Abstract
Deletions on the long arm of chromosome 6 are frequently found in acute lymphoblastic leukemia (ALL) and non-Hodgkin's lymphomas (NHL). We have used polymerase chain reaction analysis to study loss of heterozygosity of 16 microsatellite markers on chromosome 6 in 74 ALL and 54 NHL patients. Our results show that deletions of 6q in ALL are more frequent than what has been reported in previous studies, occurring in at least 32% of the patients. The corresponding figure for NHL patients is 7%. Our results define a region of minimal deletion in ALL of less than 500 kb between markers D6S1709 and D6S434. The common region of deletion in NHL is located telomeric of this region. Thus, two different tumor suppressor genes on chromosome 6q seem to be relevant for the development of lymphoid malignancies.
NONRANDOM CHROMOSOMAL deletions reflect loss of genetic material and suggest the presence of tumor suppressor genes that are inactivated during the process of malignant transformation. Chromosomal deletions on the long arm of chromosome 6 have been found in a variety of human malignancies including malignant melanoma,1 renal cell carcinoma,2 salivary gland adenocarcinoma,3 ovarian carcinoma,4 and in hematologic malignancies.
In hematologic malignancies, deletions on chromosome 6 occur most frequently in acute lymphoblastic leukemia (ALL) and non-Hodgkin's lymphomas (NHL). Cytogenetic studies have shown deletions in 4% to 13% in ALL5 and in 13% to 15% in various subtypes of NHL.6-8 Previous studies of loss of heterozygosity (LOH) have shown LOH in 7% to 13% of ALL9,10 and in 22% of NHL.11
In this study we have analyzed LOH in polymorphic microsatellite markers on chromosome 6 using polymerase chain reaction (PCR). We have screened 128 patients with ALL and NHL. Our results show a higher frequency of chromosome 6 deletions in ALL than has been reported in previous studies and define a region of minimal deletion localized at D6S283. Also, our results suggest that a different region located more telomeric is deleted in NHL.
MATERIAL AND METHODS
Patients.
Seventy-four children with diagnoses of ALL and 54 patients with NHL were investigated. The distribution of immunphenotypes among the ALL samples were 58 pre-B ALL, 14 T-ALL, and 2 clones with B-ALL. In the NHL group, 32 patients had chronic lymphocytic leukemia (CLL), 10 had centroblastic-centrocytic NHL, 3 had hairy-cell leukemia, and 3 patients had high-grade NHL. Diagnosis was made on bone marrow (BM) and/or lymphnode biopsies and in most cases confirmed with flow cytometry.
Cell separations and DNA extraction.
ALL samples were collected from peripheral blood or BM and mononuclear cells were seperated on a Lymphoprep gradient (Nycomed, Oslo, Norway). In cases where fluorescence-activated cell sorter (FACS) analysis showed a significant number of normal cells, malignant cells were purified by positive or negative panning using monoclonal antibodies for CD7, 10, or 19.12 CLL cells collected from peripheral blood were separated on Lymphoprep. T and B cells were separated through erythorocyte (E)-rosette sedimentation or by separation on nylon wool.13 The remaining NHL samples were collected from BM or lymphonode biopsies. After these procedures the cell populations were greater than 90% pure for ALL and greater than 95% pure for NHL.
Normal DNA was obtained from peripheral blood in cases where FACS analysis showed no malignant cells in blood, or from peripheral blood or BM after remission had been obtained. In some cases malignant cells were excluded from peripheral blood by negative panning followed by a 2-week culture of normal T cells as previously described.14For CLL cases, normal cells were also obtained from granulocytes or from T cells after E-rosette separation. DNA was extracted according to standard procedures as previously described.12
PCR analysis.
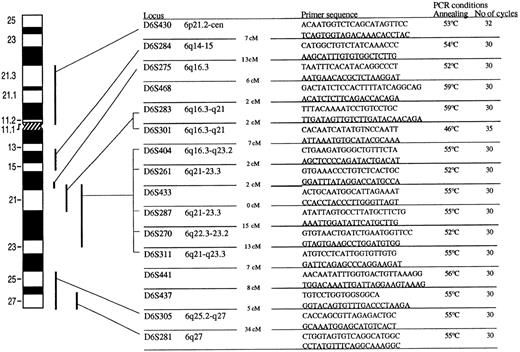
Twenty microsatelite markers from chromosome 6 were used to screen the patients for 6q deletions. The PCR conditions and sequences for PCR primers are shown in Fig 1.One of the primers was labeled with γ35S-dATP by T4 polynucleotide kinase. After PCR amplification the products were electrophoresed on 6% denaturing polyacrylamide gel. The gels were transferred to Whatman 3MM paper (Maidstone, UK), dried, and autoradiographed on radiograph film.
Primarsequence and cytogenetic location of 16 microsatellite markers used in the study. The distance in centimorgans between the markers is indicated. The PCR conditions included denaturation in 95° for 5 minutes. Denaturation temperature was always 94°C for 30 to 60 seconds. Annealing time was 30°C and the temperature is shown for every microsatelite marker. Elongation temperature was 72°C for 30 to 40 seconds. The number of cycles is shown for every marker. Amplification was always concluded with 10-minute final extension at 72°C. Marker D6S15433 and D6S1709, located between D6S468 and D6S283, and marker D6S434 and D6S1028, located between D6S283 and D6S301, are not shown in the figure.
Primarsequence and cytogenetic location of 16 microsatellite markers used in the study. The distance in centimorgans between the markers is indicated. The PCR conditions included denaturation in 95° for 5 minutes. Denaturation temperature was always 94°C for 30 to 60 seconds. Annealing time was 30°C and the temperature is shown for every microsatelite marker. Elongation temperature was 72°C for 30 to 40 seconds. The number of cycles is shown for every marker. Amplification was always concluded with 10-minute final extension at 72°C. Marker D6S15433 and D6S1709, located between D6S468 and D6S283, and marker D6S434 and D6S1028, located between D6S283 and D6S301, are not shown in the figure.
Sharply descreased intensity or abscence of one allele was interpreted as LOH. Cases with decreased intensity but not complete loss of one allele in all microsatellite markers were not evaluated as LOH.
Southern blot analysis and DNA probes.
Southern blotting and hybridization was performed as previously described.13 Scanning densitometry was used to quantitate the band intensity. The 149b19 probe obtained from UK Human Genome Mapping Project Resource Centre was assigned immediately centromeric to the D6S468 marker on chromsome 6. The BCL-1 probe located at 11q13 was used as a control of DNA quantity.
Chromosome analysis.
RESULTS
ALL.
LOH of at least one microsatellite marker on chromosome 6 was found in 24 of 74 analyzed patients (32%) and the distribution was similar in the different immunophenotypic ALL groups. Six patients had LOH in every evaluable mircrosatellite marker, suggesting loss of one complete chromosome. The 18 patients with limited deletions on chromosome 6 are shown in Fig 2. A region of minimal deletion (RMD) was found involving marker D6S283. The RMD was limited by marker D6S1709 on the centromeric side (retained in patients 11, 26, 33, 55, and 66) and by marker D6S434 on the telomeric side (retained in patients 11, 20, and 66). The distance between the markers has been estimated to be less than 1 cM.16
Eighteen ALL patients with LOH on chromosome 6. Six ALL patients with LOH in very evaluable marker on chromosome 6 are not shown in the figure. Filled box (▪) indicates retention of two alleles. White box (□) indicates LOH. Diagonally striped box (▧) indicates that the marker was not informative and reversed stripes (▨) that the marker was not evaluable or not done. MSI indicates microsatellite instability.
Eighteen ALL patients with LOH on chromosome 6. Six ALL patients with LOH in very evaluable marker on chromosome 6 are not shown in the figure. Filled box (▪) indicates retention of two alleles. White box (□) indicates LOH. Diagonally striped box (▧) indicates that the marker was not informative and reversed stripes (▨) that the marker was not evaluable or not done. MSI indicates microsatellite instability.
In one patient (no. 13) with LOH from marker D6S275 to D6S301, there was also an LOH in marker D6S305 with retention of two alleles in at least five intervening microsatellite markers.
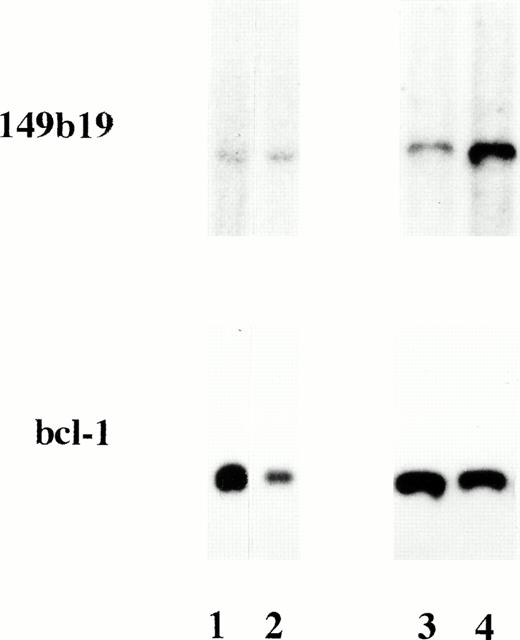
To confirm LOH data, Southern blot hybridizations using DNA from two ALL patients with 6q LOH and one case without detectable LOH were performed. The 149b19, located immediately centromeric of the RMD, showed hemizygous deletions, detected as a reduction of signal intensity by approximately 50%, in the two ALL patients with LOH, whereas no deletion was detected in the patient without LOH (Fig3).
Autoradiographs of Msp I—digested DNA, hybridized to the 149b19 probe on chromosome 6 and aBCL-1 probe. ALL patient 56 (lane 1) and patient 64 (lane 3) had LOH on chromosome 6 in the PCR analysis. Lane 2 contains normal DNA and lane 4 contains DNA from an ALL patient without LOH. There is a reduced signal in DNA from patients 56 and 64, indicating a hemizygous deletion of the 149b19 marker.
Autoradiographs of Msp I—digested DNA, hybridized to the 149b19 probe on chromosome 6 and aBCL-1 probe. ALL patient 56 (lane 1) and patient 64 (lane 3) had LOH on chromosome 6 in the PCR analysis. Lane 2 contains normal DNA and lane 4 contains DNA from an ALL patient without LOH. There is a reduced signal in DNA from patients 56 and 64, indicating a hemizygous deletion of the 149b19 marker.
NHL.
Four out of 54 patients (7%) had LOH shown by microsatelite analysis (Fig 5). Two patients, L3 with a high-grade follicular B-cell lymphoma and L4 with a high-grade T-cell lymphoma, had limited deletions extending from marker D6S301 and D6S261, respectively. Both patients retained marker D6S301, which was found to be the border for the RMD in ALL. In patients L1 and L2, both with CLL, the microsatelite analysis showed extensive deletions on chromosome 6. Patient L2 retained the most distal marker D6S281. These patients were evaluated cytogenetically showing del(6)(q16-21) in patient L1 and del(6)(q21) in patient L2. An additional CLL patient showed an aberrant chromosome 6 with a translocation t(6;6)(q23;q27), but the microsatellite analysis did not reveal any LOH.
Four NHL patients with LOH on chromosome 6. Filled box (▪) indicates retention of two alleles. White box (□) indicates LOH. Diagonally striped box (▧) indicates that the marker was not informative or reverse stripes (▨) that the marker was not evaluable or not done.
Four NHL patients with LOH on chromosome 6. Filled box (▪) indicates retention of two alleles. White box (□) indicates LOH. Diagonally striped box (▧) indicates that the marker was not informative or reverse stripes (▨) that the marker was not evaluable or not done.
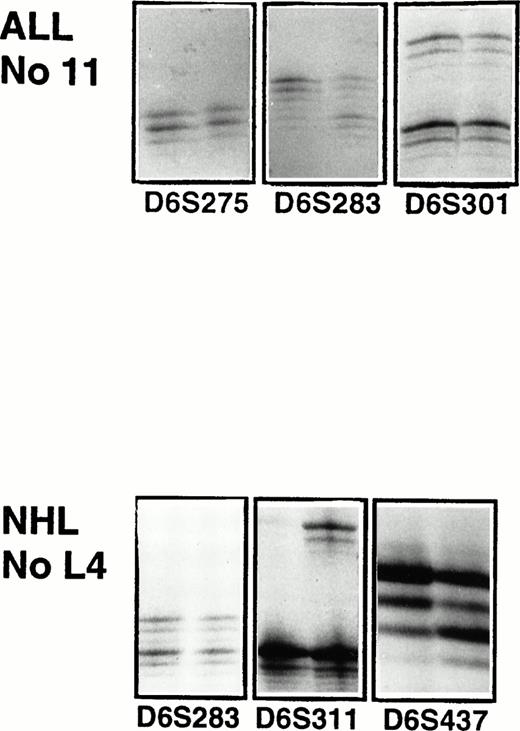
Microsatellite analysis of patient 11 with ALL and patient L4 with high-grade T-cell NHL. For each microsatellite marker the PCR products of the malignant sample is shown to the left and of the normal sample to the right. For ALL patient 11, there is LOH in marker D6S283. Patient L4 has a LOH in marker D6S311 and D6S437.
Microsatellite analysis of patient 11 with ALL and patient L4 with high-grade T-cell NHL. For each microsatellite marker the PCR products of the malignant sample is shown to the left and of the normal sample to the right. For ALL patient 11, there is LOH in marker D6S283. Patient L4 has a LOH in marker D6S311 and D6S437.
DISCUSSION
Our study shows that deletions on chromosome 6 are a more frequent phenomenon in ALL than has been reported in previous studies. Using PCR of microsatelite markers we found deletions in 32% of analyzed cases. The deletions clustered around marker D6S283 and our results indicate a minimal region of deletion limited by marker D6S1709 and D6S434. The cytogenetic localization of D6S283 is 6q16-q21. In the published radiation hybrid map for chromosome 6, marker D6S283 and D6S434 are localized on the same fragment, indicating that they are closely related. Also, markers D6S1709 and D6S283 are localized on the same PI-derived artificial chromosome (PAC) clone, suggesting the distance between these two markers to be less than 100 kb. These data suggest that the minimal region of deletion defined by this study does not exceed 500 kb. This region is likely to harbour a tumor suppressor gene relevant for ALL.
Most published studies are based on cytogenetic analysis which is often complicated in ALL because of poor yield of mitoses and fuzzy-appearing chromosomes. The frequency of 6q deletions in cytogenetic studies has been 4% to 14%.5,6,17 Our study, using PCR to overcome the problems associated with cytogenetic analysis of ALL, shows that deletions of chromosome 6 are more common. The PCR method is based on an evaluation of signal intensity of heterozygous alleles and one possible explanation for alteration in signal intensity could be extra copies of chromosome 6. In the patients with deletions in this study this possibility could be excluded, as a result of the fact that the LOH did not include all of chromosome 6, or of the complete or almost complete disappearance of one of the bands. In previous LOH studies of childhood ALL, deletions of 6q have been found in 7% to 13% of the patients.9 10
Patient 13 had LOH in two separate regions with retention of two alleles in the intervening microsatellite markers. The larger deletion included the common region of deletion around marker D6S283. The second deleted region could be a result of genetic instability rather than an indication of a second cluster for deletions on chromosome 6 in ALL.
In the NHL group, deletions were found in 4 out of 54 patients (7%). In two high-grade lymphomas, the deleted markers were located telomeric on 6q, and two alleles were retained of the microsatellite markers in the RMD found in ALL. However, in two CLL patients there were extensive deletions that were not distinctly separated from the RMD discovered in ALL. These results suggest that there is at least one additional tumor suppressor gene located on chromosome 6 that is inactivated during the development of NHL entities.
In previous reports based on cytogenetics, the incidence of abnormalities on chromosome 6 has been 13% to 15% in various subtypes of NHL.6-8 In CLL, deletions on chromosome 6 seem to be less frequent than in other NHL entities, occurring in 4% of CLL cases.18 Molecular studies have shown 6q deletions in 19% of acquired immunodeficiency syndrome–related NHL19 and in 22% of other NHL entities.11
The low frequency of 6q deletions in this series can partly be explained by the large proportion of CLL patients (60%). However, another possibility could be that chromosome 6 deletions are limited to a subclone of the malignant cells and that the number of cells with two alleles of the microsatellite markers thus will be amplified using a PCR method.
In previous cytogenetic and molecular studies it has been possible to associate specific histological subtypes with specific deleted regions on 6q. Deletions in 6q21 are associated with high-grade lymphomas. Deletions of 6q25-27 are associated with intermediate-grade lymphomas. Additionally, deletions in 6q23 are commonly found in low-grade NHL without t(14;18).11,20 It has been suggested that the cluster of deletions in 6q21 frequently found in high-grade lymphomas overlaps to the deleted region found in ALL.11 21 However, in our material the two high-grade lymphomas were found to have deletions in a region distinctly separated from the RMD found in ALL.
In a recent study using micro—fluorescence in situ hybridization it was shown that what seem to be terminal 6q deletions by cytogenetic analysis can be interstitial deletions or nonreciprocal translocations.22 A new RMD at 6q11 was suggested for follicular lymphomas. Our study supports the finding that deletions on 6q can be misinterpreted on cytogenetic analysis. In patients L1 and L2 the cytogenetic analysis indicated terminal deletions with breakpoints at 6q16-21 and 6q21, respectively. However, the PCR analysis indicated that patient L2 had an interstitial deletion from marker D6S283 (6q16.3-21) to D6S270 (6q22.3-23.3) and retention of two alleles in D6S281 (6q27). In patient L1 there was LOH in all evaluable markers on 6q on PCR, indicating a more extensive loss than what was revealed by cytogenetics.
This study defines a region of less than 500 kb on chromosome 6 that is likely to harbor a tumor suppressor gene relevant for the development of ALL. We also confirm that a second region of chromosome 6 seems to be deleted in NHL. However, in NHL the picture is still complex and more molecular studies are needed to clarify what the recurrent breaks on 6q represent.
ACKNOWLEDGMENT
The skillful technical assistance of Elisabeth Anderbring, Birgitta Stellan, and Kristina Friberg is gratefully acknowledged.
Address reprint requests to Mats Merup, MD, PhD, Department of Medicine, Karolinska Institute at Huddinge University Hospital, 141 86, Huddinge, Sweden.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal