Abstract
CD44 is a ubiquitous cell-surface glycoprotein that displays many variant isoforms (CD44v) generated by alternative splicing of exons 2v to 10v. The expression of variant isoforms is highly restricted and correlated with specific processes, such as leukocyte activation and malignant transformation. We have herein studied CD44v expression in acute myeloid leukemia (AML) and, for comparison, in normal myelopoiesis. Protein expression of total CD44 and of CD44-3v, -6v, and -9v isoforms has been measured using specific monoclonal antibodies and flow cytometry. The composition of variant exon transcripts has been analyzed by semi-quantitative reverse transcriptase-polymerase chain reaction followed by Southern hybridization with exon-specific probes. Our data show that (1) CD44-6v isoforms are expressed on 12.0% ± 2.5% of normal CD34+ cells; this expression is sharply upregulated through monopoiesis and, inversely, downregulated during granulopoiesis. Also, CD44-3v and CD44-9v isoforms are detected on 10% and 14% of normal monocytes, respectively. (2) Sixty-nine from a total of 95 AML patients display a variable proportion (range, 5% to 80%) of CD44-6v+ leukemic cells. (3) A shorter overall survival characterizes the group of AML patients displaying more than 20% of CD44-6v+ leukemic cells (8 months v 18 months, P < .02). These data suggest, for the first time, that the protein expression of CD44-6v containing isoforms may serve as a new prognostic factor in AML.
THE CD44 ANTIGEN is a highly glycosylated transmembrane protein encoded by a unique 20-exon gene on chromosome 11 in humans.1 It displays multiple isoforms. The so-called standard isoform CD44s (90 kD) is the most common one and is encoded by 10 standard exons (1s to 10s). A substantial number of so-called variant isoforms, CD44v (110 to 250 kD), have also been described.2 They are generated by the alternative splicing of nine variant exons (2v to 10v) in multiple combinations, and characterized by additional sequences inserted into the extracellular domain of the molecule.3
CD44 is thought to play an important role in myelopoiesis because anti-CD44 monoclonal antibodies (MoAbs) fully inhibit in vitro long-term hematopoiesis on pre-established stroma.4,5 In addition, it has been shown that CD44 mediates the adhesion of CD34+ hematopoietic progenitors (HPC) to hyaluronan,6-8 its best-known ligand,9-11 which is present in the hematopoietic extracellular matrix.7 12-15
The standard CD44 isoform is expressed on all types of mature blood cells,11,16,17 on the majority of mononuclear bone marrow (BM) precursors,18,19 and on all CD34+HPC.20,21 The level of its expression varies according to hematopoietic cell lineage and stage of differentiation. For example, it is high on monocytic cells, intermediate on polymorphonuclear cells (PMN) and on CD34+ HPC, and low on erythroid cells and platelets. The variant isoforms CD44-6v and CD44-9v have been detected on monocytes, macrophages, lymphocytes, and dendritic cells.2,22-27 Inflammation, mitogens, and inflammatory cytokines have been shown to upregulate CD44-3v, 6v, and CD44-9v expression in monocytes and lymphocytes.22-26,28-30As far as myeloid cells are concerned, only CD44-10v has been shown on a few human BM myeloid precursors.19 The expression of CD44-6v has been also recently reported on HPC in the rat.31
Several CD44 variant isoforms are overexpressed in malignant hematopoietic cells. For example, CD44-6v or CD44-9v isoforms are upregulated in non-Hodgkin's lymphoma (NHL) and myeloma,26,27,29,32 and CD44-10v expression is enhanced in chronic myeloid leukemia (CML) and acute myeloid leukemia (AML).19,33 Moreover, the expression of CD44-6v and CD44-9v correlates with an unfavorable clinical evolution in lymphoma and myeloma,27 32 suggesting a role of these CD44 variant isoforms in hematopoietic malignancies.
In the present study our aim has been to evaluate the role of CD44 variant isoforms in the pathophysiology and clinical evolution of AML. AML is a heterogenous disease, characterized by the accumulation of leukemic cells in BM and peripheral blood (PB). These cells display cytological and antigenic features of immature granulocytic and/or monocytic cells. On the basis of these features, AML has been classified into distinct subtypes: myeloblastic (M1/M2), promyelocytic (M3), myelomonocytic (M4), and monoblastic (M5).34 35
We have analyzed the expression of 3v-, 6v-, and 9v-containing CD44 isoforms on circulating leukemic cells using flow cytometry; we have determined the exon composition of these isoforms by reverse transcriptase-polymerase chain reaction (RT-PCR); and we have correlated the results with clinical characteristics. For comparison we have also analyzed the CD44v expression pattern in normal myeloid cells at various stages of maturation. We show that in normal subjects the expression of CD44-6v isoforms is restricted to CD34+ cells and to monocytic cells. We also show that CD44-6v is expressed on AML leukemic cells and, most importantly, that its expression level is correlated with the overall survival of AML patients treated by conventional chemotherapy.
MATERIALS AND METHODS
AML Patients and Clinical Data
Leukemic cells from 95 patients with AML were collected at diagnosis (82 patients) or at relapse (13 patients) from the following hospitals: Leiden University Hospital (Leiden, The Netherlands; 45 samples), Hotel-Dieu (Paris, France; 21 samples), Henri Mondor (Creteil, France; 13 samples), The Royal Victoria Infirmary (Newcastle, UK; 9 samples), Hôpital Paul-Brousse (Villejuif, France; 3 samples), Hôpital St Louis (Paris, France; 2 samples), and Institut Gustave-Roussy (Villejuif, France; 2 samples). The main clinical features of these patients are summarized in Table 1. All patients displayed more than 60% blasts in PB. The patients' treatments are indicated in Table 1. They include (1) conventional induction chemotherapy,36 ie, administration of anthracycline plus cytarabine, which may be followed by allograft, and (2) palliative chemotherapy. Two M3 patients also received all-trans retinoic acid. The patients' survival was followed-up for at least 2 years after complete remission.
Clinical Characteristics of AML Patients
| AML FAB Type . | No. of Patients . | Age in Years Median (range) . | Sex (male/ female) . | WBC/μL Median (range) . | Chemotherapy of De Novo AML Patients . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conventional-150 . | Conventional-150 + Allograft . | Palliative . | Undetermined No. . | |||||||||
| Total . | De Novo . | No. . | CR . | No. . | CR . | No. . | CR . | |||||
| Myeloblastic (M1/M2) | 51 | 44 | 51 (18-83) | 26/25 | 40 (4-417) | 28 | 18 | 3 | 2 | 5 | 0 | 8 |
| Promyelocytic (M3) | 8 | 8 | 39 (22-72) | 4/4 | 20 (2-85) | 4 + 2 Patients also treated with ATRA | 3 | 1 | 1 | 0 | 0 | 1 |
| Myelomonocytic (M4) | 13 | 9 | 49 (14-62) | 10/3 | 52 (8-166) | 7 | 4 | 2 | 1 | 0 | 0 | 0 |
| Monoblastic (M5) | 17 | 17 | 39 (15-70) | 8/9 | 107 (9-242) | 9 | 4 | 4 | 3 | 4 | 0 | 0 |
| Undefined or M7 | 6 | 4 | 37 (33-60) | 3/3 | 7 (0.9-190) | 3 | 1 | 0 | 0 | 1 | 0 | 0 |
| Total | 95 | 82 | 48 (14-83) | 51/44 | 47 (0.9-417) | 53 | 30 | 10 | 7 | 10 | 0 | 9 |
| AML FAB Type . | No. of Patients . | Age in Years Median (range) . | Sex (male/ female) . | WBC/μL Median (range) . | Chemotherapy of De Novo AML Patients . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conventional-150 . | Conventional-150 + Allograft . | Palliative . | Undetermined No. . | |||||||||
| Total . | De Novo . | No. . | CR . | No. . | CR . | No. . | CR . | |||||
| Myeloblastic (M1/M2) | 51 | 44 | 51 (18-83) | 26/25 | 40 (4-417) | 28 | 18 | 3 | 2 | 5 | 0 | 8 |
| Promyelocytic (M3) | 8 | 8 | 39 (22-72) | 4/4 | 20 (2-85) | 4 + 2 Patients also treated with ATRA | 3 | 1 | 1 | 0 | 0 | 1 |
| Myelomonocytic (M4) | 13 | 9 | 49 (14-62) | 10/3 | 52 (8-166) | 7 | 4 | 2 | 1 | 0 | 0 | 0 |
| Monoblastic (M5) | 17 | 17 | 39 (15-70) | 8/9 | 107 (9-242) | 9 | 4 | 4 | 3 | 4 | 0 | 0 |
| Undefined or M7 | 6 | 4 | 37 (33-60) | 3/3 | 7 (0.9-190) | 3 | 1 | 0 | 0 | 1 | 0 | 0 |
| Total | 95 | 82 | 48 (14-83) | 51/44 | 47 (0.9-417) | 53 | 30 | 10 | 7 | 10 | 0 | 9 |
Abbreviations: FAB, French-American-British classification of AML34,35; CR, complete remission. CR has been determined by cytological examination of BM aspirate.
Conventional chemotherapy refers to treatment with anthracycline and cytarabine.36 Two from the 6 M3 patients have been also treated with ATRA (all-trans retinoic acid).44These two patients have not been considered for the disease-free survival studies. The same was for patients with undefined or M7 AML.
Isolation of Leukemic Cells
Low-density mononuclear cells from PB were separated by density gradient centrifugation on Ficoll-hypaque density 1.077 g/mL (Pharmacia LKB, Uppsala, Sweden) and resuspended in RPMI 1640 plus 10% fetal calf serum (FCS). Preliminary experiments showed that cell freezing and thawing did not change the antigenic profile of these cells. Therefore, AML low-density mononuclear cells were cryopreserved in 25% FCS plus 10% dimethylsulfoxide in RPMI 1640 culture medium, and stored in liquid nitrogen until use. Four hours before CD44 analysis, cells were thawed at room temperature in RPMI 1640 culture medium containing 50% FCS, then washed twice in RPMI 1640 plus 2% FCS. B and T lymphocyes were removed from all samples by specific immunoadsorption on Dynabeads (Dynal, Oslo, Norway) coated with MoAbs to CD2 and CD19, according to the manufacturer's recommendations. Monocytes were removed from myeloblastic and promyelocytic AML (M1 to M3), using Dynabeads coated with MoAb to CD14. Cell suspensions containing more than 95% of AML blasts were thus obtained. The absence of CD2+ and CD19+ cells (and of CD14+ cells in M1 to M3 AML) was verified by flow cytometry (Fig1).
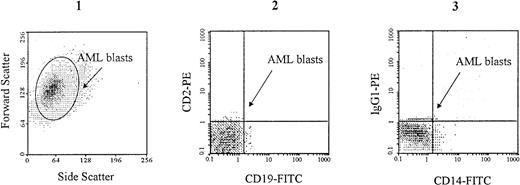
Specific gating of AML blasts. Cell suspensions containing more than 95% of AML blasts were prepared by removing CD2+ CD19+ cells (lymphocytes) using specific immunoadsorption on Dynabeads coated with specific MoAbs as described in Materials and Methods. In the example shown here, which is from an M1 AML patient, CD14+ monocytes have also been removed. AML blasts, which are characterized by large forward and side scattering values37 (1), are gated in the CD2neg, CD19neg (2) and CD14neg (3) windows.
Specific gating of AML blasts. Cell suspensions containing more than 95% of AML blasts were prepared by removing CD2+ CD19+ cells (lymphocytes) using specific immunoadsorption on Dynabeads coated with specific MoAbs as described in Materials and Methods. In the example shown here, which is from an M1 AML patient, CD14+ monocytes have also been removed. AML blasts, which are characterized by large forward and side scattering values37 (1), are gated in the CD2neg, CD19neg (2) and CD14neg (3) windows.
Isolation of Normal Myeloid Cells
BM and circulating myeloid cells from distinct lineages and at different stages of maturation are intermingled. Therefore, for both RT-PCR and fluorescence-activated cell sorter (FACS) analysis, populations enriched with CD34+ cells and granulocytic precursors were isolated from BM; because circulating mature monocytes and PMN could be easily identified on the basis of specific cell-surface antigen expression (CD14 and CD15, respectively) and light-scattering properties,18 they were directly labeled and analyzed by flow cytometry, as detailed below; they were isolated only for RT-PCR studies. Monocytic precursors were too scarce for being isolated as a pure population for RT-PCR and therefore they were only analyzed by flow cytometry (see section “Flow Cytometric Analysis”).
Normal Immature Hematopoietic Cells (CD34+ cells) and CD34neg Granulocytic Precursors (GP)
Samples from normal BM were obtained following informed consent from donors receiving hip surgical intervention (Dr J. de Thomasson, Clinique Chirurgicale de Choisy, Paris, France) or BM cell donors for allograft (Dr J.L. Pico, Institut Gustave Roussy, Villejuif, France). Normal donors comprised three females and two males with age ranging from 42 to 75 years, except for one 1-year-old child. In the case of donors with hip surgical intervention, hematopoietic cells were extracted from normal BM by grounding the trabeculae with a potter and vortexing the samples several times in phosphate-buffered saline (PBS). Light-density mononuclear cells were separated by density gradient centrifugation on Ficoll-hypaque density 1.077 g/mL (Pharmacia LKB), resuspended in RPMI 1640 plus 10% FCS and processed for the isolation of two distinct cell populations, respectively, enriched in CD34neg granulocytic precursors and CD34+cells. Mature monocytes were removed by adhesion on plastic for 2 hours at 37°C and after two washes with PBS. The most differentiated precursors and stromal cells were discarded by adhesion on soy bean agglutinin CELLector flasks (Applied Immune Sciences Inc, Menlo Park, CA).
CD34+ cells were recovered by adhesion on ICH3 anti-CD34 MoAb-coated CELLector flasks (Applied Immune Sciences Inc), according to the manufacturer's instructions. Flow cytometric analysis indicated that all samples contained more than 95% CD34+ cells. The nonadherent CD34neg cell population was highly enriched in immature granulocytic cells by incubation with Dynabeads coated with MoAbs to CD2, CD19, CD14, and CD71 for depleting lymphocytes, monocytic cells, and erythroblastic cells. Microscopic observation of May-Grünwald Giemsa–stained cytosmears indicated that this cell population comprised 3% ± 1% myeloblasts, 21% ± 2% promyelocytes, 34% ± 4% myelocytes, and 35% ± 3% metamyelocytes. It was designated as CD34neg granulocytic precursors (CD34neg GP). We confirmed by flow cytometry that the purification procedure did not alter the expression level of CD44.
Normal PMN and Monocytes
Samples of heparinized PB were obtained from five informed and consenting adult donors. To recover PMN, the blood sample was first diluted with PBS containing 3% gelatin (Plasmion; Rhone-Poulenc, Lyon, France), with a ratio of 4:1 (vol/vol); the red blood cells were pelleted by low-speed centrifugation (50g for 1 minute, at 20°C), and the nucleated cells, recovered in the upper phase, were layered on Ficoll-hypaque density 1.077 g/mL (Pharmacia LKB) and centrifuged at 500g for 30 minutes, at 20°C. The pellet comprised more than 98% granulocytes. For isolating monocytes, light-density mononuclear cells were recovered by centrifugation on Ficoll-hypaque as described above and suspended in RPMI 1640 medium plus 10% FCS (5 × 106 cells/mL). Monocytes were collected by adhesion for 2 hours on plastic (T-flask Costar, Cambridge, MA), at 37°C with 5% CO2 in air. At least 90% of plastic adherent cells were shown to express the monocyte-specific antigen CD14.
MoAbs
The MoAb F10-44-2 (IgG2a),22 which is directed at an epitope in the CD44 common region, cannot discriminate between CD44s and CD44v-containing isoforms. Its binding reflects the total amount of CD44 on the cell surface. It was conjugated to phycoerythrin (PE) and used at 5 μg/mL; this MoAb was provided by Serotec (Oxford, UK). MoAbs directed at CD44 variant epitopes were specific for 3v (BBA11, IgG2b), 6v (VFF-18, IgG1), and 9v (FW11-24, IgG1). BBA11 was obtained from R&D SystemsEurope (Abingdon, UK) and used at 1 μg/mL. VFF-18 was purchased from Bender MedSystems (Vienna, Austria) and used at 10 μg/mL, and FW11-24 was a hybridoma supernatant,22 used undiluted. Fluorescein isothiocyanate (FITC)-conjugated MoAb to CD14 (IgG2b, 5 μg/mL) provided by Coulter (Coulter Immunology, Hialeah, FL) was used to identify monocytic cells. FITC-conjugated anti-CD15 MoAb (IgM, 1 μg/mL; Becton Dickinson, San Jose, CA) was used to characterize cells from the granulocytic lineage. Anti-CD34 MoAb (HPCA-2, IgG1, 1 μg/mL) conjugated to FITC was obtained from Becton Dickinson. Antibodies were used at saturating concentrations. Murine IgG1 and IgG2a conjugated with either FITC or PE were from Immunotech (Marseille, France). PE-conjugated goat F(ab′)2 anti-mouse IgG (H + L) (PE-GAM) was purchased from Southern Biotechnologies Associates (Birmingham, AL) and used in a 1:50 dilution.
Immunofluorescent Staining and Flow Cytometry of Normal and Leukemic Cells
Gating of normal and leukemic cells.
PMN and mature monocytes were stained directly in the heparinized blood samples, after red blood cells lysis using 0.8% ammonium chloride, and they were identified by specific electronic gating18 and on the basis of specific cell-surface antigen expression (CD15 and CD14, respectively). CD34+ cells were isolated as described and identified by staining with an FITC-conjugated MoAb directed to CD34, as well as by specific scatter features. Monocytic precursors were identified in the CD34neg BM population by staining with an FITC-conjugated MoAb to CD14, and by specific scatter features.18 Granulocytic precursors were isolated in the CD34neg BM population as described above, and identified by staining with an FITC-conjugated MoAb to CD15, and by specific scatter features.18 Suspensions containing more than 95% of AML blasts were prepared. As described above, these blasts were characterized both by specific scatter parameters37 and by a CD2neg, CD19neg phenotype, as shown in Fig 1. In addition, in myeloblastic (M1/M2) and promyelocytic (M3) AML, leukemic blasts were also characterized by a CD14negphenotype (Fig 1).
Immunofluorescent staining and flow cytometry.
For labeling, cells were suspended in RPMI 1640 containing 0.2% bovine serum albumin (BSA) and 0.02% NaN3 (label medium) at 106 cells/mL; incubated at 4°C for 30 minutes with 5 μg/mL PE-conjugated MoAb F10-44-2, which labeled both CD44s and CD44v (total CD44); and washed twice with label medium. CD44v expression was analyzed after successive incubation with 5% normal human serum (AB type), unconjugated MoAb, 5% goat serum, and GAM-PE. The MoAb binding was measured by flow cytometry relative to isotype-matched control antibodies, using a Profile II flow cytometer (Coulter Immunology) equipped with a 15-mW argon laser and calibrated using a panel of fluorescent beads (Immunobrite; Coulter, Hialeah, FL). Under these conditions, the fluorescence intensity of total CD44 on HL60 and MO7e cells, respectively, used as negative and positive controls for variant expression, was constant. Adjustment of the crossover fluorescence was obtained by compensation of the two single-stained samples to limit superposition of the fluorochrome emission spectra. Each measurement was performed on 5,000 cells.
The staining intensity of total CD44 and CD44 variant isoforms was measured as the relative mean fluorescence intensity (MFI) compared with isotypic controls. The relative MFI of CD44v was much lower than the one of total CD44 (maximum of 6). For this reason, the amount of CD44v isoforms was also evaluated by the percentage of labeled cells over the background (cells incubated with isotype-matched control antibodies). It was considered as negative (−) when less than 5% of the cells were labeled, positive (+) for 5% to less than 20% of labeled cells (similar to normal CD34+ cells, see Results), and highly positive (++) when more than 20% of the cells were CD44v+ (similar to normal monocytes, see Results).
CD34+CD44-6v+ and CD34+CD44-6vneg cells were sorted on a FACSvantage (Becton Dickinson) equipped with an INNOVA70-4 Argon ion laser (Coherent Radiation, Palo Alto, CA) tunned at 488 nm and operating at 500 mW. Highly diffusive and/or too large objects were rejected. Positivity or negativity for the CD44-6v among the CD34+ cells was determined using control cells labeled with FITC-conjugated anti-CD34 MoAb (FITC-HPCA2) and PE-conjugated IgG1. Compensation was set up with single-stained samples.
Clonogenic Features of CD34+CD44-6v+ and CD34+CD44-6vneg Cells
CD34+CD44-6v+ and CD34+CD44-6vneg cells were cultured, in triplicate, in 35-mm non–tissue culture grade Petri dishes (Falcon; reference 1008; Becton Dickinson, Plymouth, UK) containing 1 mL of complete methylcellulose medium purchased from StemCell Technologies Inc (Vancouver, British Columbia, Canada) (0.8% methylcellulose in Iscove's modified Dulbecco's medium [IMDM], 30% FCS, 1% deionized BSA, 10−4 mol/L 2-mercaptoethanol). Colony-stimulating factors were provided as 5% of phytohemagglutinin-human leukocyte-conditioned medium (Hemostim H2400; StemCell Technologies Inc, Vancouver, Canada). Human erythropoietin purified from human urine was added at 3 U/mL (StemCell Technologies Inc). Plates were incubated at 37°C in a fully humidified atmosphere containing 5% CO2 in air. Colony-forming units monocytic (CFU-M), granulocytic (CFU-G), granulo/monocytic (CFU-GM), and granulocytic/erythroid/monocytic/megakaryocytic (CFU-GEMM), and burst-forming units-erythroid (BFU-E) were scored at day 16 following the standard criteria. Their number was expressed as the percentage of total colonies obtained.
Preparation of RNA and cDNA/Reverse-Transcriptase Polymerase Chain Reaction (RT-PCR)
Variant exon expression was analyzed at the RNA level on enriched cell populations, prepared as described above. Both normal myeloid cells (five samples per cell type) and AML leukemic cells from 70 patients (from all French-American-British [FAB] types) were analyzed. Total cellular RNA was prepared and semi-quantitative RT-PCR performed as described previously.27 Briefly, total cellular RNA was prepared using 105 purified cells, followed by cDNA synthesis. cDNA amounts were equilibrated semi-quantitatively for the housekeeping gene hypoxanthine phosphoribosyltransferase (HPRT) as a marker transcript. Equilibrated amounts of cDNA were taken for CD44-specific PCR amplification using primers homologous to CD44s regions close to the variant sequences. The cDNA from CD44s lacking all variant exons thus yielded a product of 67 bp. The exon-specific probes were labeled with 33P-dATP (Megaprime labeling kit; Amersham, Zürich, Switzerland), and blots were hybridized and washed under stringent conditions (0.2 × standard sodium citrate [SSC], 0.1% sodium dodecyl sulfate, 65°C). Blots were exposed to Kodak BioMax MR films (Eastman Kodak Co, Rochester, NY) at room temperature for 12 to 48 hours. Staining intensity was scored semiquantitatively on a scale of − to ++++ (−, negative; ±, faint, not clearly above background; +, weakly positive; ++, moderately positive; +++ and ++++, strongly positive).
Statistical Analysis
The mean MFI value ± 1 SE from the distinct cell populations were compared by the Kruskal-Wallis test. The comparisons two by two were performed by Student's t-test depending on the number of cases. The statistical significance between the presence of CD44 isoforms and the clinical parameters was determined by Fisher's exact test or chi-squared test depending on the number of cases. The probability of overall survival was computed according to the Kaplan-Meier method.38 Statistical comparisons between curves were based on log-rank tests. Relationship between prognostic factors was determined using Cox's proportional hazard model for covariate analysis of censored data.39 All survival analyses were performed by using the statistical package BMDP (BMDP Statistical Software, Los Angeles, CA). The correlation between the expression of CD44v isoforms and the following clinical parameters were investigated: overall survival rate (alive v deceased), rate and duration of complete remission, and rate of relapse. In addition, correlation of CD44v expression and AML subtype, age, number of circulating leukemic blasts, and percentage of CD34+leukemic cells were also analyzed. Survival rate, age, and white blood cell number per microliter (WBC/μL) were expressed as median.
RESULTS
Flow Cytometric Analysis of CD44 Isoform Expression
Normal myeloid cells.
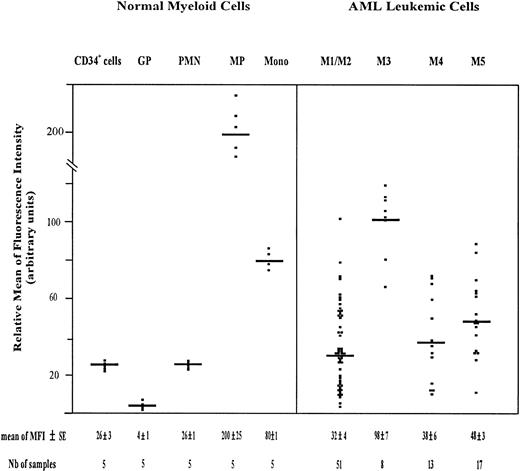
The amount of total CD44 was measured using PE-conjugated F10-44-2 MoAb. This MoAb, which is directed to an epitope in the constant domain of the molecule, binds both standard and variant CD44 isoforms. The highest expression of CD44 was observed on monocytic cells, including monocytic precursors (MP, CD34neg CD14+ cells) and mature monocytes (relative MFI = 200 ± 25 and 80 ± 1, respectively). CD34+ cells and PMN displayed also a significant amount of CD44 (relative MFI of 26 ± 3 and 26 ± 1, respectively), whereas maturing granulocytic precursors (GP, CD34neg CD15+ cells) expressed total CD44 only weakly (relative MFI = 4 ± 1) (Fig2). The GP population comprised 3% ± 1% myeloblasts, 21% ± 2% promyelocytes, 34% ± 4% myelocytes, and 35% ± 3% metamyelocytes.
Scatter plot of total CD44 expression on normal myeloid cells and on AML leukemic cells. Normal Myeloid Cells: CD34+ cells (very immature hematopoietic cells); GP, granulocytic precursors (CD34−, CD15+ BM cells); PMN, polymorphonuclear cells; MP, monocytic precursors (CD34−, CD14+ BM cells); Mono, mature monocytes (circulating CD14+ cells). AML Leukemic Cells: from AML patients with the following FAB types.34,35 M1/M2, myeloblastic AML; M3, promyelocytic AML; M4, myelomonocytic AML; M5, monoblastic AML. Mature monocytes and PMN from PB were identified according to their forward angle and side scatter.37CD34+ cells and MP were double-stained with (1) an FITC-conjugated MoAb directed to either CD34, which is specific for very immature hematopoietic cells, or CD14, a monocyte-specific antigen and (2) a PE-conjugated anti-CD44 MoAb F10-44-2, which binds to an epitope located in the constant part of the CD44 molecule. Negative controls were cells labeled with FITC and PE-conjugated isotype-matched control antibodies. The MFI was determined by flow cytometry, relative to cells labeled with PE-conjugated IgG2a (negative controls), as described in Materials and Methods. Each symbol refers to the MFI value per patient, and the horizontal bars indicate the mean MFI value in each category of patients. The normal BM cells and the leukemic cell populations were isolated as described in Materials and Methods. The gating of leukemic cells is shown in Fig 1.
Scatter plot of total CD44 expression on normal myeloid cells and on AML leukemic cells. Normal Myeloid Cells: CD34+ cells (very immature hematopoietic cells); GP, granulocytic precursors (CD34−, CD15+ BM cells); PMN, polymorphonuclear cells; MP, monocytic precursors (CD34−, CD14+ BM cells); Mono, mature monocytes (circulating CD14+ cells). AML Leukemic Cells: from AML patients with the following FAB types.34,35 M1/M2, myeloblastic AML; M3, promyelocytic AML; M4, myelomonocytic AML; M5, monoblastic AML. Mature monocytes and PMN from PB were identified according to their forward angle and side scatter.37CD34+ cells and MP were double-stained with (1) an FITC-conjugated MoAb directed to either CD34, which is specific for very immature hematopoietic cells, or CD14, a monocyte-specific antigen and (2) a PE-conjugated anti-CD44 MoAb F10-44-2, which binds to an epitope located in the constant part of the CD44 molecule. Negative controls were cells labeled with FITC and PE-conjugated isotype-matched control antibodies. The MFI was determined by flow cytometry, relative to cells labeled with PE-conjugated IgG2a (negative controls), as described in Materials and Methods. Each symbol refers to the MFI value per patient, and the horizontal bars indicate the mean MFI value in each category of patients. The normal BM cells and the leukemic cell populations were isolated as described in Materials and Methods. The gating of leukemic cells is shown in Fig 1.
The expression of variant CD44 proteins (CD44v) was investigated using variant-specific MoAbs (described in Materials and Methods).
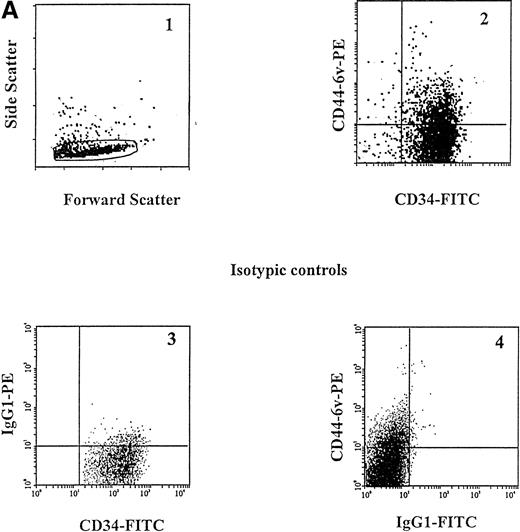
A small proportion of CD34+ cells (12.0% ± 2.5%) was found to display CD44-6v isoforms (relative MFI of 2 ± 0, Fig 3A and B, and Fig4). We sorted CD34+CD44-6v+ and CD34+CD44-6vneg cells and analyzed their clonogenic features, using a methylcellulose assay: the CD34+CD44-6v+ cell population comprised more CFU-M (44% ± 7% v 24% ± 5%, P < .05, Student's t-test) and less BFU-E (10% ± 2% v31% ± 8%, P < .05, Student's t-test), than the CD34+CD44-6vneg cells. CFU-G were in equal number in CD34+CD44-6v+ and CD34+CD44-6vneg cell populations (Table2).
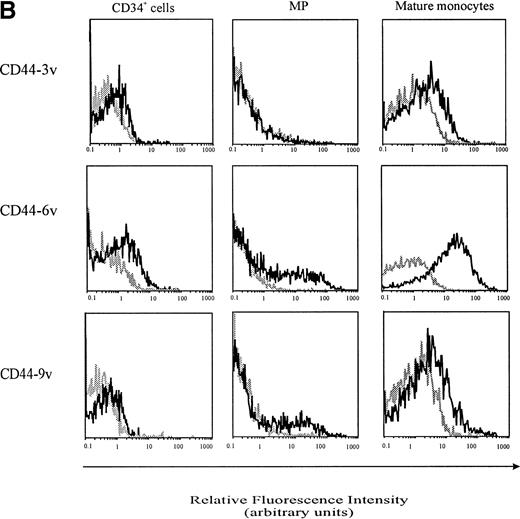
Cell-surface expression of CD44-3v, -6v, and -9v containing isoforms on normal myeloid cells. (A) Expression of CD44-6v on CD34+ cells. CD34+ cells were isolated from normal BM by specific immunoadsorption, as described in Materials and Methods. Histogram 1 defines the morphologic gate. Cells were double-stained with the anti-6v MoAb VFF18 plus GAM-PE and FITC-conjugated HPCA-2 MoAb directed to CD34 (histogram 2). Quadrant limits defining positivity and negativity have been set up using cells labeled with PE-conjugated and FITC-conjugated control antibodies (histograms 3 and 4), as described in Materials and Methods. Adjustment of the crossover fluorescence was obtained by compensation of the two single-stained samples to limit superposition of the fluorochrome emission spectra. (B) CD34+ cells and monocytic cells. CD34+ cells, very immature hematopoietic cells; MP, monocytic precursors (CD34neg, CD14+ BM cells); Mature monocytes, circulating CD14+ cells. Mature monocytes from PB were identified according to their forward angle and side scatter.37 CD34+ cells and MP were double-stained with (1) an FITC-conjugated MoAb directed to CD34, which defines very immature hematopoietic cells, and CD14, a monocyte-specific antigen, respectively and (2) MoAbs directed at CD44-3v (BBA11), -6v (VFF-18), and -9v (11-24), plus PE-conjugated goat anti-mouse IgG. Negative controls (gray lines) were cells labeled with FITC and PE-conjugated isotype-matched control antibodies. Flow cytometric analysis was performed as described in Materials and Methods. Four independent experiments gave similar results. HPC were negative for CD44-3v and CD44-9v proteins.
Cell-surface expression of CD44-3v, -6v, and -9v containing isoforms on normal myeloid cells. (A) Expression of CD44-6v on CD34+ cells. CD34+ cells were isolated from normal BM by specific immunoadsorption, as described in Materials and Methods. Histogram 1 defines the morphologic gate. Cells were double-stained with the anti-6v MoAb VFF18 plus GAM-PE and FITC-conjugated HPCA-2 MoAb directed to CD34 (histogram 2). Quadrant limits defining positivity and negativity have been set up using cells labeled with PE-conjugated and FITC-conjugated control antibodies (histograms 3 and 4), as described in Materials and Methods. Adjustment of the crossover fluorescence was obtained by compensation of the two single-stained samples to limit superposition of the fluorochrome emission spectra. (B) CD34+ cells and monocytic cells. CD34+ cells, very immature hematopoietic cells; MP, monocytic precursors (CD34neg, CD14+ BM cells); Mature monocytes, circulating CD14+ cells. Mature monocytes from PB were identified according to their forward angle and side scatter.37 CD34+ cells and MP were double-stained with (1) an FITC-conjugated MoAb directed to CD34, which defines very immature hematopoietic cells, and CD14, a monocyte-specific antigen, respectively and (2) MoAbs directed at CD44-3v (BBA11), -6v (VFF-18), and -9v (11-24), plus PE-conjugated goat anti-mouse IgG. Negative controls (gray lines) were cells labeled with FITC and PE-conjugated isotype-matched control antibodies. Flow cytometric analysis was performed as described in Materials and Methods. Four independent experiments gave similar results. HPC were negative for CD44-3v and CD44-9v proteins.
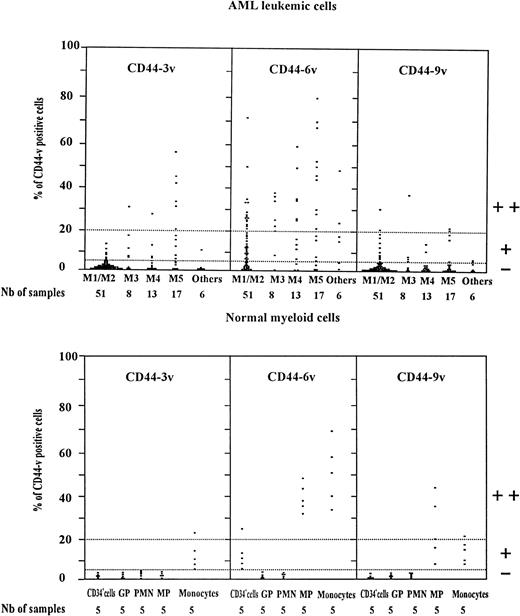
Scatter plot of CD44v protein expression on AML leukemic cells and normal myeloid cells. Normal myeloid cells: CD34+ cells (very immature BM hematopoietic cells). GP, granulocytic precursors (CD34−, CD15+ BM cells); PMN, polymorphonuclear cells; MP, monocytic precursors (CD34neg, CD14+ BM cells); Monocytes: mature monocytes (circulating CD14+ cells). AML leukemic cells from AML patients with the following FAB types.34,35 M1/M2, myeloblastic AML; M3, promyelocytic AML; M4, myelomonocytic AML; M5, monoblastic AML. CD34+ cells, GP, and MP were identified by double-staining with (1) an FITC-conjugated MoAb directed to CD34, CD15 for GP and CD14 for MP, respectively, (2) unconjugated MoAbs directed to variant epitopes 3v, 6v, and 9v plus PE-goat anti-mouse (PE-GAM). Leukemic cells were identified by specific electronic gating.37 Flow cytometric analysis was performed as described in Materials and Methods. The staining intensity of variant CD44 isoforms was evaluated by measuring the percentage of labeled cells over the background (cells incubated with isotype-matched control antibodies). Each symbol (▪) refers to the percent CD44v cells per patient. Expression of CD44v was considered as negative (−) when less than 5% of the cells were labeled, low (+) when 5% to less than 20% of cells were labeled, and high (++) when more than 20% of the cells were CD44v+.
Scatter plot of CD44v protein expression on AML leukemic cells and normal myeloid cells. Normal myeloid cells: CD34+ cells (very immature BM hematopoietic cells). GP, granulocytic precursors (CD34−, CD15+ BM cells); PMN, polymorphonuclear cells; MP, monocytic precursors (CD34neg, CD14+ BM cells); Monocytes: mature monocytes (circulating CD14+ cells). AML leukemic cells from AML patients with the following FAB types.34,35 M1/M2, myeloblastic AML; M3, promyelocytic AML; M4, myelomonocytic AML; M5, monoblastic AML. CD34+ cells, GP, and MP were identified by double-staining with (1) an FITC-conjugated MoAb directed to CD34, CD15 for GP and CD14 for MP, respectively, (2) unconjugated MoAbs directed to variant epitopes 3v, 6v, and 9v plus PE-goat anti-mouse (PE-GAM). Leukemic cells were identified by specific electronic gating.37 Flow cytometric analysis was performed as described in Materials and Methods. The staining intensity of variant CD44 isoforms was evaluated by measuring the percentage of labeled cells over the background (cells incubated with isotype-matched control antibodies). Each symbol (▪) refers to the percent CD44v cells per patient. Expression of CD44v was considered as negative (−) when less than 5% of the cells were labeled, low (+) when 5% to less than 20% of cells were labeled, and high (++) when more than 20% of the cells were CD44v+.
Clonogenic Features of CD34+CD44-6v+ and CD34+ CD44-6V−Cells
| . | CFU-M (%) . | CFU-G (%) . | CFU-GM (%) . | BFU-E (%) . | CFU-GEMM (%) . |
|---|---|---|---|---|---|
| CD34+ CD44-6v+ cells | 44* ± 7 | 19 ± 5 | 22 ± 5 | 10 ± 2 | 4 ± 1 |
| CD34+ CD44-6vneg cells | 24* ± 5 | 18 ± 7 | 20 ± 7 | 31 ± 8 | 7 ± 1 |
| . | CFU-M (%) . | CFU-G (%) . | CFU-GM (%) . | BFU-E (%) . | CFU-GEMM (%) . |
|---|---|---|---|---|---|
| CD34+ CD44-6v+ cells | 44* ± 7 | 19 ± 5 | 22 ± 5 | 10 ± 2 | 4 ± 1 |
| CD34+ CD44-6vneg cells | 24* ± 5 | 18 ± 7 | 20 ± 7 | 31 ± 8 | 7 ± 1 |
CD34+ CD44-6v+ and CD34+CD44-6vneg were sorted as detailed in Materials and Methods, and seeded at 2.103 cells/mL complete methyl cellulose medium supplemented with stimulating factors as described; CFU-M, CFU-GM, BFU-E, and CFU-GEMM were scored at day 16 (methyl cellulose assay). Results were expressed as the percentage of each type of colonies. All cultures were made in triplicate. Data are the means ± SE of three independent experiments.
The difference between data marked is statistically significant (Student's t-test, P < .05).
CD44-6v was also detected on about half of the monocytic cells (relative MFI: 5 ± 1, Figs 3B and 4). Considering that the level of CD44-6v expression was significantly higher on monocytic cells than on CD34+ cells, we defined as strongly positive (++) the level of CD44-6v on normal monocytic cells and as positive (+) the one on normal CD34+ cells (Fig 4).
In addition, CD44-9v was found on 25% of monocytic precursors and 14% of mature monocytes (relative MFI: 3 ± 2), whereas CD44-3v was only detected on 10% of mature monocytes (Figs 3B and 4). No significant expression of CD44v isoforms could be evidenced on GP or on PMN (<5% of positive cells, Fig 4).
AML leukemic cells.
Flow cytometric analysis of CD44 isoform expression was performed in AML cells from PB. In 10 patients, analysis was performed on both blood and BM blasts: a similar CD44 expression pattern was observed (comparison not shown, data from blood are shown, Figs 2 and 4). As detailed in Materials and Methods, the labeling was performed on cell suspension containing more than 95% AML blasts, after removal of PMN, lymphocytes, and monocytes, the latter for myeloblastic (M1/M2) and (M3) promyelocytic AML types. In addition, the AML blasts were specifically gated as described.37
Total CD44 was expressed on AML leukemic cells (Fig 2). The mean MFI value was similar in M1/M2, M4, and M5 AML (32 ± 4, 38 ± 6, and 48 ± 3, respectively). It was higher in M3 AML (98 ± 7).
CD44-6v expressing leukemic cells were detected in 69 of 95 patients (Fig 4). The proportion of leukemic cells labeled above the background (cells labeled with isotype-matched control antibodies) ranged between 5% and 80% in 34 of 51 M1/M2 cases (median value, 20%), in 7 of 8 M3 patients (median value, 23%) and in 24 of 30 M4/M5 patients (median value, 43%). More than 20% of CD44-6v expressing leukemic cells (mean relative MFI value of 3 ± 1, 4 ± 1, and 5 ± 1, respectively) were displayed by 15 M1/M2, 6 M3, and 16 M4/M5 patients as well as on 2 of 6 patients with undefined or M7 AML type. By reference to the high level of CD44-6v expression on normal monocytes, as described above, these patients were designated as CD44-6v++. The intensity of CD44-6v expression positively correlated with the level of total CD44 (P < .01, chi-squared test, n = 82, samples collected at relapse were not included).
A weak expression of CD44-3v was observed in 6 of 51 M1/M2 patients, 4 M3, and 9 M4/M5 cases. Six M5 patients were strongly CD44-3v+ (Fig 4). Only a low expression of CD44-9v was evidenced in 11 M1/M2, 4 M3, and 6 M4/M5 patients.
Correlation of CD44 Isoform Expression With the Clinical Outcome of Patients
To determine whether CD44-6v isoform expression influences the clinical outcome of AML patients, the rate of first complete remission was compared in CD44-6v++ and CD44-6v+/neg groups of patients undergoing induction chemotherapy. Because treatment may modify CD44-6v expression,40 only patients with de novo AML have been included in this statistical study (n = 63, Table 1): the remission rate was 69% for CD44-6v++ group (n = 23) and 58% for CD44-6v+/neg cases (n = 40); the difference was not statistically significant (P = .33, chi-squared test).
The median overall survival of CD44-6v++ and CD44-6v+/neg cases has been compared using the Kaplan-Meier method.38 To analyze a homogenous group of patients, this statistical analysis has been restricted to patients treated by conventional induction chemotherapy and with a well-defined FAB AML type. Consequently, the two M3 patients treated with all-trans retinoic acid, as well as the three patients with undefined FAB AML type, were excluded from the analysis (Table 1).
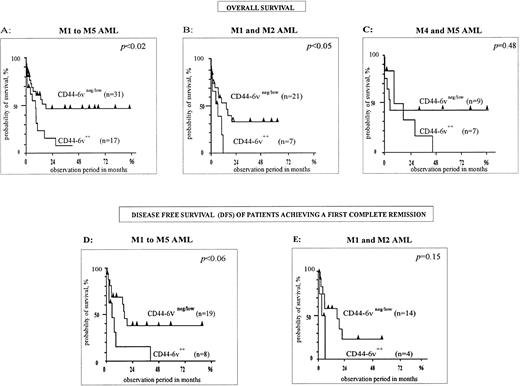
First, M1 to M5 FAB types have been analyzed together (n = 48, Fig 5A). The median overall survival of CD44-6v++ patients was 8 months, compared with 18 months for CD446v+/neg cases. The difference was statistically significant, with P < .02 (Kaplan-Meier method38). Moreover, the disease-free survival (DFS), measured for patients achieving a first complete remission, was shorter in the CD44-6v++ group of patients than in the CD44-6v+/neg group (6 months v 17 months, P < .06, n = 29, Kaplan-Meier method,38 Fig 5D).
Correlation of CD44-6v isoform expression with survival of AML patients. Survival curves of AML patients according to the expression of CD44-6v on leukemic cells at diagnosis, have been computed using the Kaplan-Meier method.38 Statistical comparisons between curves were based on log-rank tests. (A, B, and C) Overall survival curves of AML patients treated by conventional chemotherapy36 (anthracyclin plus cytosin arabinosine). (D and E) Disease-free survival curves of AML patients induced in complete remission by conventional chemotherapy. M1/M2, myeloblastic AML; M4, myelomonocytic AML; M5, monoblastic AML. Significant numbers have been obtained by grouping M4 and M5 patients for overall survival, but not for disease-free survival analysis.
Correlation of CD44-6v isoform expression with survival of AML patients. Survival curves of AML patients according to the expression of CD44-6v on leukemic cells at diagnosis, have been computed using the Kaplan-Meier method.38 Statistical comparisons between curves were based on log-rank tests. (A, B, and C) Overall survival curves of AML patients treated by conventional chemotherapy36 (anthracyclin plus cytosin arabinosine). (D and E) Disease-free survival curves of AML patients induced in complete remission by conventional chemotherapy. M1/M2, myeloblastic AML; M4, myelomonocytic AML; M5, monoblastic AML. Significant numbers have been obtained by grouping M4 and M5 patients for overall survival, but not for disease-free survival analysis.
Second, M1/M2 AML (n = 28) have been separately analyzed. A strong CD44-6v expression was clearly associated with a shorter overall survival. Indeed, this latter was 5.6 months in the group of CD44-6v++ patients, and 14.6 months in the group of CD44-6v+/neg patients (P < .04, Kaplan-Meier method,38 Fig 5B). Moreover, the DFS was 3 months and 16 months for CD44-6v++ and CD44-6v+/neg groups, respectively (P = .15, n = 18, Kaplan-Meier method,38 Fig 5E).
Statistical significance was not achieved in the group of myelomonocytic plus monoblastic AML (M4/M5, P = .48, n = 16, Kaplan-Meier method,38 Fig 5C).
The expression of CD44-6v was not correlated with the patients' age (P = .3, n = 82, Student's t-test), or with the number of circulating blasts (P = .4, n = 82, Student'st-test), which are both important prognostic factors in AML. An eventual correlation with karyotypic abnormalities could not be investigated because karyotype analyses were available only in a minority of cases.
Finally, there was no correlation between patients' survival and the expression of total CD44 (P = .6, n = 48, Kaplan-Meier method38).
Determination of the CD44v Exon Composition in AML Leukemic Cells and in Normal Myeloid Cells
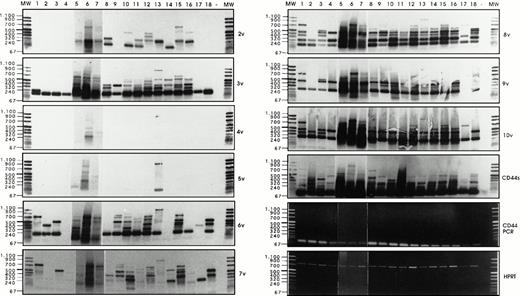
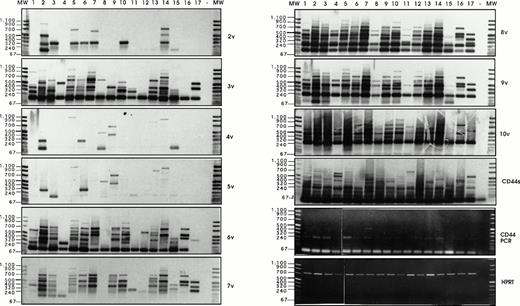
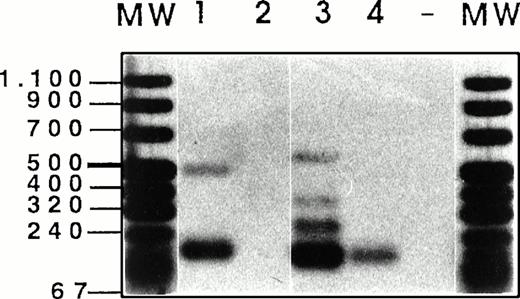
Semi-quantitative RT-PCR analysis has been performed, followed by Southern blotting and detection with exon-specific probes, as described by Stauder et al.27 Using HPRT controls, equal amounts of cDNAs (Fig 6)have been analyzed per RT-PCR reaction, and the composition of CD44 isoforms has been determined for each sample.
Analysis of CD44v exon composition in normal myeloid and AML leukemic cells. RNA and cDNA were prepared as described in Materials and Methods. Using primers for HPRT, the amounts of cDNA were equilibrated to this internal standard. A CD44-specific PCR was performed and the PCR products were blotted and hybridized with33P-labeled exon-specific probes indicated at each blot. Represented are the autoradiographs and the photos taken from the CD44 and HPRT-PCR reaction products as indicated. Negative controls (−) were performed by omitting the cDNA. (1) Normal myeloid cells: CD34+ cells, blot A, lanes 1, 2, 3, CD34−granulocytic precursors GP (blot A, lane 4), PMN (blot A, lane 5), and monocytes (blot A, lanes 6 and 7). RT-PCR has been performed on five distinct samples of PMN, monocytes, GP, and CD34+ cells. Similar results have been obtained (data not shown). (2) Leukemic cells from AML patients with the following FAB types: M1 (blot A, lanes 8 through 14), M2 (blot A, lanes 15 through 18), M3 (blot B, lanes 1 through 4), M4 (blot B, lanes 5 through 11) and M5 (blot B, lanes 12 through 16). The sample shown on blot B lane 17 is from a patient with acute lymphoid leukemia. (3) Control experiment: Lymphocytes (lane 1) or monocytes (lane 3) were isolated from a patient with a myeloblastic AML (M1) and mixed in a ratio 5:95, with HL60 cells that express variant exons only very weakly (CD44vneg/low). The presence of 5% of lymphocytes from the AML patient did not result in the expression of any detectable 6v signal (lane 2), and only a faint expression of CD44-6v in the directly spliced version resulted from the presence of 5% of monocytes from the AML patient (lane 4). {/ANNT;4224n;;0n;0n}3
Analysis of CD44v exon composition in normal myeloid and AML leukemic cells. RNA and cDNA were prepared as described in Materials and Methods. Using primers for HPRT, the amounts of cDNA were equilibrated to this internal standard. A CD44-specific PCR was performed and the PCR products were blotted and hybridized with33P-labeled exon-specific probes indicated at each blot. Represented are the autoradiographs and the photos taken from the CD44 and HPRT-PCR reaction products as indicated. Negative controls (−) were performed by omitting the cDNA. (1) Normal myeloid cells: CD34+ cells, blot A, lanes 1, 2, 3, CD34−granulocytic precursors GP (blot A, lane 4), PMN (blot A, lane 5), and monocytes (blot A, lanes 6 and 7). RT-PCR has been performed on five distinct samples of PMN, monocytes, GP, and CD34+ cells. Similar results have been obtained (data not shown). (2) Leukemic cells from AML patients with the following FAB types: M1 (blot A, lanes 8 through 14), M2 (blot A, lanes 15 through 18), M3 (blot B, lanes 1 through 4), M4 (blot B, lanes 5 through 11) and M5 (blot B, lanes 12 through 16). The sample shown on blot B lane 17 is from a patient with acute lymphoid leukemia. (3) Control experiment: Lymphocytes (lane 1) or monocytes (lane 3) were isolated from a patient with a myeloblastic AML (M1) and mixed in a ratio 5:95, with HL60 cells that express variant exons only very weakly (CD44vneg/low). The presence of 5% of lymphocytes from the AML patient did not result in the expression of any detectable 6v signal (lane 2), and only a faint expression of CD44-6v in the directly spliced version resulted from the presence of 5% of monocytes from the AML patient (lane 4). {/ANNT;4224n;;0n;0n}3
Normal myeloid cells.
Exon 6v was expressed in CD34+ cells (Fig 6A, lanes 1 through 3), PMN (lane 5), and monocytes (lanes 6 and 7). Only granulocytic precursors (GP) did not express this variant exon (lane 4). In CD34+ cells, 6v was found in a directly spliced version (ie, the variant exon flanked directly by the framework of CD44 standard, size 196 bp). In these cells, 6v was also combined with other variant exons, such as 7v to 10v (724 bp, lane 1), 10v (400 bp, lane 2), and 7v to 9v (520 bp, lane 3); a single exon 6v-containing combination was observed in each CD34+ cell sample. In contrast, 6v was contained in a great variety of high-molecular-weight isoforms in each sample of PMNs (lane 5) and monocytes (lanes 6 and 7). The large 6v-containing isoforms were most abundant in samples of PMN and monocytes shown in lanes 5 and 6.
Exon 3v was expressed in the directly spliced version in CD34+ cells and GP (size 193 bp, lanes 1 to 4). In contrast, a great variety of high-molecular-weight transcripts containing exon 3v in combination with exons 8v, 9v, and 10v were abundantly produced in monocytes and PMN (lanes 5 to 7). In CD34+ cells and GP, a few transcripts containing 8v + 9v (262 bp, lanes 1 to 4) and 8v + 9v + 10v (466 bp, lanes 1, 3 and 4) were also synthesized.
AML leukemic cells.
CD44-6v transcripts were found to be expressed in all AML patients (except in one patient, who displayed only a weak CD44-6v protein expression; Fig 6A, lane 14). In a few patients (18 of 70), CD44-6v transcripts were only in the directly spliced version (size 196 bp), as shown in Fig 6A, lanes 8 and 16, and Fig 6B, lanes 8 and 11. In half of the AML patients (39 of 70), 6v transcript was also combined with other CD44 variant exons in a variety of large-size transcripts, as shown in Fig 6A, lanes 9, 10, 12, 15, and 18 and Fig 6B, lanes 1 to 3, 5 to 7, 9, 10, 13 through 16). These large transcripts combined in particular 6v + 7v (325 bp), 6v + 8v to 10v (595 bp), and 6v to 10v (724 bp). They were very abundant in 15 of 70 patients (scored in semi-quantitative score: ++ to ++++, Table 3).
Expression of CD44-Isoforms in Normal Myeloid and in AML Leukemic Cells
| Cells . | CD44 Isoforms . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total CD44 . | CD44-3v . | CD44-6v . | CD44-9v . | ||||||
| Cell Type . | No. of Samples . | RT-PCR . | Immunostaining . | RT-PCR . | Immunostaining . | RT-PCR . | Immunostaining . | RT-PCR . | Immunostaining . |
| Normal | |||||||||
| CD34+ cells | 5 | ++ | ++ | + | − | + | + | + | − |
| GP | 5 | −/+ | − | + | − | −/+ | − | + | − |
| PMN | 5 | +++ | ++ | +++ | − | ++ | − | +++ | − |
| MP | 5 | ND | ++ | ND | − | ND | ++ | ND | ++ |
| Monocytes | 5 | +++ | ++ | ++++ | + | +++ | ++ | ++++ | + |
| Leukemic | |||||||||
| AML | 95 | ++ to ++++ | − to ++ | +++ | − to + | +++ | − to ++ | +++ | − to + |
| Cells . | CD44 Isoforms . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total CD44 . | CD44-3v . | CD44-6v . | CD44-9v . | ||||||
| Cell Type . | No. of Samples . | RT-PCR . | Immunostaining . | RT-PCR . | Immunostaining . | RT-PCR . | Immunostaining . | RT-PCR . | Immunostaining . |
| Normal | |||||||||
| CD34+ cells | 5 | ++ | ++ | + | − | + | + | + | − |
| GP | 5 | −/+ | − | + | − | −/+ | − | + | − |
| PMN | 5 | +++ | ++ | +++ | − | ++ | − | +++ | − |
| MP | 5 | ND | ++ | ND | − | ND | ++ | ND | ++ |
| Monocytes | 5 | +++ | ++ | ++++ | + | +++ | ++ | ++++ | + |
| Leukemic | |||||||||
| AML | 95 | ++ to ++++ | − to ++ | +++ | − to + | +++ | − to ++ | +++ | − to + |
These cells were prepared as described in Materials and Methods. The RT-PCR were performed as described 32 and scored semi-quantitatively on a scale of − to ++++: −, negative; +/−, faint, not clearly above background; +, weakly positive; ++, moderately positive; +++ and ++++, strongly positive. Total CD44 and CD44v-containing isoforms were immunostained using specific MoAbs as described. Cells of interest were analyzed by flow cytometry using both specific electronic gating (PMN, monocytes, HPC, leukemic cells), and double-staining with an MoAb directed to a lineage-specific antigen (CD34 for CD34+ cells, CD14 for monocytic cells, CD15 for granulocytic cells). Immunostainings of total CD44 and of the CD44v-containing isoforms were also scored semi-quantitatively on a scale of − to ++: −, negative (<5% of positive cells); +, positive (5% to 20% of positive cells); ++, strongly positive (>20% of positive cells).
Abbreviations: AML, leukemic cells from AML patients; ND, not done.
Strong signals for 3v, 8v, and 10v transcripts were also obtained in all AML patients, both in the directly spliced version (193 bp, 187 bp, and 271 bp, respectively) and in multiple combinations with other exons. For example, 3v was also found to be combined with 2v (322 bp), and 10v was frequently associated in an 8v + 9v + 10v complex (466 bp). Exon 9v was always associated with 8v (262 bp). 4v and 5v were rarely expressed.
We asked whether the strong variant mRNA signals may be produced by contaminating lymphocytes or monocytes, which could have been incompletely removed by the immunoabsorption procedure. To answer this question, semi-quantitative RT-PCR analysis was performed on a cell mixture containing monocytes and lymphocytes from a patient with a myeloblastic AML (M1 type), mixed in a ratio 5:95, with HL60 cells, that express variant exons only very weakly (CD44vneg/low). As shown in Fig 6C, the presence of 5% of lymphocytes from the AML patient did not result in the expression of any detectable 6v signal (lane 2), and the presence of 5% of monocytes from the AML patient lead only to a faint expression of CD44-6v in the directly spliced version (lane 4). This faint signal contrasted with the abundance of 6v transcripts (both directly spliced and combined with other exons) in the AML blast sample from this patient (Fig 6A, lane 12). This mixing experiment indicates that the bulk of the 6v transcripts detected by semi-quantitative RT-PCR analysis in AML blast samples were synthesized by the leukemic cells themselves.
To summarize, large CD44 transcripts combining 3v, 6v, and 8v to 10v are much more abundant in AML leukemic cells and in normal monocytes than in CD34+ cells and in GP. However, contrasting with the prognostic importance of 6v protein expression in AML, the composition of 6v-containing transcripts did not correlate with the survival rate of AML patients.
DISCUSSION
In this study we report on the expression of CD44 variant isoforms in normal myeloid cells and in AML patients, and its correlation to clinical data. Using antibodies to CD44-3v, -6v, and -9v, flow cytometry, and RT-PCR analysis of variant-specific transcripts, we have shown that CD44-6v is expressed on 12% of normal CD34+ HPC and on 50% of monocytic cells, whereas it could not be detected on granulocytic cells. We also demonstrated that AML leukemic cells display CD44-6v and, to a lesser extent, CD44-3v and -9v isoforms. Furthermore, we give evidence that a strong expression of CD44-6v correlates with a shorter survival of AML patients treated with conventional chemotherapy.
Because of the high reproducibility of CD44-6v isoform expression through more than seven experiments, it seems relevant to attest that this variant isoform is displayed by about 12% of normal human CD34+ cells (Fig 3A). Cell-sorting experiments indicate that the CD34+ CD44-6v+ cell population is enriched in progenitors committed to the monocytic lineage (Table 2). In addition, CD44-6v is strongly expressed on monocytic precursors (CD34neg CD14+ cells). This suggests that monocytic differentiation may be associated with an upregulation of CD44-6v expression, and that this isoform may be particularly involved in monopoiesis. Inversely, CD34+ CD44-6v+ cells are depleted from BFU-E, suggesting that CD44-6v is not much expressed on erythroid progenitors. Finally, granulocytic committed progenitors are found in equal number in CD34+CD44-6v+ and CD34+CD44-6vneg cell populations. Their differentiation into granulocytic precursors (CD34negCD15+ cells) is probably associated with a downregulation of CD44-6v because granulocytic precursors do not display CD44-6v.
In PMN, 6v-containing transcripts are abundant, although CD44-6v protein cannot be detected by FACS analysis. Activation of PMN during the in vitro isolation procedure may lead to the synthesis of 6v-specific transcripts, but it may be too short for final protein expression on the membrane. Arguing in favor of this hypothesis, we have observed significant changes in the level of CD10 and CD11b antigens, which have been shown to be surface markers for neutrophil activation.41 42
In AML, CD44-6v is more frequently and more strongly expressed than CD44-3v and CD44-9v, with 39 CD44-6v++ patients, versus 8 CD44-3v++ and 2 CD44-9v++ cases (Fig 4). In contrast to this low frequency in 3v and 9v protein expression, a great amount of 3v and 9v specific transcripts were detected in nearly all AML leukemic cell samples (Fig 6). Similarly, 6v protein could not be detected on the blasts from up to 26 AML patients, despite a significant amount of 6v specific transcripts (Fig 6, except in one case, 6A lane 14). As a hypothesis, the accessibility of variant epitopes to the specific MoAbs may be prevented by a particular conformation of the CD44v proteins, or by changes in the glycosylation pattern of the molecules, due in particular to the insertion of variant sequences.43 Using another MoAb to CD44-6v isoform, the FW11-9,22 similar data have been obtained, indicating that the present results are not restricted to the particular MoAb (VFF18) used here.
The overall survival of the AML patients is clearly related to the protein expression level of CD44-6v containing isoforms with a value of 8 months in the CD44-6v++ group compared with 18 months in the CD44-6v+/neg group (P < .02, n = 48, Fig5A). The level of this expression is unlikely to influence the achievement of the first complete remission, since the rate of first complete remission did not significantly differ in the CD44-6v++ and CD44-6v+/neg groups of patients (P = .33, see Results). On the contrary, the level of 6v containing CD44 molecules may accelerate the relapse, because the disease-free survival of patients achieving a first complete remission was significantly shorter in the CD44-6v++ group than in the CD44-6v+/neg group (Fig 5D). In this regard it would be of interest to determine whether CD44-6v has a role in stimulating the proliferation of residual leukemic cells.
Because CD44-6v may be differently involved in the evolution of the different FAB types of AML, we have separately analyzed the influence of its expression in M1/M2 (myeloblastic) and M4/M5 (myelomonocytic and monoblastic) leukemic cells.
We have found that a strong CD44-6v protein expression is also correlated with a shorter overall survival in M1/M2 (Fig 5B), suggesting a role for CD44-6v in the pathology of these AML. Interestingly, CD44-6v expression is strikingly higher in myeloblastic leukemic cells than in normal granulocytic precursors (Fig 4). This suggests that CD44-6v is overexpressed in M1/M2 AML compared with the normal cellular counterpart, and that this dysregulation in CD44-6v gene expression may play a part in the pathophysiology of this type of AML. The overexpression of 6v specific transcripts in M1/M2 blasts compared with normal granulocytic precursors (Fig 6A) also supports the hypothesis of a CD44-6v gene dysregulation in these AML.
Total CD44 is also overexpressed in M1/M2 AML compared with the normal granulocytic precursors (Fig 2). However, its overexpression is not correlated with the clinical evolution (P = .6, n = 48), underlining that the various isoforms of CD44 constitute a family of molecules endowed with specific functions.2
No correlation between CD44-6v expression and overall survival has been found in M4/M5 AML (Fig 5C). Because this analysis has been performed on only 16 patients, and although the P value is very far from the statistical significance (P = .48), further studies dealing with more cases are necessary for better evaluation of the involvement of CD44-6v in M4/M5 AML. It is remarkable that the CD44-6v expression pattern is broadly similar in M5 leukemic cells and in normal monocytic precursors that are their putative normal cell counterparts. Both cell types strongly express CD44-6v containing proteins (Figs 3B and 4), and high-molecular-weight 6v-containing transcripts (Fig 6), indicating a comparable composition of CD44-6v containing isoforms. A leukemic amplification of a CD44-6v+subset of monoblastic cells may account for the very high frequency of CD44-6v+ blasts in several M5 AML patients.
Although the clinical evolution of patients with promyelocytic AML (M3) is generally favorable,44 the outcome of the eight M3 AML patients from our cohort has not been better than that of other FAB types. Indeed, four M3 patients rapidly died because of AML, and complete remission has been achieved in only three of them (the clinical evolution of one patient has not been documented; Table 1). Accordingly, these M3 cases are likely to represent a selection of poor-prognosis patients, all the more since most of them display more than 10,000 WBC/μL, a feature associated with an unfavorable clinical evolution in this type of AML. Because of the low number of M3 cases, no survival analysis has been performed. However, it is noticeable that 4 of 5 CD44-6v++ cases died, whereas 2 CD44-6v+/neg patients are still in complete remission.
How CD44-6v overexpression participates in the leukemic process is still elusive because the specific functions of CD44-6v containing isoforms are unknown so far. CD44-6v overexpression may participate in the release of BM leukemic cells into the circulation. Indeed, it is noteworthy that the exon 6v-encoded peptidic sequence displays several potential sites of glycosylations.2 Since glycosylation of CD44 has been shown to negatively regulate its binding to hyaluronan,45,46 an overexpression of CD44-6v containing isoforms may diminish the CD44-mediated adhesion of early myeloid cells to hyaluronan6-8 and, thereby, to BM stroma. Interestingly, it is known that adhesion of immature myeloid cells to the BM stroma, which involves CD44,4,5,47 is crucial for regulating their proliferation and their differentiation along the granulocytic and the monocytic lineages. Therefore, by decreasing the myeloid cell-stroma interaction, overexpression of 6v-containing CD44 molecules may impair stroma-dependent control of both proliferation (as suggested above) and differentiation of myeloid cells, thus perhaps contributing to the freezing of myelopoiesis in AML. Overexpression of CD44-6v isoforms may also confer affinity to a specific ligand distinct from hyaluronan and favor an extra-hematopoietic localization of the leukemic cells, as observed in certain AML patients.36
In conclusion, this study is the first report that the protein expression of CD44-6v containing isoforms on leukemic cells may influence the survival of AML patients. Accordingly, it suggests that CD44-6v may serve as a new prognostic factor in AML, and that analysis of its expression may contribute to improve the management of new therapeutical approaches aimed at AML patients with adverse prognostic factors.48
ACKNOWLEDGMENT
We thank Prof J.P. Vernant, Prof C. Chomienne (Hôpital St Louis, Paris, France), and Dr J.L. Pico (Institut G. Roussy, Villejuif, France) for generously supplying 18 AML samples; Dr L. Coulombel (INSERM 362, Institut G. Roussy, Villejuif, France) for the flow cytometric analysis and sorting experiments of CD34+ cells; and R.S. Charrad for the cloning experiments. We are grateful to Drs M. Salio and J. Fraser McBlane (Basel Institute for Immunology, Basel, Switzerland), for helpful comments and suggestions on the manuscript. We are indebted to V. Anquez (Basel Institute for Immunology, Basel, Switzerland) and J. Chaker (Inserm U268) for their technical assistance; to A.M. Gonzales (Department of Hematology, Hôpital Hôtel Dieu, Paris, France), I. Starrenburg and the technicians of the Laboratory of Hematology, Section Hematomorphology and Immunotyping (University Hospital, Leiden, The Netherlands) for their help with the collection of the materials; to P.S. Beaussier (Institut Gustave Roussy, Villejuif, France) for providing normal bone marrow samples; and to N. Vriz for editorial assistance.
Supported by INSERM, ANRB-Vaincre le Cancer, Assistance Publique Grant No. AOB94057, and ARC Grant No. 6923. The Basel Institute for Immunology was founded and is supported by Hoffmann-La Roche Inc., Basel, Switzerland.
Address reprint requests to F. Smadja-Joffe, PhD, Unité de Différenciation Hématopoiétique Normale et Leucémique, INSERM U268, Hôpital Paul-Brousse, 14, Avenue P.V. Couturier, 94807 Villejuif Cedex, France.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal