p15INK4b gene is an inhibitor of cyclin-dependent kinase (CDK) 4 and CDK6 whose expression is induced by transforming growth factor (TGF)β. Recent reports suggest frequent methylation of the p15INK4b gene promoter in leukemias, and it has been proposed that this methylation could be necessary for leukemic cells to escape TGFβ regulation. We investigated the methylation status of p15INK4b gene in 53 myelodysplastic syndromes (MDS) cases, including nine that had progressed to acute myeloid leukemia (AML), using a recently described sensitive method where polymerase chain reaction (PCR) is preceded by bisulfite modification of DNA (methylation specific PCR). p15INK4b methylation was observed in 20 of 53 (38%) of the cases. Twenty of the 24 patients with greater than 10% bone marrow blasts had p15INK4bmethylation (including all nine patients who had progressed to AML) as compared with none of MDS patients with <10% bone marrow blasts. No correlation between karyotypic abnormalities and methylation status was found. Patients with p15INK4b methylation had a worse prognosis, but the prognostic significance of p15INK4bmethylation was no more found by multivariate analysis, due to its strong correlation to the percentage of marrow blasts. In 10 MDS cases, sequential DNA samples were available. In five of them, methylation of the p15INK4b gene was detected at leukemic transformation, but not at diagnosis. Our results showed that methylation of the p15INK4b gene in MDS is correlated with blastic bone marrow involvement and increases with disease evolution toward AML. It suggests that proliferation of leukemic cells might require an escape of regulation of the G1 phase of the cell cycle, and possibly of TGFβ inhibitory effect.
MYELODYSPLASTIC syndromes (MDS) are clonal stem cell disorders characterized by ineffective hematopoiesis leading to blood cytopenias and by a high incidence of progression to acute myeloid leukemia (AML). MDS are heterogeneous and include refractory anemia (RA) and refractory anemia with ring sideroblasts (RARS), with low incidence of progression toward AML, refractory anemia with excess blasts (RAEB), and refractory anemia with excess blasts in transformation (RAEB-t), which often progress to AML, and chronic myelomonocytic leukemia (CMML) with an intermediate risk of progression.1 MDS, therefore, represents an excellent model of leukemic development with a progressive increase of blastic bone marrow involvement, but genetic events that lead to this evolution are still not identified. Chromosomal abnormalities have been described in MDS and they are generally associated with resistance to chemotherapy and short survival, but few molecular abnormalities, such as C-fms, N-ras, and p53 mutations, have been identified in MDS.2-4
p15INK4b and p16INK4a proteins are cell cycle regulators involved in the inhibition of G1 phase progression.5 p15INK4b associates with cyclin-dependent kinase (CDK) 4 and 6, and cyclin D-CDK4/6 complexes and inhibits their kinase activities.6,7 Transforming growth factor β (TGFβ) inhibitory effects on normal hematopoietic progenitors are mediated by many different mechanisms including downregulation of CDK4 synthesis and increase in p15INK4bgene expression and p15INK4b protein stability.8-10 It has been previously reported that loss of sensitivity of leukemic cell lines to growth inhibition by TGFβ is correlated with inactivation of p15INK4b. Recent studies have shown that p15INK4b gene is inactivated by 5′ CpG island methylation of the promoter region in most AML and acute lymphoblastic leukemia (ALL).11-13
To investigate the role of p15INK4b gene methylation in the progression of MDS, we analyzed the methylation status of p15INK4b gene in 53 MDS cases, using the recently described methylation specific PCR (MSP) method.14 This method, which is more sensitive than Southern Blot analysis, seemed particularly useful in MDS, where the blastic bone marrow involvement is variable and sometimes very low.
MATERIALS AND METHODS
Patients.
We investigated the methylation status of the p15INK4B gene in 53 de novo MDS cases, including 10 RA, two RARS, five RAEB-t, 18 RAEB, nine CMML, and nine AML evolved from MDS (MDS-AML). Their characterization is summarized in Table 1. In 10 MDS (two RAEB-t, seven RAEB, and one CMML with 26%, 23%, 15%, 12%, 12%, 11%, 7%, 9%, 8%, and 4% marrow blasts on first analysis, respectively), sequential DNA samples were available. Twenty-five cases of de novo AML were also studied. Genomic DNA was extracted from mononuclear cells after bone marrow aspirates and Ficoll-Hypaque sedimentation. T cells from a RAEB case with 17% bone marrow blasts were isolated using magnetic Dynabeads-CD2 (Dynal, Oslo, Norway). Controls included normal DNA obtained from healthy volunteers after informed consent, DNA from K562 cell line (American Type Culture Collection [ATCC], Rockville, MD, no. CCL243), which is homozygously deleted for the p15INK4b gene, and positive control DNA extracted from Raji cell line (ATCC, no. CCL86), previously reported to have p15INK4B gene methylation.11
Patient Characteristics and Methylation Status of MDS
| FAB Type . | No. of Patients . | Median Age . | Gender . | Median BM Blast % (range) . | Abnormal Karyotype . | No. of Methylated Cases (%) . |
|---|---|---|---|---|---|---|
| RA | 10 | 61 (33-72) | 3 M, 7 F | 2 (1-3) | 4/8 (2 ND) | 0 (0) |
| RARS | 2 | 55 (51-60) | 0 M, 2 F | 1 | 0/2 | 0 (0) |
| RAEB | 18 | 63 (16-80) | 10 M, 8 F | 11 (6-19) | 6/15 (3 ND) | 7 (39) |
| RAEB-t | 5 | 61 (38-70) | 3 M, 2 F | 25 (21-28) | 3/4 (1 ND) | 4 (80) |
| CMML | 9 | 73 (51-87) | 6 M, 3 F | 4 (1.5-4) | 0/7 (2 ND) | 0 (0) |
| AML evolved from MDS | 9 | 60.5 (60-73) | 4 M, 5 F | 64 (30-85) | 2/8 (1 ND) | 9 (100) |
| Total | 53 | 61.5 (16-87) | 26 M, 27 F | 11 (1-85) | 15/44 (9 ND) | 20 (38) |
| FAB Type . | No. of Patients . | Median Age . | Gender . | Median BM Blast % (range) . | Abnormal Karyotype . | No. of Methylated Cases (%) . |
|---|---|---|---|---|---|---|
| RA | 10 | 61 (33-72) | 3 M, 7 F | 2 (1-3) | 4/8 (2 ND) | 0 (0) |
| RARS | 2 | 55 (51-60) | 0 M, 2 F | 1 | 0/2 | 0 (0) |
| RAEB | 18 | 63 (16-80) | 10 M, 8 F | 11 (6-19) | 6/15 (3 ND) | 7 (39) |
| RAEB-t | 5 | 61 (38-70) | 3 M, 2 F | 25 (21-28) | 3/4 (1 ND) | 4 (80) |
| CMML | 9 | 73 (51-87) | 6 M, 3 F | 4 (1.5-4) | 0/7 (2 ND) | 0 (0) |
| AML evolved from MDS | 9 | 60.5 (60-73) | 4 M, 5 F | 64 (30-85) | 2/8 (1 ND) | 9 (100) |
| Total | 53 | 61.5 (16-87) | 26 M, 27 F | 11 (1-85) | 15/44 (9 ND) | 20 (38) |
Abbreviations: ND, not determined; BM, bone marrow.
MSP.
This method, initially described by Herman et al,14consists of two steps: modification of DNA by sodium bisulfite converting all unmethylated, but not methylated cytosines to uracil; and subsequent amplification with primers specific for methylated versus unmethylated DNA.
Bisulfite modification.
DNA (2 μg) in a volume of 40 μL was denatured by addition of 10 μL 1 mol/L NaOH (final concentration 0.2 mol/L) for 10 minutes at 37°C. Next, 30 μL of 10 mmol/L hydroquinone (Sigma, St Louis, MO) and 520 μL of 3 mol/L sodium bisulfite (Sigma) at pH 5 were added and mixed and samples were incubated under mineral oil at 50°C for 16 hours. To meet strict anaerobic conditions, all of these manipulations were performed under nitrogen in an anaerobic chamber. Modified DNA was purified using the Wizard purification resin and the Vacuum Manifold (Promega, Madison, WI), and eluted into 50 μL H2O. Final desulphonation was achieved by adding 25 μL of 1 mol/L NaOH (final concentration 0.3 mol/L) at room temperature for 5 minutes. After ethanol precipitation and dilution in 50 μL H2O, samples were stored at −20°C before use.
PCR amplification and sequencing.
MSP, based on sequence differences resulting from bisulfite modification, was performed with the primer sets designed by Herman et al.14 Briefly, sequences of those primers recognize the region of the p15INK4B gene, which contain frequent cytosine to distinguish unmodified from modified DNA and CpG pairs near the 3′ end of the primers to ensure maximal discrimination between methylated and unmethylated DNA. The multiple mismatches in these primers allow amplification only from the intended template. Primer sets were provided from Eurogentec (Seraing, Belgium). The PCR reaction contained 1× PCR buffer (6.5 mmol/L MgCl2, 10 mmol/L Tris HCl, pH 9, 50 mmol/L KCl, 0.1% triton X100, 0.2 mg/mL bovine serum albumin [BSA]), deoxynucleotide triphosphates (dNTPs; each at 0.5 mmol/L), 20 pmol of each primer, 5% dimethyl sulfoxide (DMSO), 1 U of Taq polymerase (Oncor-Appligene, Gaithersburg, MD), and bisulfite modified DNA (100 to 200 ng) in a final volume of 80 μL. Reactions were hot started using Ampliwax 100 beads (Perkin Elmer, Foster City, CA). For each reaction, a lower layer containing PCR buffer, dNTP, DMSO, diluted in a final volume of 30.5 μL, was pipetted in a reaction tube and AmpliWax 100 beads were added. After incubation at 85°C for 3 minutes and 25°C for 3 minutes, leading to formation of a solid wax layer, an upper layer containing Taq polymerase, MgCL2, PCR buffer, 100 ng modified DNA, diluted in 49.5 μL was added. Amplification were performed in a MJ research MiniCycler thermal cycler (MJ Research, Wattertown, MA). A first step of denaturation at 95°C for 4 minutes then 98°C for 30 seconds, was followed by 35 cycles of amplification (30 seconds at 95°C, 30 seconds at 60°C, 30 seconds at 72°C), and by a final 10-minute extension at 72°C. Controls without DNA were performed for each set of PCR reactions. PCR products (20 μL) were loaded on 2% agarose gels stained with ethidium bromide and visualized under ultraviolet (UV) illumination directly and with a gel scan software analysis system (Bio-Print, Marne la Vallée, France).
To confirm the specificity of MSP, five PCR products obtained after amplification with primer pairs specific for methylated DNA (p15M) or unmethylated DNA (p15U) were sequenced. Ampligens purified with Centricon-100 Concentrator columns were sequenced with the ABI Prism dRhodamine Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer) according to manufacturer's recommendations, and samples were analyzed with the ABI Prism 310 automatic sequencer (Perkin Elmer). Resulting sequences were compared with wild-type sequence (Genbank accession number, S75756) using Sequence Navigator software (Perkin Elmer).
Statistical analysis.
Comparisons were made with Fisher's exact test and the χ2 test. Multivariate analysis was performed with a Cox model. All of these analyses were performed on SPSS 6.2 analysis software (SPSS, Chicago, IL).
RESULTS
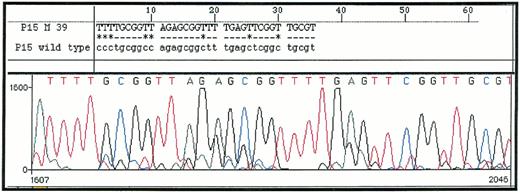
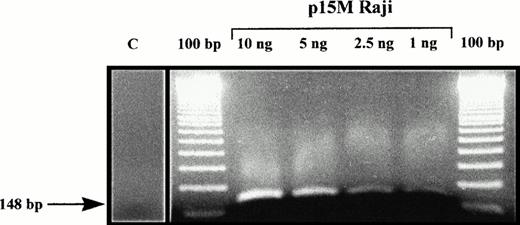
MSP of 25 de novo AML demonstrated p15INK4b methylation, as shown by amplification with p15M primer pair in 18 (72%) of the patients analyzed. Those samples could also be amplified with p15U primer pair, due to sample contamination by nonblastic cells (whose percentage ranged from 5% to 19% cells in bone marrow samples). Sequencing of five p15M PCR products (from one AML and four MDS) confirmed that all cytosines had been converted in thymidines except in CpG dinucleotides, where cytosines remained unchanged (Fig 1). Normal unmethylated DNA from the K562 cell line could not be amplified with p15M primer pair, but the Raji cell line, which is methylated for the p15INK4b gene, gave a strong amplification. Dilution of the Raji cell line showed that detection of as little as 1 ng of methylated DNA was possible, confirming the sensitivity of MSP (Fig 2). All of these results were concordant with previous reports with MSP and/or p15INK4b gene methylation in AML.11,12 14
DNA sequence of p15M PCR product from a patient with RAEB (upper sequence) as compared with Genbank sequence of p15INK4b gene (lower sequence). All cytosines are converted in thymidines except in methylated CpG dinucleotides.
DNA sequence of p15M PCR product from a patient with RAEB (upper sequence) as compared with Genbank sequence of p15INK4b gene (lower sequence). All cytosines are converted in thymidines except in methylated CpG dinucleotides.
Amplification with p15M primer pair of Raji cell line DNA, diluted in negative control DNA from a healthy volunteer. C, amplification with p15 primer pair of a control DNA from a healthy volunteer.
Amplification with p15M primer pair of Raji cell line DNA, diluted in negative control DNA from a healthy volunteer. C, amplification with p15 primer pair of a control DNA from a healthy volunteer.
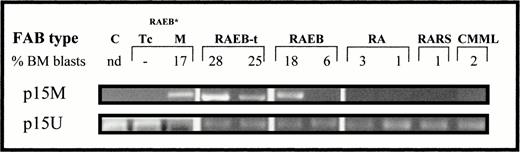
In the 53 patients with MDS, amplification with p15M primer pair was seen in 20 patients (Fig 3). Methylation was exclusively seen in patients with at least 10% bone marrow blasts (20 of 24 v 0 of 29 of those with less than 10% marrow blasts,P < .001). In patients with at least 10% marrow blasts, 11 of 15 of those that were still in MDS phase (<30% marrow blasts) were methylated, as compared with nine of nine of those that had progressed to AML (>30% marrow blasts)(P = .6). Amplification of DNA from T cells isolated from a patient with 17% bone marrow blasts showed no evidence of methylation (Fig 3). Nine of the 30 cases with normal karyotype or “favorable karyotype” (isolated del5q or del20q) had p15INK4b methylation, as compared with five of the 14 cases with other cytogenetic findings (P = .73).
Amplification of DNA from several MDS cases with p15M or p15U primer pair and from T (Tc) and mononuclear bone marrow (BM) cells isolated from a RAEB case*.
Amplification of DNA from several MDS cases with p15M or p15U primer pair and from T (Tc) and mononuclear bone marrow (BM) cells isolated from a RAEB case*.
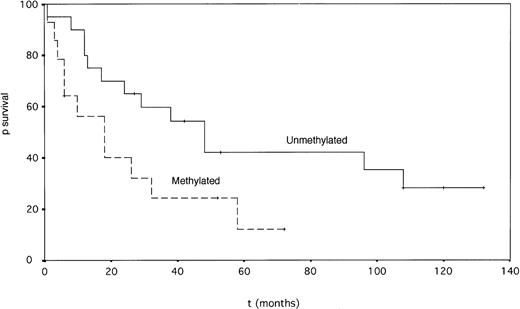
Patients with p15INK4b gene methylation had a significantly shorter survival than unmethylated cases (median 18 v 48 months, P = .049, log-rank test) (Fig 4). The percentage of bone marrow blasts and French-American-British (FAB) type also had prognostic value for survival by univariate analysis (Table 2). In a Cox multivariate analysis, the percentage of marrow blasts emerged as the only prognostic factor for survival, p15INK4b methylation status being strongly correlated with this parameter. Twelve patients (nine methylated and three unmethylated) were treated with intensive chemotherapy. Five patients achieved complete remission (CR), and four methylated patients had failure; two patients achieved partial remission (PR) and one unmethylated patient had failure.
Actuarial survival of MDS patients according to the p15INK4b gene methylation status (P = .049, log-rank test.)
Actuarial survival of MDS patients according to the p15INK4b gene methylation status (P = .049, log-rank test.)
Prognostic Value of p15ink4b Methylation and Other Pretreatment Factors in 53 Cases of MDS
| . | No. . | Median Actuarial Survival (mo) . | P . |
|---|---|---|---|
| Univariate analysis | |||
| p15INK4b methylation | .049 | ||
| Unmethylated | 33 | 48 | |
| Methylated | 20 | 18 | |
| FAB type | .05 | ||
| RA + RARS | 12 | 42 | |
| RAEB + RAEB-t | 23 | 18 | |
| CMML | 9 | 21 | |
| Karyotype | .18 | ||
| Normal or favorable* | 30 | 26 | |
| Other single and complex ab | 14 | 24 | |
| Bone marrow blasts | .032 | ||
| <10% | 29 | 48 | |
| >10% | 24 | 16 | |
| Cox multivariate analysis | |||
| Bone marrow blasts | .026 | ||
| p15INK4b methylation | .4 | ||
| FAB type | .6 |
| . | No. . | Median Actuarial Survival (mo) . | P . |
|---|---|---|---|
| Univariate analysis | |||
| p15INK4b methylation | .049 | ||
| Unmethylated | 33 | 48 | |
| Methylated | 20 | 18 | |
| FAB type | .05 | ||
| RA + RARS | 12 | 42 | |
| RAEB + RAEB-t | 23 | 18 | |
| CMML | 9 | 21 | |
| Karyotype | .18 | ||
| Normal or favorable* | 30 | 26 | |
| Other single and complex ab | 14 | 24 | |
| Bone marrow blasts | .032 | ||
| <10% | 29 | 48 | |
| >10% | 24 | 16 | |
| Cox multivariate analysis | |||
| Bone marrow blasts | .026 | ||
| p15INK4b methylation | .4 | ||
| FAB type | .6 |
*Favorable: isolated del 5q or del20q (1 patient had del20q and no patient had del 5q).
In 10 patients, sequential DNA samples were available. In five of them, p15M amplification was negative at diagnosis, but became positive after leukemic transformation. (Table 3). Four patients were found methylated on first analysis, but they already had relatively high bone marrow blast counts (15% to 26%). The remaining patient was unmethylated both at diagnosis and at leukemic transformation.
Sequential Analysis of p15INK4b Methylation in 10 MDS Cases
| . | Patient No. . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . |
| First analysis | ||||||||||
| FAB type | RAEB | RAEB | RAEB | RAEB-t | RAEB | CMML | RAEB | RAEB | RAEB-t | RAEB |
| % bone marrow blasts | 7 | 11 | 8 | 26 | 9 | 4 | 12 | 12 | 23 | 15 |
| Methylation status | U | U | U | M | U | U | U | M | M | M |
| Second analysis | ||||||||||
| Interval between first and second analysis (mo) | 17 | 18 | 4 | 4 | 11 | 7 | 11 | 3 | 8 | 14 |
| % Bone marrow blasts | 66 | 35 | 74 | 69 | 67 | 65 | 80 | 35 | 28 | 40 |
| Methylation status | M | M | M | M | U | M | M | M | M | M |
| Third analysis | ||||||||||
| Interval between second and third analysis (mo) | 5 | 12 | 2 | |||||||
| % bone marrow blasts | 41* | 27* | 23* | |||||||
| Methylation status | M | M | M | |||||||
| . | Patient No. . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . |
| First analysis | ||||||||||
| FAB type | RAEB | RAEB | RAEB | RAEB-t | RAEB | CMML | RAEB | RAEB | RAEB-t | RAEB |
| % bone marrow blasts | 7 | 11 | 8 | 26 | 9 | 4 | 12 | 12 | 23 | 15 |
| Methylation status | U | U | U | M | U | U | U | M | M | M |
| Second analysis | ||||||||||
| Interval between first and second analysis (mo) | 17 | 18 | 4 | 4 | 11 | 7 | 11 | 3 | 8 | 14 |
| % Bone marrow blasts | 66 | 35 | 74 | 69 | 67 | 65 | 80 | 35 | 28 | 40 |
| Methylation status | M | M | M | M | U | M | M | M | M | M |
| Third analysis | ||||||||||
| Interval between second and third analysis (mo) | 5 | 12 | 2 | |||||||
| % bone marrow blasts | 41* | 27* | 23* | |||||||
| Methylation status | M | M | M | |||||||
Abbreviations: U, unmethylated; M, methylated.
Third analysis performed after intensive chemotherapy, which yielded only transient reduction in the bone marrow blast count.
DISCUSSION
The progressive increase of bone marrow blast involvement in MDS suggests that a fraction of clonal dysplastic cells loses its cell cycle regulation and differentiation during evolution of the disease. p16INK4a gene is frequently inactivated in cancer, by homozygous deletion in ALL (especially T-ALL) and some solid tumors, and by point mutation or methylation in other solid tumors.15-17 p16INK4a deletion, point mutation, and methylation, on the other hand, are not seen in AML and MDS.17-19 On the contrary, methylation of CpG islands of the p15INK4b promoter region has been recently reported to occur at high frequency both in myeloid and lymphoid leukemias (88% of AML and 71% of ALL), but not in lymphoma, chronic myeloid leukemia, and in solid tumors.11,12 20
In this report, we analyzed p15INK4b gene hypermethylation by a recently described MSP method.14 The sensitivity of this method was 1%. This is higher than the sensitivity obtained with Southern blot analysis, generally in the range of 5%, a technique which also requires relatively large amounts of DNA. Such a high sensitivity is probably important to obtain in MDS, where the percentage of marrow blasts is often very low. Specificity of the MSP technique, which we confirmed by direct sequencing, is probably higher than that of classical PCR-based methylation assay, where amplification from primers flanking restriction site can be performed only if DNA cleavage has been prevented by methylation. This latter method requires a complete restriction, as any uncleaved DNA will be amplified by PCR yielding a false positive result.
In this study, we observed a high incidence of p15INK4bmethylation in MDS (38%) confirming results from Uchida et al19 (p15INK4b methylation in 16 of 32 MDS cases analyzed). Methylation was found in bone marrow mononuclear cells, but not in T lymphocytes from a patient with 17% bone marrow blasts. Uchida et al observed p15INK4b methylation in 14 of 18 MDS patients with excess of blast, but not in T lymphocytes, and only in one of 12 RA and RARS. However, their study was made by Southern blot and PCR-based methylation assay, which appear to have lower sensitivity and specificity, respectively, than the MSP method used in the present work. We found p15INK4b methylation exclusively in MDS with >10% bone marrow blasts. The high sensitivity of MSP ruled out the possibility of a p15INK4bmethylation restricted to blast cells in MDS cases with fewer than 10% marrow blasts. Thus, p15INK4b methylation appears to be almost exclusively found in MDS with an excess of blasts. Precise marrow blast counts, in MDS with an excess of marrow blasts, particularly RAEB, were not available in the report of Uchida et al. In our study, 10% marrow blasts was an important figure, as no cases of p15INK4b methylation were seen below that threshold. Interestingly, several scoring systems for prognosis in MDS have shown that the 10% blasts threshold had an important clinical significance.21 22 Indeed, most MDS with marrow blasts below 5% and many of those with marrow blasts between 5% and 10% have a relatively mild course, with unfrequent progression to AML. On the contrary, MDS with greater than 10% blasts are generally associated with severe blood cytopenias, very frequent transition to AML, and short survival. Thus, the presence of p15INK4bmethylation appears to carry an important clinical significance in MDS and had prognostic significance in univariate analysis in our study. However, because of its high correlation with marrow blast percentage, p15INK4b methylation lost its prognostic value for survival in multivariate analysis, where only blast bone marrow involvement emerged as an independent prognostic factor. The small number of unmethylated patients treated by intensive chemotherapy precluded the study of any correlation between response to treatment and methylation status.
By studying 10 patients where sequential DNA was available, we observed in five patients that p15INK4b methylation was acquired during disease progression. Four patients were methylated both at diagnosis and at leukemic transformation, but they already had high bone marrow blast count at diagnosis. Only one remained unmethylated both at diagnosis and leukemic transformation. Uchida et al have also reported progression in the methylation status in four of 10 patients studied sequentially. All of these data support the hypothesis that p15INK4b methylation and progression toward AML are linked in MDS and that p15INK4b methylation is acquired during disease progression.
Whether p15INK4b methylation in MDS is a specific event or a simple consequence of a global hypermethylation activity remains to be determined. Indeed, hypermethylation of the calcitonin gene has also been described in MDS, suggesting a more global increased methylation activity in this group of disorders.23,24 However, the calcitonin gene is close to the cell cycle inhibitor gene p57KIP2, which has been found to be imprinted.25 26 Moreover, p15INK4b methylation has also been reported in AML and ALL, and leukemic cell lines that are nonsensitive to TGFβ inhibition generally have p15INK4bmethylation. Finally, Uchida et al in MDS, and other groups in AML and ALL, did not find any methylation of the p16INK4a gene, which is adjacent to the p15INK4b gene on 9p21 chromosome. These findings strongly suggest that p15INK4b inactivation is important and perhaps necessary for the proliferation and/or differentiation block of marrow blast cells in myeloid malignancies. Selective methylation of the p15INK4b gene could be a specific mechanism of immature bone marrow cells to escape to G1 phase regulation by TGFβ inhibitory effect.
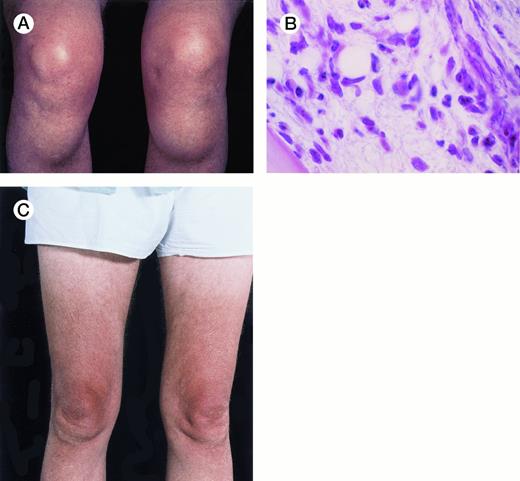
Unusual presentation of relapsed hairy cell leukemia. The patient is a 64-year-old man with hairy cell leukemia diagnosed in 1982 and had been in partial remission after treatment with splenectomy (1982), intermittent courses of α-interferon (1987-1994), and pentostatin (1994). In May 1996, he developed bilateral bony masses in the tibia, each about 6 cm in diameter (A), and a lump in the mucosa of the lower lip. He also had generalized lymphadenopathy, hepatomegaly of 5 cm, a hemoglobin level of 9.8 g/dL, a platelet count of 91 × 109/L, and a leukocyte count of 12 × 109/L with 70% hairy cells. Marrow trephine biopsy showed substantial replacement of normal hematopoietic tissue by hairy cells (B) and reticulin fibrosis. The buccal mucosal mass proved, on biopsy, to be a deposit of leukemic cells. Five weeks after a 7-day course of 2-chlorodeoxyadenosine, the tibial masses had resorbed (C). Repeat marrow biopsy 3 weeks later showed few hairy cells and a normal amount of reticulin. At follow-up 11 weeks after chemotherapy, the hematologic parameters were a hemoglobin level of 12.8 g/dL, a platelet count of 199 × 109/L, a white blood cell count of 6.5 × 109/L, a granulocyte count of 3.1 × 109/L, and a lymphocyte count of 2.9 × 109/L. There were no hairy cells in the peripheral blood film. (Courtesy of I.E. Okpala, MRCPath, FWACP, Central Sheffield University Hospitals, Royal Hallamshire Hospital, Glossop Road, Sheffield, UK, and J. Leslie, FRCPath, Norfolk & Norwich Hospital, Norwich, UK.) {/ANNT;;;left;0n}
Unusual presentation of relapsed hairy cell leukemia. The patient is a 64-year-old man with hairy cell leukemia diagnosed in 1982 and had been in partial remission after treatment with splenectomy (1982), intermittent courses of α-interferon (1987-1994), and pentostatin (1994). In May 1996, he developed bilateral bony masses in the tibia, each about 6 cm in diameter (A), and a lump in the mucosa of the lower lip. He also had generalized lymphadenopathy, hepatomegaly of 5 cm, a hemoglobin level of 9.8 g/dL, a platelet count of 91 × 109/L, and a leukocyte count of 12 × 109/L with 70% hairy cells. Marrow trephine biopsy showed substantial replacement of normal hematopoietic tissue by hairy cells (B) and reticulin fibrosis. The buccal mucosal mass proved, on biopsy, to be a deposit of leukemic cells. Five weeks after a 7-day course of 2-chlorodeoxyadenosine, the tibial masses had resorbed (C). Repeat marrow biopsy 3 weeks later showed few hairy cells and a normal amount of reticulin. At follow-up 11 weeks after chemotherapy, the hematologic parameters were a hemoglobin level of 12.8 g/dL, a platelet count of 199 × 109/L, a white blood cell count of 6.5 × 109/L, a granulocyte count of 3.1 × 109/L, and a lymphocyte count of 2.9 × 109/L. There were no hairy cells in the peripheral blood film. (Courtesy of I.E. Okpala, MRCPath, FWACP, Central Sheffield University Hospitals, Royal Hallamshire Hospital, Glossop Road, Sheffield, UK, and J. Leslie, FRCPath, Norfolk & Norwich Hospital, Norwich, UK.) {/ANNT;;;left;0n}
Supported by the Ligue Contre le Cancer (Comité du Nord and Comité du Pas de Calais), the Association de Recherche sur le Cancer, and The Centre Hospitalier Universitaire of Lille, France.
Address reprint requests to Bruno Quesnel, MD, PhD, Service des maladies du sang, CHU Lille, 1 Place de Verdun, 59037 Lille, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal