at codon 80 in two transformed FCLs (Table 1). Interestingly, no gene deletions or mutations were found in the 5 additional aggressive lymphomas, 4 de novo LCLs and one ALCL, with lack of protein expression, suggesting that other genetic alterations could be implicated in the inactivation of the gene.
Western blot analysis of p16INK4a in indolent (A) and aggressive (B) lymphomas. Two transformed FCLs (3834 and 1700), one LBL (2286), and two diffuse large B-cell lymphomas (9630 and 15136) show a loss of protein expression. The Southern blot analysis of cases 17000 (T-FCL), 2286 (LBL), 9630 (DLCL), and 15136 (DLCL) is shown in Fig 1. Tumors 2286 and 9630 showed a homozygous deletion of p16INK4a gene (Fig 1), whereas tumors 17000 and 15136 with germline configuration in the Southern blot analysis had a stop codon (Fig 2) and hypermethylation of the gene (Fig 4), respectively.
Western blot analysis of p16INK4a in indolent (A) and aggressive (B) lymphomas. Two transformed FCLs (3834 and 1700), one LBL (2286), and two diffuse large B-cell lymphomas (9630 and 15136) show a loss of protein expression. The Southern blot analysis of cases 17000 (T-FCL), 2286 (LBL), 9630 (DLCL), and 15136 (DLCL) is shown in Fig 1. Tumors 2286 and 9630 showed a homozygous deletion of p16INK4a gene (Fig 1), whereas tumors 17000 and 15136 with germline configuration in the Southern blot analysis had a stop codon (Fig 2) and hypermethylation of the gene (Fig 4), respectively.
Methylation analysis.
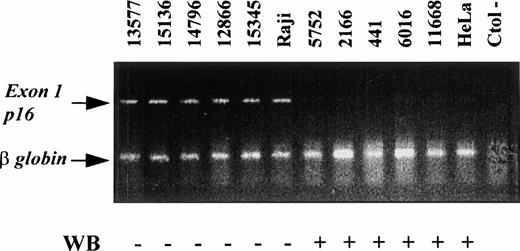
To know if the lack of protein expression in these later five aggressive lymphomas with no evidence of deletions or mutations could be caused by hypermethylation of the gene, DNA from these cases was digested with the methylation sensitive restriction enzymes SacII and Sma I and amplified with a multiplex PCR including primers for p16INK4a exon 1 and β-globin as an internal control. All these cases showed the expected 340-bp band of exon 1 of p16INK4a and the β-globin band at 110 bp, indicating that all these cases were methylated at exon 1 (Fig4). To confirm these results, we also studied the methylation status in five more cases (one FCL, two MCLs, one blastoid-MCL, and one LCL progressed from a CLL) with p16 protein expression. No hypermethylation of the gene could be shown in these cases.
Methylation status of the p16INK4a exon 1 in NHL analyzed by PCR and comparison with the results of the protein expression by Western blot analysis (WB). Products of p16INK4a exon 1 and the internal control β-globin are indicated. The amplification of exon 1 after digestion with SmaI indicates that this restriction site was methylated. Cases with p16INK4a exon 1 amplification showed loss of protein expression by Western blot whereas protein expression was detected in cases in which p16INK4a exon 1 was not amplified.
Methylation status of the p16INK4a exon 1 in NHL analyzed by PCR and comparison with the results of the protein expression by Western blot analysis (WB). Products of p16INK4a exon 1 and the internal control β-globin are indicated. The amplification of exon 1 after digestion with SmaI indicates that this restriction site was methylated. Cases with p16INK4a exon 1 amplification showed loss of protein expression by Western blot whereas protein expression was detected in cases in which p16INK4a exon 1 was not amplified.
DISCUSSION
The genetic basis underlying histological progression in NHLs is not well known. p53 mutations are a common alteration in aggressive tumors6 and they are also associated with morphological progression in different types of indolent lymphomas.4,7-11In addition, c-myc rearrangements have been implicated in the transformation of occasional FCLs and CLLs.26,27 However, alterations of these genes occur in 10% to 50% of the tumors indicating that other mechanisms must be also involved in this process. The strong inhibitory action of p16INK4a gene on cell cycle progression,28 its frequent inactivation in advanced stages of different solid tumors,29-31 and the spontaneous development of B-cell lymphomas with an aggressive morphology in the INK4a knock-out mice32 suggest that this gene may also be a target in the pathogenesis of aggressive and transformed lymphomas.
In hematologic malignancies, p16INK4a gene alterations have been mainly described in lymphoid rather than myeloid neoplasms.24,33-37 However, in contrast to the high number of deletions found in acute lymphoblastic leukemias, the incidence of p16INK4a homozygous deletions and mutations observed in NHLs has been low, ranging from 0% to 15% of the cases. In most of these studies,18,24,38-40 the NHLs examined were part of a larger series of hematologic neoplasms with no further information on the histological type of the lymphomas or the clinical evolution of the patients. Some other studies have found p16INK4a deletions in occasional diffuse lymphomas.16,17 However, the association between p16INK4a alterations and aggressive variants of the tumors was not clear in other series in which the incidence of p16INK4a alterations were similar in low- and high-grade lymphomas.19,20 In this study, we have detected p16INK4a gene alterations in 33% of primary high-grade and 33% of transformed lymphomas, but only in three (5%) histologically indolent tumors indicating that inactivation of this gene may participate in the development of primary aggressive and progressed lymphomas. In addition, two of the indolent cases with p16INK4a alterations were a typical MCL and a grade II FCL that followed an aggressive clinical evolution. In a previous study on MCLs, we found p16INK4a deletions and loss of gene expression only in blastoid variants of this lymphoma.21 We have now expanded the study and found p16INK4a homozygous deletions with loss of protein expression in one case that was morphologically a typical MCL. However, this patient had no response to therapy and died in 19 months, an overall survival similar to that observed in the blastoid variants of MCL. The FCL also had a homozygous deletion of the p16INK4a gene with loss of protein expression and followed an aggressive evolution with transformation to an LCL. The third low-grade lymphoma with p16INK4aalteration was a typical CLL with a hemizygous missense mutation, protein expression, and a clinical behavior similar to other typical CLLs. These findings suggested that complete abrogation of p16 expression in morphologically indolent lymphomas may confer to the tumor a proliferative growth advantage similar to that observed in aggressive lymphomas.
Whether p16INK4a gene alterations are involved in histological progression of indolent lymphomas is not well known. Biallelic loss of the p16INK4a gene has been reported in isolated cases of lymphoblastic transformation of chronic myeloid leukemias,41 chronic and acute phases of adult T-cell leukemias,42 and sporadic peripheral lymphomas.17 18 However, very few cases of transformed lymphomas have been included in previous series. In this study, we have observed that p16INK4a gene inactivation, either by biallelic deletions or, less frequently, point mutations, is a relatively common phenomenon in progressed NHLs. Specifically, we found inactivation of the p16INK4a gene in 33% of transformed CLLs, FCLs, and blastic MCLs but only in 5% of the indolent counterparts. The incidence of p16INK4a alterations in transformed lymphomas in our study is similar to the number of p53 alterations detected previously in other series of progressed lymphomas and suggests that inactivation of this gene may also define a molecular pathway in lymphoma progression.
In three of our progressed cases, two CLLs and one FCL, we were able to study the indolent component and the subsequent transformed LCL. In the two CLLs, a p16INK4a homozygous deletion was detected in the LCL but not in the CLL phase, whereas in the FCL the homozygous deletion was present in both components, the follicular and the subsequent diffuse LCL. Although p53 mutations have been frequently described in Richter's syndrome,7 they may be related to the development of a new malignant clone rather than progression of indolent CLLs.43 In our two CLLs, the immunoglobulin heavy chain gene showed the same rearrangement pattern in the indolent and aggressive component suggesting a clonal evolution. The finding of the p16INK4a biallelic deletion in the progressed but not in the low-grade phase in these two cases suggested that this alteration was acquired during the transformation process and may have played a role in its pathogenesis. In the FCL the homozygous deletion was detected in both the indolent tumor and in the progressed LCL. The clinical evolution in this case was more aggressive than in conventional FCLs suggesting that this molecular alteration may have had an influence in this evolution. Similarly to this case, c-myc rearrangements and p53 overexpression have also been found in the indolent component of FCLs that underwent morphological transformation to LCLs8,9 26 indicating that these alterations may occur early in the development of these lymphomas and may participate in their progression.
The majority of inactivating alterations of the p16INK4agene in human tumors are homozygous deletions rather than point mutations.12,25 Concordantly, p16INK4a gene deletions, generally associated with deletions of the p15INK4b gene, were the most frequent alteration detected in our study. p16INK4a point mutations are rare in lymphoid neoplasms, ranging from 0% to 7% of the cases.20,44,45Similarly, we only detected three point mutations (3%) in our series, a missense mutation at codon 143 in a CLL and two nonsense mutations at codon 80 in two transformed FCLs. Nonsense mutations or microdeletions and insertions leading to subsequent stop codons have been frequently described in hematologic neoplasms.20,44-46 In solid tumors, nonsense mutations of the p16INK4a gene are also relatively frequent (30%) compared with the number of these mutations in the p53 gene (8%).25 It has been postulated that this difference may reflect different mechanisms of inactivation of these proteins. Missense mutations in the p53 gene frequently disrupt its function, whereas it is possible that p16INK4a may tolerate some missense mutations without impairing the normal function of the protein.25,47,48 In this respect, the only missense mutation in our study was seen in an indolent CLL, whereas the two nonsense mutations were present in two transformed FCLs. This missense mutation (codon 143) was outside the ankyrin repeat motifs of the gene and located in a region that is not required for the protein to bind and inhibit CDK4,47,48 suggesting that this mutation was most probably not functionally significant. Interestingly, 5 out of 6 mutations previously described in indolent lymphomas (CLLs and an FCL) were missense mutations,20,44 whereas 7 out of 10 mutations in T-ALL or high-grade lymphomas were nonsense mutations or microdeletions and insertions with subsequent stop codons.20,36,45 46
The p16INK4a gene expression in lymphoproliferative disorders has been less well examined than the structure of the gene.21,49 In this study we detected loss of p16 protein expression in 16 of the 104 (15%) lymphomas examined. Loss of protein expression in our tumors was clearly associated with disruptive alterations of the gene in 11 cases, 9 with homozygous deletions and 2 with nonsense mutations. However, 5 additional aggressive lymphomas showed loss of protein expression without clear anomalies of the gene in the Southern or SSCP analyses. These 5 tumors showed hypermethylation of exon 1, suggesting that this mechanism was leading to the inactivation of the gene in these tumors. Hypermethylation of 5′ CpG islands of the p16INK4a gene has been recently described as an alternative inactivating mechanism of this gene in different human tumors including hematologic malignancies.14,19,34,50 In this respect, Herman et al34 have recently shown hypermethylation of the p16INK4a gene in 5 of 6 high-grade lymphomas but only in 1 of 6 low-grade tumors. These findings suggest that p16INK4ahypermethylation may also be a mechanism involved in the development of aggressive lymphomas. Hypermethylation of the p15INK4b gene has also been described in acute lymphoblastic leukemias and rarely in NHLs.34 49 However, the number of mature lymphomas included in these studies is very scarce. It will be interesting to analyze whether mutations and/or hypermethylation of this gene may also be involved in the progression of indolent lymphomas.
In conclusion, our findings indicate that the p16INK4a gene may be inactivated by homozygous deletions, point mutations, and hypermethylation in NHLs. This inactivation is a relatively infrequent phenomenon in low-grade tumors, but it is associated with aggressive variants and may also define an alternative molecular pathway in the histological transformation of indolent lymphomas.
ACKNOWLEDGMENT
The authors thank Dr Manuel Serrano for the gift of the p16INK4a probe and his comments on the manuscript and Dr Miguel A. Piris for the helpful discussions on the project.
M.P. and F.C. contributed equally to this study.
Supported by Grants SAF 96/61 from CICYT, 96SGR56 from CIRIT, and Maraton-TV3 Cancer. M.P., F.C., S.B., and P.J. were fellows supported by Maraton-TV3 Cancer (M.P.), Hospital Clinic (F.C.), and Spanish Ministerio de Educacion y Cultura (S.B., P.J.).
Address reprint requests to Elias Campo, Laboratory of Pathology, Hospital Clinic, Villarroel 170, 08036- Barcelona, Spain.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal