The BCL-2 gene family regulates the susceptibility to apoptotic cell death in many cell types during embryonic development and normal tissue homeostasis. Deregulated expression of anti-apoptotic BCL-2 can be a primary aberration that promotes malignancy and also confers resistance to chemotherapeutic agents. Recently, studies ofBax-deficient mice have indicated that the pro-apoptotic BAX molecule can function as a tumor suppressor. Consequently, we examined human hematopoietic malignancies and found that approximately 21% of lines possessed mutations in BAX, perhaps most commonly in the acute lymphoblastic leukemia subset. Approximately half were nucleotide insertions or deletions within a deoxyguanosine (G8) tract, resulting in a proximal frame shift and loss of immunodetectable BAX protein. Other BAX mutants bore single amino acid substitutions within BH1 or BH3 domains, demonstrated altered patterns of protein dimerization, and had lost death-promoting activity. Thus, mutations in the pro-apoptotic molecule BAX that confer resistance to apoptosis are also found in malignancies.
THE Bcl-2 GENE FAMILY regulates cellular responsiveness to a wide variety of death-inducing stimuli, including growth factor deprivation, glucocorticoids, antireceptor antibody, γ- and UV irradiation, and chemotherapeutic agents.1,2 Death antagonists, including BCL-2, BCL-XL, MCL-1, and A1, provide protection, whereas death agonist members, including BAX, BAK, BAD, and BCL-XS, increase sensitivity to death-inducing signals. The ratio of death agonists to antagonists determines the susceptibility to death stimuli.3 A prominent feature of the BCL-2 family is their capacity to form both homodimers and heterodimers.3-5Several conserved domains, entitled BH1, BH2, and BH3, participate in the formation of various dimer pairs as well as the regulation of cell death.6-8 Recently, the multidimensional NMR and x-ray crystallographic structure of a BCL-XL monomer indicated that BH1, BH2, and BH3 correspond to α helices that are closely juxtaposed to form a hydrophobic pocket.9 Mutational analysis has indicated the importance of BH1 and BH2 domains for the anti-apoptotic function of BCL-2 and BCL-XL as well as their binding to BAX.5,6 The BH3 domains of BAK and BAX appear critical for promoting cell death and dimerization with BCL-XL or BCL-2.7,8 Moreover, several distantly related pro-apoptotic molecules, BIK and BID, possess only a BH3 domain.10-12 These lend strength to the argument that BH3 represents the critical death domain. Besides BH1, BH2, and BH3 domains, a fourth N-terminal α helical domain entitled BH4 is conserved between BCL-2 and BCL-XL and is vital for death-repressor function.13 14
Deregulated expression of BCL-2 family members has been noted in several types of human malignancies and may affect clinical outcome.BCL-2 was discovered on chromosome 18 at the site of translocation with the Ig heavy chain locus (IgH) in follicular lymphoma. Increased expression of BCL-2 in cases of non-Hodgkin's lymphoma, myeloid leukemia, and prostate cancer has been associated with a poor prognosis.1,2,15 Reduced BAX RNA and protein expression has been noted in metastatic breast cancer16,17 and correlates with a poor response to chemotherapy and shorter overall survival.17 ElevatedBCL-2/BAX RNA ratios have been associated with progression of disease in B-chronic lymphocytic leukemia18 and low-grade urinary bladder cancer.19
Bax-deficient mice indicate that several normal developmental cell deaths depend on Bax.20 Moreover, neuronal cell death due to deprivation of neurotrophic factors proved to be dependent on Bax.21 Induction of BAX expression can be sufficient to induce apoptosis and did not require an additional death stimulus.22 Induction of BAX results in the activation of Caspases22,23 and also triggers a mitochondrial dysfunction program.22,24 Finally, experimental models using Bax-deficient mice argue that approximately half of certain p53-dependent cell deaths require BAX. The removal of BAX substantially decreased apoptosis induced by a transgene expressing a truncated T antigen (TGT121), a previously documented p53-dependent death.25 Moreover, chemotherapeutic agents induce apoptosis in embryonic fibroblasts in ap53-dependent manner. Elimination of BAX prevents approximately half of those chemotherapy-induced deaths.26
Recently, we noted that several cell lines of human hematopoietic malignancies bore mutations of the BAX gene.27 We extend this study here to a larger panel of malignant hematopoietic lines. Frameshift mutations eliminated the production of BAX. These were focused in the same simple repeat sequence found to be mutated in some colon cancers with mutator phenotypes.28 Other substitution mutations within the BH1 and BH3 domains resulted in a loss of pro-apoptotic function and altered the dimerization capabilities of BAX.
MATERIALS AND METHODS
Amplification of BAX and single-stranded conformation polymorphism (SSCP)-gel analysis.
BAX RNA from cell lines of human hematopoietic malignancies was amplified by reverse transcription-polymerase chain reaction (RT-PCR) either unlabeled or, alternatively, in the presence of 500 μmol/L dATP, dTTP, and dGTP; 125 μmol/L dCTP; and 100 μCi/mL [α-32P]dCTP (Amersham, Arlington Heights, IL; 3,000 Ci/mmol; 10 mCi/mL). The complete BAXcoding region was covered by two partially overlapping PCR reactions (Fig 1B).3 In one PCR reaction,BAX cDNA was amplified from exon 1 to the exon 4/5 boundary using forward primer A (5′-TGG ACG GGT CCG GGG AGC-3′) and reverse primer B (5′-GCA CAG GGC CTT GAG CAC C-3′). In a second reaction, BAX cDNA was amplified from exon 4 through exon 6 using forward primer C (5′-GCC CTT TTC TAC TTT GCC AGC-3′) and reverse primer D (5′-TCA GCC CAT CTT CTT CCA GAT-3′; Fig 1B). Products were analyzed by SSCP-polyacrylamide gel electrophoresis (SSCP-PAGE). For the SSCP analysis, nonradioactive PCR products were purified on Wizard PCR Prep columns (Promega, Madison, WI) and dissolved in TE-buffer. Five microliters of PCR product was mixed with an equal volume of loading buffer (20% EDTA, 20 mmol/L NaOH, 0.05% xylene cyanol, 0.05% bromophenolblue in 96% formamide) and denatured for 10 minutes at 95°C. One microliter was separated on a 4% to 15% polyacrylamide gradient gel or a 12.5% polyacrylamide gel (Pharmacia LKB Phastsystem, Piscataway, NJ). Products were visualized by silver staining. Alternatively, 2 μL of 10-fold diluted radioactive PCR products in SSCP loading buffer was separated on a 5% nondenaturating polyacrylamide gel (49:1), 5% glycerol in half-strength TBE buffer. The gel was exposed to x-ray film (Eastman Kodak, Rochester, NY).
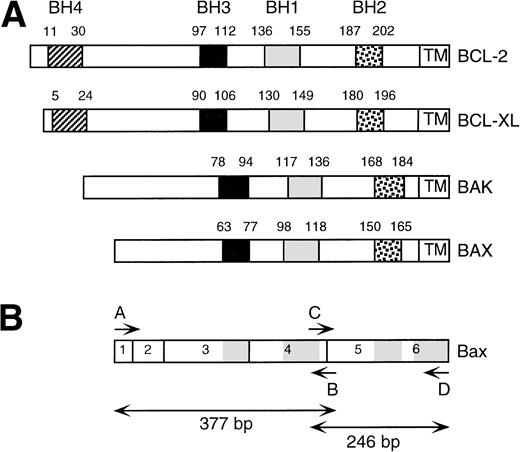
BCL-2 homology (BH) domains within BCL-2 family members. (A) Schematic representation of BCL-2, BCL-XL, BAK, and BAX and the relative positions of the BH1, BH2, BH3, and BH4 domains and transmembrane region. (B) Schematic overview of BAX open reading frame. Exons are numbered. BH1, BH2, and BH3 and transmembrane domains are shaded. Positions of PCR primers (A through D) are indicated. PCR amplification using primer pair A, B results in a 377-bp product. PCR amplification using primer pair C, D results in a 246-bp product.
BCL-2 homology (BH) domains within BCL-2 family members. (A) Schematic representation of BCL-2, BCL-XL, BAK, and BAX and the relative positions of the BH1, BH2, BH3, and BH4 domains and transmembrane region. (B) Schematic overview of BAX open reading frame. Exons are numbered. BH1, BH2, and BH3 and transmembrane domains are shaded. Positions of PCR primers (A through D) are indicated. PCR amplification using primer pair A, B results in a 377-bp product. PCR amplification using primer pair C, D results in a 246-bp product.
Immunoprecipitation.
Preparation of constructs.
EcoRI sites, an upstream Kozak sequence, and hemagglutinin influenza (HA) epitope tag were introduced by PCR using extended primers and germline and mutant BAX cDNA sequences. Products were cloned into PCRII using the TA-Cloning kit (Invitrogen, La Jolla, CA) and confirmed by DNA sequencing. Identical G67R and G108V mutations as found in Daudi and HPB-ALL, respectively, were introduced by site-directed mutagenesis (Clontech). Mutations were confirmed by DNA sequencing. BAX and mutantBAX cDNAs were cloned into pSFFV-LTR neo expression vector. For yeast two-hybrid experiments, cDNAs without their C-terminal signal-anchor sequence were cloned into the Gal4 activation domain (AD) vector pACTII and the LexA DNA binding domain (DB) vector pBTM116.5 Extended primers were used to introduce a 5′ EcoRI site and a 3′ Sal I site for cloning into pBTM116 or a 5′ Nco I site and a 3′Sma I site for cloning into pACTII. Constructs were confirmed by DNA sequencing. The constructs for BCL-XLΔC19,BCL-2ΔC22 and pACTII, and BCL-XLΔC19 in pBTM116 were previously described.5
Transfixion and Western blots.
Twenty micrograms of DNA of Xba-1 linearized pSFFV-LTR neo vector expressing HA-BAX, HA-BAXG67R, or HA-BAXG108V was transfected into FL5.12 cells or cotransfected with 1 μg of linearized pGK-hygro vector DNA into FL5.12-BCL-2 cells. Stably transfected clones were selected for neomycin resistance (2 mg/mL G418) or hygromycin resistance (1 mg/mL). Transfected clones were analyzed for HA-BAX, HA-BAXG67R, or HA-BAXG108V expression by Western blot using polyclonal antiserum N20 (Santa Cruz, Santa Cruz, CA; 1:500) as primary antibody and goat-antirabbit HRPO (Caltag Labs; 1:2,000) as the secondary antibody. BCL-2 expression was detected by the 6C8 MoAb (1:250) and a secondary goat-antihamster HRPO antibody (Caltag Labs; 1:2,000). Immunoblots were developed by enhanced chemiluminescence (ECL; Amersham).
Yeast two-hybrid analysis.
cDNAs cloned in pBTM116 or pACTII were cotransformed into yeast strain L40 (MATa trp1-901 leu2-3, 112 ade2 his3-Δ200 LYS2::(lexA0p)4-HIS3 URA3::(lexA)8-LacZ)31 using the lithium-acetate method. After transformation, yeast cells were plated on selective media (His+, Ura−, Leu−, and Trp−) and incubated at 30°C. After 2 to 4 days, yeast colonies were transferred to nitrocellulose filters and incubated for 1 minute in liquid N2. Filters were dried on Whatmann paper (Whatmann, Maidstone, UK) and stained with 5-bromo-4-chloro-3-indolyl β-D-galactoside (X-Gal) substrate solution (60 mmol/L Na2HPO4, 40 mmol/L KC1, 1 mmol/L MgSO4, 50 mmol/L β-mercaptoethanol, 1 mg/mL X-Gal, pH 7.0) for 1 to 12 hours at 37°C.
RESULTS
Substitution mutations.
Twenty-nine cell lines derived from hematopoietic malignancies of various cell types were analyzed for mutations in BAX by SSCP and sequence analysis. We found evidence for mutations in 7 cell lines (Table 1). The Burkitt lymphoma (BL) cell line Daudi expressed a mutant BAX allele that contained a G108V mutation and a wild-type allele. This mutation substitutes the central glycine within the BH1 domain (Fig 1). The plasmacytoma (PC) cell line OPM1 expressed a mutant allele with a G11E mutation and a wild-type allele. This glycine resides in the N-terminus proximal to the first predicted α helix. The acute lymphoblastic leukemia (ALL) cell line HPB-ALL expressed a G67R mutant allele as well as a wild-type allele. This mutation alters the central glycine within the BH3 domain (Fig 1A). All three cell lines expressed BAX protein by Western analysis.
Summary of Mutation Analysis
| Cell Line . | Lineage . | Origin . | SSCP . | Sequence . | Protein . |
|---|---|---|---|---|---|
| KM3 | B | B-ALL | Mutation | del (g114-12)-150 | − |
| Daudi | B | BL | Mutation | G108V | + |
| Raji | B | BL | wt | wt | ND |
| Ramos | B | BL | wt | wt | ND |
| Rpmi6666 | B | HD | wt | wt | ND |
| SUDHL6 | B | FL | wt | wt | + |
| UM1 | B | MM | wt | wt | ND |
| U266 | B | MM | wt | wt | ND |
| Rpmi8226 | B | MM | wt | wt | ND |
| IM-9 | B | MM | wt | wt | ND |
| FRAVEL | B | MM | wt | wt | ND |
| OPM1 | B | PC | Mutation | G11E | + |
| HS-sultan | B | PC | wt | wt | ND |
| MC-CAR | B | PC | wt | wt | ND |
| ARH77 | B | PCL | wt | wt | ND |
| CEM | T | T-ALL | Mutation | del (g114-121)-150 | + |
| Jurkat | T | T-ALL | 2 mutations | del/ins (g114-121)-151 | − |
| JM-150 | T | T-ALL | 2 mutations | del/ins (g114-121)-152 | − |
| HPB-ALL | T | T-ALL | Mutation | G67R | + |
| GH1 | T | T-ALL | wt | wt | ND |
| PEER | T | T-ALL | wt | wt | ND |
| HSB2 | T | T-ALL | wt | wt | ND |
| MOLT4 | T | T-ALL | wt | wt | ND |
| KCL22 | M | CML | wt | wt | ND |
| KYO | M | CML | wt | wt | ND |
| K562 | M | CML-E | wt | wt | + |
| BV173 | M | CML-L | wt | wt | + |
| HL60 | M | APL | wt | wt | ND |
| Kazumi | M | AML-M2 | wt | wt | ND |
| Cell Line . | Lineage . | Origin . | SSCP . | Sequence . | Protein . |
|---|---|---|---|---|---|
| KM3 | B | B-ALL | Mutation | del (g114-12)-150 | − |
| Daudi | B | BL | Mutation | G108V | + |
| Raji | B | BL | wt | wt | ND |
| Ramos | B | BL | wt | wt | ND |
| Rpmi6666 | B | HD | wt | wt | ND |
| SUDHL6 | B | FL | wt | wt | + |
| UM1 | B | MM | wt | wt | ND |
| U266 | B | MM | wt | wt | ND |
| Rpmi8226 | B | MM | wt | wt | ND |
| IM-9 | B | MM | wt | wt | ND |
| FRAVEL | B | MM | wt | wt | ND |
| OPM1 | B | PC | Mutation | G11E | + |
| HS-sultan | B | PC | wt | wt | ND |
| MC-CAR | B | PC | wt | wt | ND |
| ARH77 | B | PCL | wt | wt | ND |
| CEM | T | T-ALL | Mutation | del (g114-121)-150 | + |
| Jurkat | T | T-ALL | 2 mutations | del/ins (g114-121)-151 | − |
| JM-150 | T | T-ALL | 2 mutations | del/ins (g114-121)-152 | − |
| HPB-ALL | T | T-ALL | Mutation | G67R | + |
| GH1 | T | T-ALL | wt | wt | ND |
| PEER | T | T-ALL | wt | wt | ND |
| HSB2 | T | T-ALL | wt | wt | ND |
| MOLT4 | T | T-ALL | wt | wt | ND |
| KCL22 | M | CML | wt | wt | ND |
| KYO | M | CML | wt | wt | ND |
| K562 | M | CML-E | wt | wt | + |
| BV173 | M | CML-L | wt | wt | + |
| HL60 | M | APL | wt | wt | ND |
| Kazumi | M | AML-M2 | wt | wt | ND |
The presence of mutations in cell lines of hematopoietic malignancies as determined by SSCP and sequence analysis.
Abbreviations: BL, Burkitt's lymphoma; B-ALL, B-cell acute lymphoblastic leukemia; HD, Hodgkin's disease; FL, follicular lymphoma; MM, multiple myeloma; PC, plasmacytoma; PCL, plasma cell leukemia; T-ALL, T-cell acute lymphoblastic leukemia; CML, chronic myeloid leukemia; CML-E, CML with blast crisis of erythroid lineage; CML-L, CML with blast crisis of lymphoid lineage; APL, acute promyelocytic leukemia; AML-M2, acute myeloid leukemia of M2 subtype.
Deletion of 1 guanine residue in guanine stretch (nt114-121).
JM derived from Jurkat line.
Deletion of 1 guanine stretch (nt114-121) on first allele and insertion of 1 guanine residue in guanine stretch (nt114-121) on second allele. Results are based on at least two independent SSCP or DNA sequence analyses.
Frameshift mutations.
SSCP analysis also noted a distinct, altered pattern common to four cell lines (KM3, CEM, Jurkat, and JM*; Table 1). DNA sequence analysis showed the same single nucleotide deletion (G)7 in a simple tract of 8 deoxyguanosine residues (G)8 (nt 114-121) encompassing codons 38 to 41 of human BAX (Table 1). This deletion was consistently detected in 3 or more independently amplified RT-PCR products from each cell. DNA sequence of four or five independent clones from these RT-PCR products demonstrate the same (G)7 deletion. SSCP and DNA sequence analysis of the pre-B–cell ALL KM3 and the T-cell ALL CEM only detected the (G)7 deletion and no wild-type allele, indicating that the mutation was hemizygous or homozygous. PCR amplification of the third exon of BAX from the genomic DNA of KM3 also failed to show a wild-type allele (not shown). The (G)7 deletion frameshift generated a proximal stop codon. Consistent with this finding, no immunodetectable protein was observed in KM-3 or CEM (Table 1).
The T-cell ALL line Jurkat and its derivative JM32 both displayed an additional alteration on SSCP that, upon DNA sequencing, proved to be a single nucleotide insertion (G)9 in the same (G)8 (nt 114-121) tract (Table 1). Approximately half of the independent clones from the RT-PCR products of Jurkat and JM demonstrated the G(9) insertion, whereas the others demonstrated the G(7) deletion. The G(9)insertion frameshift also generates a proximal stop codon consistent with the lack of immunodetectable protein in Jurkat and JM cells (Table1) and the presence of two mutated BAX alleles.
Protein dimerization.
The BAXG67R and BAXG108V mutants found in HPB-ALL and Daudi reside in the conserved BH3 and BH1 domains, respectively, and consequently were tested for dimerization capacity with BCL-2 family members by means of yeast two-hybrid analysis (Table 2). Although the binding of BAXG67R to wild-type BAX appeared unaffected, it demonstrated enhanced dimerization with itself, BAXG67R. In contrast, the capacity to heterodimerize with BCL-2 or BCL-XL was lost. BAXG108V did not form homodimers with itself, BAXG108V, but bound to wild-type BAX. BAXG108V demonstrated enhanced binding to BCL-2 and BCL-XL. Thus, both mutations demonstrated altered, yet very distinct dimerization characteristics.
Yeast Two-Hybrid Interactions
| . | pACTII . | BAX . | G67R . | G108V . | BCL-2 . | BCL-XL . |
|---|---|---|---|---|---|---|
| BAX | − | + | + | + | + | + |
| G67R | − | + | +++ | ND | − | − |
| G108V | − | + | ND | − | ++ | ++ |
| BCL-XL | − | + | − | + | ND | ND |
| . | pACTII . | BAX . | G67R . | G108V . | BCL-2 . | BCL-XL . |
|---|---|---|---|---|---|---|
| BAX | − | + | + | + | + | + |
| G67R | − | + | +++ | ND | − | − |
| G108V | − | + | ND | − | ++ | ++ |
| BCL-XL | − | + | − | + | ND | ND |
Yeast two-hybrid analysis of dimerization capacity of wild-type BAX versus BAXG67R and BAXG108V. AD-fusion constructs are indicated across the top lane: control vector pACTII, BAXΔC19, BAXG67RΔC18, BAXG108VΔC18, BCL-2ΔC22, BCL-XLΔC19. DB-fusion constructs are indicated in the left hand column: BAXΔC18, BAXG67RΔC18, BAXG108VΔC18, BCL-XLΔC19. Protein interactions as measured by X-gal substrate conversion are indicated as “−” for no interaction or “+” for clear positive. Interactions that were enhanced are indicated as “++” or “+++.” Interactions were tested in two to four independent experiments.
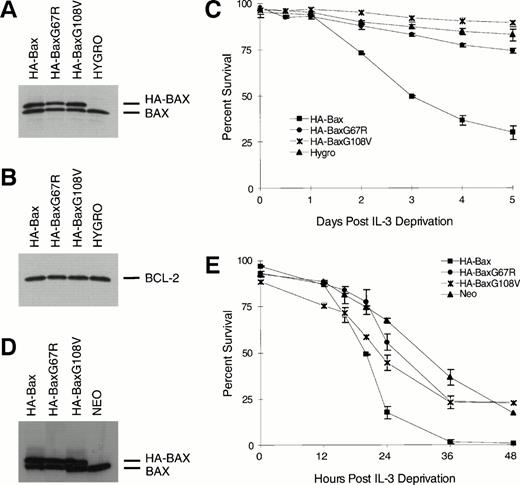
We next examined whether these BAX mutants demonstrated altered dimerization within mammalian cells. Stably transfected FL5.12-BCL-2 clones expressing comparable amounts of hemagglutinin epitope-tagged (HA) molecules HA-BAX, HA-BAXG67R, or HA-BAXG108V were generated (Fig2). When BCL-2 was immunoprecipitated from lysates, it coprecipitated HA-BAX and the endogenous BAX (Fig 2A, lane 1). HA-BAXG108Vbut not HA-BAXG67R was also coprecipitated with BCL-2, confirming the interactions suggested in yeast two-hybrid (Fig 2A, lanes 2 and 3). A consistently higher ratio of HA-BAXG108V/endogenous BAX compared with HA-BAX/endogenous BAX in BCL-2 immunoprecipitates is consistent with the enhanced binding of BAXG108V noted by yeast two-hybrid analysis (Table 2). In the reciprocal experiment, HA-BAX, HA-BAXG67R, and HA-BAXG108V were immunoprecipitated with anti-HA MoAb 12CA5 and all heterodimerized to endogenous BAX. As before, HA-BAX and HA-BAXG108V but not HA-BAXG67R coprecipitated BCL-2, confirming the pattern of interactions within mammalian cells (Fig 2).
Altered dimerization of BAXG67R and BAXG108V in vivo compared with wild-type BAX. (A) Immunoprecipitation of BCL-2 from 0.25% NP-40 lysates of35S methionine/cysteine-labeled FL5.12-BCL-2 cells (Hygro) or HA-BAX, HA-BAXG67R ,or HA-BAXG108V stably transfected clones using antihuman BCL-2 MoAb 6C8 and SDS-PAGE.3 (B) Immunoprecipitation and sodium dodecyl sulfate-PAGE analysis using anti-HA MoAb 12CA5 from 0.25% NP-40 lysates of 35S methionine/cysteine-labeled FL5.12-BCL-2 cells (Hygro) or HA-BAX, HA-BAXG67R, or HA-BAXG108V stably transfected clones.
Altered dimerization of BAXG67R and BAXG108V in vivo compared with wild-type BAX. (A) Immunoprecipitation of BCL-2 from 0.25% NP-40 lysates of35S methionine/cysteine-labeled FL5.12-BCL-2 cells (Hygro) or HA-BAX, HA-BAXG67R ,or HA-BAXG108V stably transfected clones using antihuman BCL-2 MoAb 6C8 and SDS-PAGE.3 (B) Immunoprecipitation and sodium dodecyl sulfate-PAGE analysis using anti-HA MoAb 12CA5 from 0.25% NP-40 lysates of 35S methionine/cysteine-labeled FL5.12-BCL-2 cells (Hygro) or HA-BAX, HA-BAXG67R, or HA-BAXG108V stably transfected clones.
Functional analysis of BAX mutants.
To address the functional consequence of the BH1 and BH3 mutations, they were assessed in an interleukin-3 (IL-3) deprivation assay using the FL5.12 line. Clones of FL5.12 (Neo) or FL5.12-BCL-2 expressing comparable amounts of HA-BAX, HA-BAXG67R, or HA-BAXG108V were identified by immunoblots (Fig 3A, B, and D). Addition of wild-type HA-BAX but not HA-BAXG67R or HA-BAXG108V was capable of promoting cell death in FL5.12-BCL-2 cells that were protected by BCL-2 (Fig 3C). Similarly, HA-BAXG67A and HA-BAXG108V did not substantially alter the survival of native FL5.12 cells, whereas wild-type HA-BAX clearly enhanced apoptosis (Fig 3E).
BAXG67R and BAXG108V have lost cell death-promoting activity. (A) Western blot analysis using anti-BAX polyclonal antiserum N20 on lysates of FL5.12-BCL-2 cells (Hygro) or HA-BAXα, HA-BAXG67R, or HA-BAXG108Vstably transfected clones. (B) Western blot analysis of the same lysates as in (A) using antihuman BCL-2 MoAb 6C8. (C) Viability assay. Transfected clones described in (A) were deprived of IL-3 and viability was determined by trypan blue exclusion at 0, 0.5, 1, 2, 3, 4, and 5 days after IL-3 withdrawal and plotted as the mean percentage of survival ± SEM. (D) Western blot analysis using anti-Bax polyclonal antiserum N20 on lysates of FL5.12 cells (Neo) or HA-BAX, HA-BAXG67R, or HA-BAXG108V stably transfected clones. (E) Viability assay. Transfected clones described in (D) were deprived of IL-3 and viability was determined by trypan blue exclusion at 0, 12, 16, 20, 24, 36, and 48 hours after IL-3 deprivation and plotted as the mean percentage of survival ± SEM.
BAXG67R and BAXG108V have lost cell death-promoting activity. (A) Western blot analysis using anti-BAX polyclonal antiserum N20 on lysates of FL5.12-BCL-2 cells (Hygro) or HA-BAXα, HA-BAXG67R, or HA-BAXG108Vstably transfected clones. (B) Western blot analysis of the same lysates as in (A) using antihuman BCL-2 MoAb 6C8. (C) Viability assay. Transfected clones described in (A) were deprived of IL-3 and viability was determined by trypan blue exclusion at 0, 0.5, 1, 2, 3, 4, and 5 days after IL-3 withdrawal and plotted as the mean percentage of survival ± SEM. (D) Western blot analysis using anti-Bax polyclonal antiserum N20 on lysates of FL5.12 cells (Neo) or HA-BAX, HA-BAXG67R, or HA-BAXG108V stably transfected clones. (E) Viability assay. Transfected clones described in (D) were deprived of IL-3 and viability was determined by trypan blue exclusion at 0, 12, 16, 20, 24, 36, and 48 hours after IL-3 deprivation and plotted as the mean percentage of survival ± SEM.
DISCUSSION
Prolonged cell survival with resistance to apoptosis can be a primary oncogenic event. Transgenic mice bearing a Bcl-2-Ig minigene that recapitulated the t(14;18) found in human follicular lymphoma display B-cell hyperplasia that progresses to high-grade lymphoma.33 Evidence is emerging that a principal contribution from the loss of p53 function is the elimination of a death pathway.34,35 Recent evidence suggests that BAX, a pro-apoptotic member of the BCL-2 family, can also qualify as a tumor suppressor. Bax-deficient mice display cellular expansions of neurons, lymphocytes, ovarian granulosa cells, and spermatogonia, reflecting the survival of cells that avoided developmental death.20 TGT121 transgenic mice that express a truncated T antigen that inhibits Rb but leaves p53 intact displayed an accelerated progression to malignancy upon a Bax-deficient background.25 It is of note that heterozygous Bax(+/−) mice also displayed an earlier onset of malignancy, suggesting that alteration of a single BAX allele could be of functional significance. An increase in focus formation was also documented in Bax-deficient versus wild-type fibroblasts when transfected with RAS and E1A.26 These experimental models argue that Bax can also be considered a tumor suppressor and that loss of this pro-apoptotic molecule promotes tumorigenesis.
We found mutations in BAX in approximately 21% (6 of 28 independent lines with confirmation of the mutation in Jurkat in its derivative JM) of human hematopoietic malignancy lines. No BAXalterations were noted in 35 normal individuals or in 8 Epstein-Barr virus–transformed lymphoblastoid lines, indicating that the observed alterations of BAX are not common polymorphisms or associated with immortalization. Approximately half of the mutations were frameshifts confined to a single mononucleotide (G)8tract (nt114-121). The existence of both insertions (G)9and deletions (G)7 within the same leukemia (Jurkat and JM) favors a biallelic aberration and argues that the elimination ofBAX is a selective advantage. Lack of detectable BAX protein in other leukemias with (G)7 deletions is compatible with a homozygous abnormality or perhaps loss of the second allele. Recently, Rampino et al28 described the presence of frameshift mutations in the identical (G)8 tract of BAX in about 50% of human colon adenocarcinomas with the microsatellite mutator phenotype (MMP). Some sporadic cancers and almost all cancers associated with the hereditary nonpolyposis colorectal cancer syndrome (HNPCC) accumulate mutations in microsatellites of nucleotide repeats due to defects in human DNA mismatch repair genes, including hMSH2, hMLH1, hPMS1, and hPMS2.36Msh-2–deficient mice progress to a precursor T-cell lymphoblastic lymphoma.37 Moreover, mutations have been noted in hMSH2 within human lymphoblastic lymphoma37 and in hMLH1 in a panel of lymphoid leukemia cell lines.38 This includes the CEM cell line,38 which demonstrated a frameshift mutation ofBAX in this study (Table 1). In total, these studies indicate that a subset of lymphoblastic leukemia/lymphoma have a mutator phenotype and that BAX may represent one target.
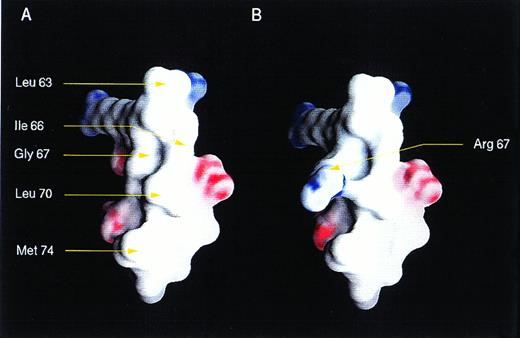
In addition to the frameshift mutations, we found missense mutations, including BAXG67R in the BH3 and BAXG108V in the BH1 domain. Both mutations demonstrated abnormal dimerization characteristics, but were nearly opposite in their patterns. This finding argues that the death agonist activity of BAX may not strictly correlate with the capacity to form any single set of homodimer or heterodimer pairs. Molecular modeling of the BAX BH3 α2-helix showed a classic amphipathic α helix. The G67R mutation would introduce a charged residue onto the hydrophobic face of this helix (Fig 4). NMR analysis of wild-type and mutant peptides of the BH3 α2-helix of BAK indicated critical interactions with BCL-XL through both hydrophobic and electrostatic interactions.39 The substitution of the central glycine in this α2-helix to arginine noted here has a much greater affect than an alanine substitution analyzed for BAK.39 The impact of the G67R mutation in BAX provides additional evidence that BH3 domains are critical for pro-apoptotic molecules.
Three-dimensional structure of BH3 region of BAX. Views of a modeled surface of the BH3 amphipathic α helix of BAX, calculated and displayed using GRASP.9 The G67R substitution as occurs in HPB-ALL is displayed at right. The surface is colored deep blue (23KBT) in the most negative, with linear interpolation for values inbetween. The model was generated using the protein building module (BUILDER) of INSIGHT II (Biosyn, San Diego, CA) and minimized using DISCOVER, the forefield simulation mode.
Three-dimensional structure of BH3 region of BAX. Views of a modeled surface of the BH3 amphipathic α helix of BAX, calculated and displayed using GRASP.9 The G67R substitution as occurs in HPB-ALL is displayed at right. The surface is colored deep blue (23KBT) in the most negative, with linear interpolation for values inbetween. The model was generated using the protein building module (BUILDER) of INSIGHT II (Biosyn, San Diego, CA) and minimized using DISCOVER, the forefield simulation mode.
The G108V substitution in the BH1 α5-helix of BAX also resulted in the loss of pro-apoptotic activity. While eliminating mutant/mutant dimerization, it, if anything, enhanced heterodimerization with BCL-2 or BCL-XL. Previous substitution of this central glycine to alanine in BCL-2 (G145A) and in BCL-XL (G159A) eliminated both their heterodimerization with BAX and their anti-apoptotic activity.5,6 However, the comparable substitution in BAX (G108A) had no effect (unpublished observations). Thus, the strong effect of the G108V substitution was somewhat unexpected. The G108 residue resides in the long hydrophobic α5-helix felt to be part of the transmembrane helical cores responsible for the ion channel activity of BCL-XL40 and in similar approaches also for BAX.41 42 This provides an alternative role for this residue beyond protein interaction.
Oncogenes that promote proliferation contribute to cancer through gain-of-function alterations, whereas growth-inhibitory tumor-suppressor genes contribute principally through loss-of-function mutations. The gain-of-function alteration of the BCL-2-Igtranslocation overexpressed the anti-apoptotic molecule BCL-2 in follicular lymphoma. This suggested that pro-apoptotic molecules could contribute to oncogenesis by loss-of-function mutations. The discovery of BAX mutations in a subset of colon carcinomas and in hematopoietic malignancies here provide such evidence. This adds evidence in human tumors to the prospective experiments usingBax-deficient mice. The loss of BAX function would confer resistance to programmed cell death within hematopoietic cells and could contribute to malignancy in several ways. Extended cell survival and resistance to apoptosis would enable cells to withstand additional genetic alterations. In this context, loss of BAX function could be a primary oncogenic aberration for which the Bax-deficient mice provide evidence. BAX mutations could also contribute to tumor progression or the establishment of cell lines. Finally, chemotherapy could have selected for the loss of BAX as BAX deficiency would confer chemoresistance.
ACKNOWLEDGMENT
The authors thank Stephen Elledge for the gift of the pACTII vector. The pBTM116 vector was constructed by Paul Bartel and Stanley Fields. We thank Mary Pichler for secretarial assistance.
Address reprint requests to Stanley J. Korsmeyer, MD, Division of Molecular Oncology, Howard Hughes Medical Institute, Washington University School of Medicine, 660 S Euclid, Box 8022, St Louis, MO 63110.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal