Abstract
It is now accepted from studies in animal models that hematopoietic stem cells emerge in the para-aortic mesoderm-derived aorta-gonad-mesonephros region of the vertebrate embryo. We have previously identified the equivalent primitive hematogenous territory in the 4- to 6-week human embryo, under the form of CD34+CD45+Lin− high proliferative potential hematopoietic cells clustered on the ventral endothelium of the aorta. To characterize molecules involved in initial stem cell emergence, we first investigated the expression in that territory of known early hematopoietic regulators. We herein show that aorta-associated CD34+ cells coexpress the tal-1/SCL, c-myb, GATA-2, GATA-3, c-kit, and flk-1/KDR genes, as do embryonic and fetal hematopoietic progenitors later present in the liver and bone marrow. Next, CD34+CD45+ aorta-associated cells were sorted by flow cytometry from a 5-week embryo and a cDNA library was constructed therefrom. Differential screening of that library with total cDNA probes obtained from CD34+embryonic liver cells allowed the isolation of a kinase-related sequence previously identified in KG-1 cells. In addition to emerging blood stem cells, KG-1 kinase is also strikingly expressed in all developing endothelial cells in the yolk sac and embryo, which suggests its involvement in the genesis of both hematopoietic and vascular cell lineages in humans.
EARLY IN THE embryogenesis of higher vertebrates, hematopoietic stem cells (HSCs) arise in situ in the extraembryonic yolk sac mesoderm and produce locally a transient wave of primitive nucleated red blood cells. Thereafter, migrating HSCs seed the successively emerging hematopoietic organ rudiments of the embryo, where they give rise to multilineage differentiated blood cells. The last blood-forming tissue anlage to be colonized is that of the bone marrow, in which hematopoiesis becomes definitively stabilized at postnatal stages. Consequently, the yolk sac was long considered as the original and only provider of self-renewing stem cells for life-long hematopoiesis.1,2 The demonstration in birds3,4and in amphibians5,6 that HSCs responsible for definitive hematopoiesis do not emigrate from extraembryonic tissues but rather originate within the splanchnopleural mesoderm of the embryo proper prompted the search for an equivalent intraembryonic source of HSCs in mammals. Indeed, the para-aortic splanchnopleura (p-SP) in the early mouse embryo and derived aorta-gonad-mesonephros (AGM) territory were found to harbor, in parallel with the yolk sac, pluripotential hematopoietic cells before the onset of fetal liver colonization.7-9 When dissected out before the establishment of circulation, the p-SP but not the yolk sac gave rise to multipotential hematopoietic progenitors in vitro.10 In contrast, after circulation connected the yolk sac with the embryo, stem cells endowed with T- and B-lymphoid potential as well as true long-term repopulating activity were detected in both the p-SP and yolk sac.10-13 Together, these data strongly suggested that, independently of the yolk sac, a wave of HSCs arise within the splanchnopleural mesoderm of the embryo between the presomitic and liver colonization stages. Accordingly, transient clusters of CD34+, c-Kit+, Flk-1+hematopoietic cells were observed during that developmental period on the ventral aspect of the mouse aorta and omphalomesenteric artery,14-16 reminiscent of the intra-aortic and para-aortic blood cell foci at the origin of definitive hematopoiesis in birds.17 18
The molecular mechanisms that locally influence the emergence and primary expansion of HSCs from mesodermal precursors remain unclear, although key regulators of these earliest developmental steps have now been evidenced in mice. The importance of the c-kit signaling pathway was first discovered through the severe hematopoietic stem cell defects that occur in Sl and W mutant mice.19,20 Other potential early hematopoietic factors have been identified by gene targeting in mouse embryonic stem (ES) cells. c-myb knock-out causes a lethal deficit in definitive multipotential progenitors,21 and a complete block in definitive hematopoietic potential is observed in ES cells null for the expression of the GATA-2, tal-1/SCL and AML-1 transcription factors.22-26 Inactivation of the flk-1 tyrosine kinase gene prevents yolk sac blood island formation and endothelial development,27 but also precludes definitive hematopoiesis even when normal vascular structures are present.16 It is not clear whether the primary emergence or the survival and expansion of HSCs are disturbed in these experiments. However, the low numbers of HSCs retrieved in both c-myb– and GATA-2–null mutant embryos would suggest a role of these factors in stem cell proliferation rather than in the commitment of primitive mesoderm to hematopoiesis.21,22,28 On the other hand, the lack of both primitive and definitive hematopoiesis resulting from tal-1/SCL and flk-1 targeted mutations may suggest a common failure of extraembryonic and intraembryonic mesodermal precursors to differentiate into HSCs.29
In humans, hematopoiesis starts in the yolk sac at week 3 of development and shifts to the liver around week 5.30,31 In an attempt to identify the primary hematogenous territory where definitive human HSCs originate, Huyhn et al32 evidenced high proliferative potential CD34+ clonogenic progenitors within the embryo body deprived from its yolk sac and liver rudiment around week 5 of gestation. Furthermore, upon immunostaining of 4- to 6-week human embryo sections, we characterized dense clusters of CD34+ hematopoietic cells closely associated with the ventral endothelium of the aorta33 that strongly evoked the intravascular blood cell clumps previously observed in animal embryos at an equivalent developmental stage.14,16,17,34,35 With respect to spatio-temporal distribution, typical hematopoietic stem cell surface phenotype (CD45+, CD34hi, CD31+, CD38−, Lin−), and high proliferative potential in vitro, these early emerging HSCs were proposed as the intraembryonic source of definitive hematopoiesis in the human species.33
The present work was aimed at defining the molecules involved in the initial emergence and expansion of HSCs within the aortic wall of the 4- to 6-week human embryo. Transcripts expressed in HSCs from the aorta were compared with those from the liver and bone marrow of the embryo and fetus. The expression of surface receptors and transcription factors already defined at the earliest steps of mouse hematopoietic development was first investigated by hybridization on embryo sections and reverse transcription-polymerase chain reaction (RT-PCR) analysis on HSCs sorted from 12- to 29-week fetal liver and bone marrow. This study showed important similarities between successive generations of embryonic and fetal HSCs. We then searched for novel genes expressed early by the unique HSC population arising from the human p-SP. To that end, we sorted by flow cytometry rare aorta-associated CD34+ cells from a single embryo to construct a cDNA library from this primitive cell population. We then performed a differential screening of that library with amplified cDNA probes prepared from aorta-associated and embryonic liver HSCs. A novel kinase was identified whose expression pattern suggests a role in the development of both hematopoietic and endothelial cell lineages in humans.
MATERIALS AND METHODS
Human Tissues
Human embryos and fetal tissues were obtained from voluntary or therapeutic abortions performed in compliance with the French legislation, after informed consent was obtained from the parents.
Tissue Processing and Section Staining
Embryos fixed overnight at 4°C in phosphate-buffered saline (PBS), 4% paraformaldehyde (vol/vol) were rinsed in PBS, dehydrated, and included in paraffin. Five-micrometer–thick sections were immersed three times for 7 minutes in toluene, absolute ethanol, and finally 95% ethanol. After preincubation for 20 minutes in Tris-buffered saline, 0.25% Triton X100 (TBST) containing 5% fetal calf serum (FCS), sections were incubated for 1 hour at room temperature with the anti-CD34 HPCA-1 monoclonal antibody (MoAb; Becton Dickinson, San Jose, CA) diluted in TBST-2% FCS, followed by three washes in TBST. Slides were then incubated for 30 minutes with appropriately diluted rabbit antimouse IgG (DAKO, Glostrup, Denmark), rinsed again with TBST, and incubated 30 minutes with diluted mouse APAAP (antialkaline phosphatase coupled to alkaline phosphatase; DAKO). Immune reaction was shown using the DAKO Fast Red substrate system according to the manufacturer’s instructions. Endogenous alkaline phosphatase activity was inhibited by adding levamisole to the substrate solution at a final concentration of 1 mmol/L.
In Situ Hybridization
Probes.
Probes for the human tal-1/SCL, GATA-2, c-kit, flk-1/KDR, and KG-1 kinase genes were made from PCR fragments subcloned into the pGEM-T vector (Promega, Madison, WI) and corresponding to the following nucleotidic sequences in the Genbank database: tal-1/SCL: nt 4082-4979, accession no. M61108; GATA-2: nt 1861-2673, accession no.M68891; c-kit: nt 4442-5077, accession no. X06182; flk-1/KDR: nt 1593-2411, accession no. X61656; and KG-1 kinase: nt 28-922, accession no. D43636. The authenticity of the clones and their orientation were determined by sequence analysis or by mapping the position of the predicted restriction sites. A pGEM-3Z plasmid containing the coding sequence of the hGATA-3 cDNA 5′ to the zinc fingers36and a pBluescript KS plasmid containing the coding sequence of the c-myb cDNA plus 400 bp of 3′UTR (a generous gift of Dr J. Sores, Université d’Orsay, Orsay, France) were used as templates for the synthesis of the corresponding sense and antisense riboprobes.
In situ hybridization.
In situ hybridization was performed on sections from paraffin-embedded human embryos. Before hybridization, slides were deparaffinized and treated with proteinase K as previously described.37 Sense and antisense riboprobes were transcribed from T3, T7, or SP6 flanking promoters of appropriate linearized vectors. The protocol used for synthesis and hybridization of 35S-labeled riboprobes was as previously described,37 whereas synthesis and hybridization of digoxygenin (DIG)-labeled probes was performed according to Myat et al.38 In case of subsequent staining with the anti-CD34 MoAb, sections were rinsed extensively in TBST and processed as described above. Each hybridization was performed at least three times on tissue sections corresponding to at least two distinct embryos.
Isolation of Fetal HSCs
Mononuclear cells from 12- to 29-week fetal liver and bone marrow (4 distinct samples of each tissue) were obtained by sedimentation at 100g over a lymphocyte separation medium (d = 1.077 g/mL; Eurobio, Les Ulis, France) for 30 minutes at room temperature. The cells recovered at the interface were washed in PBS containing 5% FCS and 2 to 4 × 107 cells were incubated for 30 minutes on ice with a mixture of fluorescein isothiocyanate (FITC)-conjugated anti-CD34 MoAb (HPCA-2; Becton Dickinson) and phycoerythrin (PE)-conjugated anti-CD38 MoAb (Immunotech, Marseille, France) diluted 1/10 in PBS, 5% FCS. The two populations of CD34+CD38+ and CD34+CD38− cells were sorted on a FACStar Plus flow cytometer (Becton Dickinson) in the gates defined on Fig 3A. Control labeling with irrelevant IgG1-PE and -FITC were used to determine positivity for the CD34 and CD38 antigens. Dead cells and debris were eliminated by using a high forward and orthogonal light scatter window. After sorting, cells (104 to 105) were pelleted and processed for RNA extraction or kept at −80°C.
RT-PCR analysis of total cellular RNA.
Total RNA was isolated from cell pellets by the RNAzol method (Tel-Test Inc, Friendswood, TX), in 200 μL/pellet, according to the instructions of the manufacturer. RNAs were dissolved in 20 μL H2O containing 200 ng Random Primers (Promega) and heated at 70°C for 5 minutes. For first-strand cDNA synthesis, 10 μL of a mixture containing 6 μL of 5× SuperScript buffer (GIBCO-BRL, Paisley, Scotland), 1.5 μL of 10 mmol/L dNTPs (Boehringer Mannheim, Mannheim, Germany), 0.5 μL of RNA guard (Pharmacia, Uppsala, Sweden), 1 μL of 10 mmol/L dithiothreitol (DTT; GIBCO), and 1 μL of SuperScript II reverse transcriptase (GIBCO) were added and the reaction was performed at 37°C for 1 hour. One thirtieth of the reaction was used for each subsequent PCR analysis, performed in a 50 μL final volume containing 5 μL of Gene Amp 10× PCR Buffer II (Perkin Elmer, Norwalk, CT), 2.5 mmol/L MgCl2, 200 μmol/L dNTPs, and 1 U Taq polymerase (GIBCO). Approximately 1.5 pmol (50 ng) of each gene-specific primer (Table 1) was added and 35 cycles of PCR (94°C for 1 minute, 55°C for 1 minute, and 72°C for 1.5 minutes) were performed, with a final extension step of 7 minutes. Aliquots of PCR products were agarose gel electrophoresed, transferred onto N+ nylon membranes (Amersham, Amersham, United Kingdom), and hybridized with specific32P-labeled internal cDNA probes as described below.
Oligonucleotide Primers Used for RT-PCR Analysis of Fetal HSCs
| Genes . | 5′ Primer . | 3′ Primer . | Size* (bp) . |
|---|---|---|---|
| tal-1/SCl† | TTGGGGAGCCGGATGCCTTC | CTCCCGGCTGTTGGTGAA | 135 |
| c-myb | ACAGCATATATAGCAGTGACG | AGCCTGAGCAAAACCCATCAA | 659 |
| GATA-2 | CCCTAAGCAGCGCAGCAAGAC | TGACTTCTCCTGCATGCACT | 439 |
| GATA-3 | ACCCCACTGTGGCGGCGAGAT | CACAGCACTAGAGACC | 770 |
| c-kit | TGTACTGCCAGTGGATGTGCA | TCGTCATCCTCCATGATGGCG | 921 |
| flk-1/KDR | CAACAAAGCGGAGAGGAG | ATGACGATGGACAAGTACCC | 818 |
| CD38 | ATGGCCAACTGCGAGTTCAGC | GACTTTGGGGAAAAAGGCTTC | 1,056 |
| β-Actin | AACCGCGAGAAGATGACCCAG | TGCGCTCAGGAGGAGCAATGA | 663 |
| Genes . | 5′ Primer . | 3′ Primer . | Size* (bp) . |
|---|---|---|---|
| tal-1/SCl† | TTGGGGAGCCGGATGCCTTC | CTCCCGGCTGTTGGTGAA | 135 |
| c-myb | ACAGCATATATAGCAGTGACG | AGCCTGAGCAAAACCCATCAA | 659 |
| GATA-2 | CCCTAAGCAGCGCAGCAAGAC | TGACTTCTCCTGCATGCACT | 439 |
| GATA-3 | ACCCCACTGTGGCGGCGAGAT | CACAGCACTAGAGACC | 770 |
| c-kit | TGTACTGCCAGTGGATGTGCA | TCGTCATCCTCCATGATGGCG | 921 |
| flk-1/KDR | CAACAAAGCGGAGAGGAG | ATGACGATGGACAAGTACCC | 818 |
| CD38 | ATGGCCAACTGCGAGTTCAGC | GACTTTGGGGAAAAAGGCTTC | 1,056 |
| β-Actin | AACCGCGAGAAGATGACCCAG | TGCGCTCAGGAGGAGCAATGA | 663 |
Primers were selected to cross introns or checked for no amplification of same-size genomic products.
Predicted PCR product.
PCR was performed at a 1.5 mmol/L MgCl2 final concentration and 58°C annealing temperature.
Isolation of Aortic and Hepatic Embryonic HSCs
The aorta and liver were microdissected and dissociated by incubation for 1 hour at 37°C in 50 μL collagenase/dispase (Boehringer) 0.25% (vol/vol) in PBS without Ca2+ and Mg2+. After washing, the cell suspension was double-labeled for 30 minutes on ice with FITC-HPCA-2 anti-CD34 and either PE-anti-CD45 (DAKO) or PE-anti-CD38 MoAbs and sorted as above. After sorting, the cells (<60) were directly collected in thin-walled PCR tubes (Perkin Elmer) containing PBS, pelleted, resuspended in 4 μL ice-cold cell lysis buffer, and immediately processed for 3′cDNA synthesis.
Synthesis of Total cDNA and Southern Blot Analysis
The procedure for first-strand cDNA synthesis and amplification from low numbers of cells sorted by fluorescence-activated cell sorting (FACS) was derived from the protocol of Brady and Iscove39 and performed exactly as described by Dulac and Axel.40 Aliquots of amplified cDNAs were run on 1.5% agarose gels in Tris-borate buffer, denatured in NaOH/NaCl, and transferred onto Hybond N+ nylon membranes (Amersham). After prehybridization for 1 hour at 65°C in 0.5 mol/L sodium phosphate buffer (pH 7.3) containing 1% bovine serum albumin (BSA) and 4% sodium dodecyl sulfate (SDS), hybridization was performed in the same buffer overnight at 65°C by adding 106 cpm/mL of32P-labeled cDNA probe (Random Primer Labeling Kit; Stratagene, La Jolla, CA). After two washes at 65°C in 0.5× SSC, 0.1% SDS, membranes were autoradiographed on a Kodak X-OMAT film (Eastman Kodak, Rochester, NY) for 30 minutes to 24 hours.
Probes.
Probes were excised from plasmids containing the appropriate 3′cDNA sequences: tal-1/SCL and GATA-2 (idem in situ hybridization); GATA-3, 0.8-kb Pst I/BamHI fragment encompassing the final exon of the GATA-3 gene41; c-myb, 1.1-kb Xmn I-EcoRI 3′ fragment of the full-length cDNA cloned into pUC 18 (a generous gift of Dr J. Sores); G3PDH, 1.1 kb (purchased from Clontech, Palo Alto, CA). The CD34 (nt 1665-2555; accession no. M81104) and β-actin (nt 1042-1741: accession no. M10279) 3′cDNAs were PCR-amplified using human genomic DNA as a template and subcloned into the pGEM-T vector as described above.
Construction of a cDNA Library From Purified Aortic HSCs and Differential Screening
The procedure for cDNA library construction and differential screening was exactly as previously described.40 Briefly, 10 μL of cDNAs from CD34+CD45+ aorta-associated cells was submitted to an additional polymerization step (94°C for 5 minutes, 42°C for 5 minutes, and 72°C for 30 minutes), phenol/chloroform-extracted, and EcoRI-digested. After agarose gel purification, 50 ng of cDNAs was ligated into the λ ZAP II vector (Stratagene) and packaged according to the instructions of the manufacturer. The resulting library consisted of 9 × 105 plague forming units, with insert sizes comprised between 500 and 800 bp. Probes for differential screening were obtained by reamplifying for 10 cycles in the presence of 100 μCi [32P] α-dCTP, 1 μL cDNAs from CD34+CD45+ 5-week aortic HSCs (probe 1), same-stage CD34+CD45+ liver HSCs (probe 2), and 6.5-week CD34+CD38− liver HSCs (probe 3). An average of 6,000 recombinant phages were plated and two differential screenings were performed in parallel. After a 6-hour prehybridization step at 65°C in 0.5 mol/L sodium phosphate buffer, pH 7.3, containing 1% bovine serum albumin and 4% SDS, one half of replica filters (Hybond N+; Amersham) was hybridized with probes 1 and 2 and the second half was hybridized with probes 1 and 3 (107 cpm/mL, overnight at 65°C). One hundred candidate clones that exhibited specific hybridization or much brighter intensity with the probe 1 were isolated. Phage inserts were amplified by PCR using the T3 and T7 primers and the PCR products were rehybridized with the three cDNA probes on three independent Southern blots. Only clones specifically hybridizing to probe 1 were further processed for phagemid rescue as instructed by the manufacturer (Stratagene) and sequenced.
DNA Sequencing and Sequence Analysis
DNA sequencing was performed using the PRISM Ready Reaction Dye Deoxy Terminator Cycle Sequencing Kit (Perkin Elmer) on the ABJ model 377 DNA Sequencer (Applied Biosystems, Foster City, CA). Sequence comparisons with all available nucleic acids were performed using the National Center for Biotechnology Informations’ Basic Local Alignment Search Tools [BLASTN, available at the National Center for Biotechnology Information (NCBI) website].
RESULTS
Expression of Early Hematopoiesis-Regulating Factors in CD34+ Embryonic and Fetal Human HSCs
Expression in aorta-associated CD34+ cells.
In situ hybridization of digoxygenin- or 35S-labeled riboprobes on 5-week embryo cross-sections was first used to detect the expression by aorta-associated CD34+ cells of known hematopoietic transcription factors and surface receptors. To label both endothelial and hematopoietic cells in the region of interest, CD34 immunostaining was performed as a control either on adjacent sections, when radiolabeled probes were used, or directly on the same slide when labeled with a digoxygenin-labeled probe. As shown in Fig 1, the cell clusters associated with the endothelial floor of the 5-week aorta express mRNAs encoding the tal-1/SCL, c-myb, GATA-2, and GATA-3 transcription factors. We noticed a higher expression of the GATA-3 transcript in CD34+ cells that were closely adjacent to the aortic endothelium, as well as in mesodermal cells underlying the floor of the aorta (Fig 1E and F). Circulating CD34+GATA-2+ cells visible in the lumen of the aorta (Fig 1D) might reflect the ongoing process of liver colonization, which was shown by the presence of c-myb+cells scattered in the epithelial framework of the hepatic rudiment (not shown).
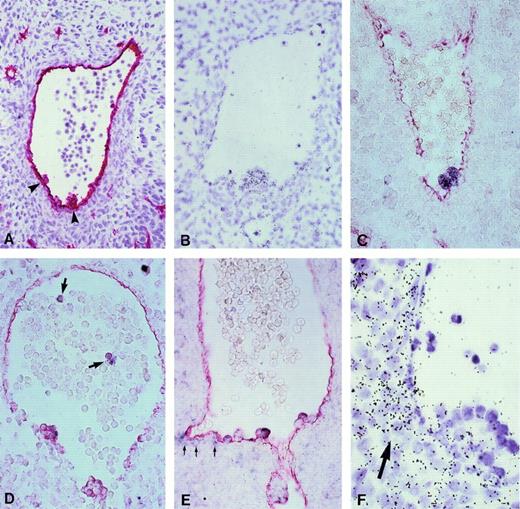
Expression of hematopoietic transcription factors in CD34+ aorta-associated cells from 5-week human embryos. (A) Immunostaining with an anti-CD34 antibody. Arrowheads point to hematopoietic cell clusters in the aorta. Hematoxylin counter-staining (original magnification × 200). (B) Hybridization on an equivalent section of a tal-1/SCL radioactive probe. All cells in the intra-aortic cluster are specifically labeled (original magnification × 260). (C through E) Hybridization of DIG-labeled riboprobes (purple staining) specific for c-myb (C),GATA-2 (D), and GATA-3 (E). Further incubation of these sections with an anti-CD34 antibody (red staining) shows the coexpression of both markers in aorta-associated blood cell progenitors (original magnification × 260). Arrows in (D) point to circulatingGATA-2+ cells in the lumen of the aorta and those in E to a faint GATA-3 signal in mesenchymal cells subjacent to the ventral endothelium of the aorta. (F) GATA-3 labeling in mesenchymal cells underlying the ventral wall of the aorta is obvious using a 35S-radiolabeled probe (arrow). Note the higher expression of GATA-3 mRNA in the innermost cells within a large hematopoietic cluster (original magnification × 650). No signals were observed upon hybridization of sense riboprobes (not shown).
Expression of hematopoietic transcription factors in CD34+ aorta-associated cells from 5-week human embryos. (A) Immunostaining with an anti-CD34 antibody. Arrowheads point to hematopoietic cell clusters in the aorta. Hematoxylin counter-staining (original magnification × 200). (B) Hybridization on an equivalent section of a tal-1/SCL radioactive probe. All cells in the intra-aortic cluster are specifically labeled (original magnification × 260). (C through E) Hybridization of DIG-labeled riboprobes (purple staining) specific for c-myb (C),GATA-2 (D), and GATA-3 (E). Further incubation of these sections with an anti-CD34 antibody (red staining) shows the coexpression of both markers in aorta-associated blood cell progenitors (original magnification × 260). Arrows in (D) point to circulatingGATA-2+ cells in the lumen of the aorta and those in E to a faint GATA-3 signal in mesenchymal cells subjacent to the ventral endothelium of the aorta. (F) GATA-3 labeling in mesenchymal cells underlying the ventral wall of the aorta is obvious using a 35S-radiolabeled probe (arrow). Note the higher expression of GATA-3 mRNA in the innermost cells within a large hematopoietic cluster (original magnification × 650). No signals were observed upon hybridization of sense riboprobes (not shown).
Lower amounts of mRNA encoding the c-kit and flk-1/KDR signal-transducing tyrosine kinases were evidenced only upon hybridization of 35S-labeled probes. Whereas the c-kit messenger appeared uniformly expressed within hematopoietic cell clusters (Fig 2A and B), the flk-1/KDR mRNA was essentially confined therein to the most basal layers of cells, closest to the vascular endothelium (Fig 2D), which itself also exhibited flk-1/KDR intense staining (Fig 2C and D), as did the whole vascular network of the embryo (not shown).
Expression of growth factor receptors by CD34+ aorta-associated cells from 5-week human embryos. (A) All cells within hematopoietic clusters (arrow) are uniformly labeled upon hybridization of a c-kit35S-labeled riboprobe (original magnification × 260). (B) higher magnification of (A; original magnification × 650). (C) Flk-1/KDR transcripts are detectable in aorta endothelial cells upon hybridization of a DIG-labeled riboprobe (arrow), whereas intra-aortic hematopoietic clusters are unlabeled (arrowhead; original magnification × 200). (D) Hybridization of a radioactive flk-1/KDR probe allows to evidence positive cells in the innermost layer within hematopoietic foci (arrowhead; original magnification × 650). No signals were observed upon hybridization of sense riboprobes (not shown).
Expression of growth factor receptors by CD34+ aorta-associated cells from 5-week human embryos. (A) All cells within hematopoietic clusters (arrow) are uniformly labeled upon hybridization of a c-kit35S-labeled riboprobe (original magnification × 260). (B) higher magnification of (A; original magnification × 650). (C) Flk-1/KDR transcripts are detectable in aorta endothelial cells upon hybridization of a DIG-labeled riboprobe (arrow), whereas intra-aortic hematopoietic clusters are unlabeled (arrowhead; original magnification × 200). (D) Hybridization of a radioactive flk-1/KDR probe allows to evidence positive cells in the innermost layer within hematopoietic foci (arrowhead; original magnification × 650). No signals were observed upon hybridization of sense riboprobes (not shown).
Expression in fetal liver and bone marrow HSCs.
The presence of hematopoiesis-associated transcripts was also examined in HSCs that later populate regular fetal blood-forming tissues, ie, liver and bone marrow. Larger and heterogenous hematopoietic populations are present in these organs. For this reason, the study was performed by RT-PCR on CD34+ cell subsets sorted by FACS from 12- to 29-week fetal liver and bone marrow. Because the expression of the CD38 cell surface molecule defines an early step of human HSC activation and commitment, CD38+ and CD38− cells were analyzed separately (Fig 3A). At least four distinct experiments were performed independently using different donor tissues. The absence of CD38+ cells within sorted CD34+CD38− subpopulations was confirmed by RT-PCR using CD38-specific primers (Fig 3B).
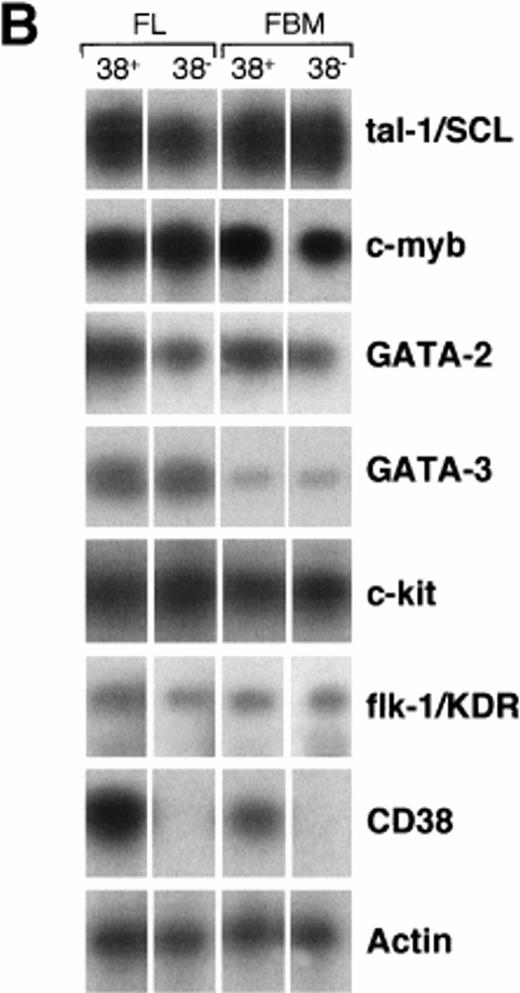
Expression of hematopoiesis-regulating factors by HSCs sorted from 12- to 29-week fetal liver (FL) and bone marrow (FBM). (A) CD34/CD38 two-color stainings of mononucleated cells from 12-week FL and 20-week FBM. The percentages of cells that fall within each of the sorting gates are indicated. The purity of recovered populations was ascertained by PCR-amplification of CD38 cDNA, as shown in (B). (B) Semiquantitative RT-PCR analysis of hematopoiesis-specific genes in the selected CD34+CD38+ and CD34+CD38− cell subsets. Each track is representative of at least four experiments performed on subpopulations sorted from different organs of various stages (see Materials and Methods). Negative control with no cDNA added was included in each PCR experiment and the product size was checked by running molecular weight markers. The amplified products were transferred to nylon membranes and hybridized with internal specific 32P-labeled cDNA probes. Autoradiography was prolonged for 18 hours for flk-1/KDR PCR products but did not exceed 2 hours for the other gene products. Signals obtained for β-actin amplification were used as reference to normalize quantitative differences between cDNA samples.
Expression of hematopoiesis-regulating factors by HSCs sorted from 12- to 29-week fetal liver (FL) and bone marrow (FBM). (A) CD34/CD38 two-color stainings of mononucleated cells from 12-week FL and 20-week FBM. The percentages of cells that fall within each of the sorting gates are indicated. The purity of recovered populations was ascertained by PCR-amplification of CD38 cDNA, as shown in (B). (B) Semiquantitative RT-PCR analysis of hematopoiesis-specific genes in the selected CD34+CD38+ and CD34+CD38− cell subsets. Each track is representative of at least four experiments performed on subpopulations sorted from different organs of various stages (see Materials and Methods). Negative control with no cDNA added was included in each PCR experiment and the product size was checked by running molecular weight markers. The amplified products were transferred to nylon membranes and hybridized with internal specific 32P-labeled cDNA probes. Autoradiography was prolonged for 18 hours for flk-1/KDR PCR products but did not exceed 2 hours for the other gene products. Signals obtained for β-actin amplification were used as reference to normalize quantitative differences between cDNA samples.
As shown in Fig 3B, the tal-1/SCL, c-myb, GATA-2, and GATA-3 transcription factor mRNAs were ubiquitously expressed among subsets of liver and bone marrow fetal progenitors. The c-kit and flk-1/KDR signal-transducing tyrosine kinase messengers were also present in both CD34+CD38+ and CD34+CD38− fetal HSCs. However, we noticed that the amount of PCR-amplified flk-1/KDR transcript was strikingly lower than that of c-kit (see legend to Fig 3) and even undetectable in some cases (not shown).
Cloning of Novel Sequences From CD34+Aorta-Associated Blood Precursor Cells
We next looked for genes that would be differentially expressed between the primitive population of blood precursor cells emerging in the wall of the aorta and HSCs that had colonized the hepatic rudiment. To that end, we constructed a 3′cDNA library from 5-week aorta-associated CD34+ cells sorted by FACS and performed a differential screening of that library with total cDNA probes prepared from aortic CD34+ cells and from liver CD34+ cells isolated from either same-stage or 6.5-week embryos.
Preparation of 3′cDNAs from aortic and hepatic CD34+ embryonic hematopoietic cells.
The dorsal aorta of a 5-week embryo was microdissected on a 10- to 12-somite length between the anterior bifurcation and posterior connection with the vitelline artery. After collagenase dissociation, the cell suspension was labeled with CD34 and CD45 antibodies and CD34+CD45+ double-stained cells were sorted by FACS from CD34+CD45− endothelial cells (Fig 4). The same protocol was used to sort 5-week CD34+CD45+ and 6.5-week CD34+CD38− cells from liver rudiments. These two populations correspond, respectively, to the liver counterpart of aortic HSCs and to the most primitive subset of HSCs present in a 1-week older hepatic tissue. Less than 60 sorted cells were estimated to be recovered from each experiment. Total cDNAs were then prepared from the three populations of hematopoietic progenitors by oligo-dT reverse transcription and subsequent PCR, according to the method originally described by Brady and Iscove39 and modified by Dulac and Axel.40 At least two distinct samples of hematopoietic cells sorted from the aorta and embryonic liver were processed for 3′cDNA amplification. The representation of these amplified cDNAs was then analyzed on Southern blots hybridized with a panel of ubiquitous and specific probes. Figure 5 shows Southern blot analysis of the amplified cDNAs selected to construct the aorta-associated HSC and perform differential screening. The reliable representation of these cDNAs was inferred from the presence of all the transcription factor transcripts previously characterized by in situ hybridization and RT-PCR, together with that of the CD34 messenger and of high amounts of mRNAs encoding both β-actin and GAPDH.
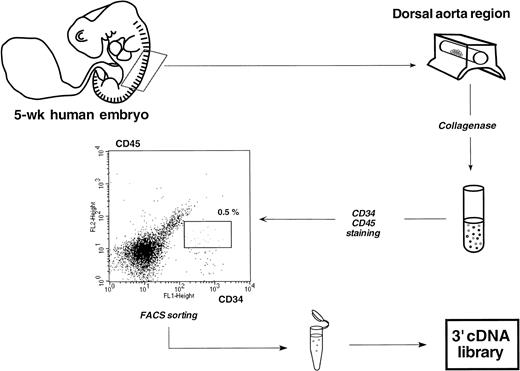
Schematic representation of the protocol used to sort rare double-stained CD34+CD45+ HSCs contained in the aorta of a single 5-week human embryo. Less than 20 double-positive cells were finally recovered from 30,000 total cells obtained after collagenase dissociation. An analogous protocol was used to sort CD34+CD45+ and CD34+CD38− HSCs from a 5- and a 6.5-week embryonic liver, respectively.
Schematic representation of the protocol used to sort rare double-stained CD34+CD45+ HSCs contained in the aorta of a single 5-week human embryo. Less than 20 double-positive cells were finally recovered from 30,000 total cells obtained after collagenase dissociation. An analogous protocol was used to sort CD34+CD45+ and CD34+CD38− HSCs from a 5- and a 6.5-week embryonic liver, respectively.
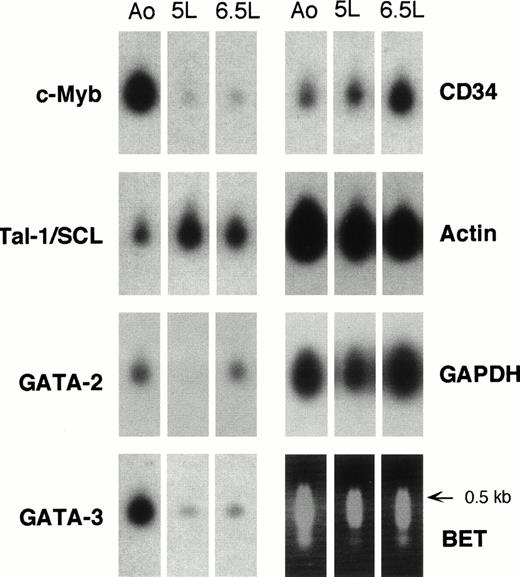
Hybridization of probes for hematopoietic and housekeeping genes to cDNA samples from purified embryonic aorta-associated and liver HSCs. The lower right panel shows the set of samples stained with ethidium bromide before blotting onto nylon filters and probing with the indicated 32P-labeled cDNA probes. Autoradiography times were 1 to 2 hours for β-actinand GAPDH and 8 to 24 hours for hematopoietic genes. Ao, 5-week CD34+CD45+ aortic cells; 5L, 5-week CD34+CD45+ liver cells; 6.5L, 6.5-week CD34+CD38− liver cells.
Hybridization of probes for hematopoietic and housekeeping genes to cDNA samples from purified embryonic aorta-associated and liver HSCs. The lower right panel shows the set of samples stained with ethidium bromide before blotting onto nylon filters and probing with the indicated 32P-labeled cDNA probes. Autoradiography times were 1 to 2 hours for β-actinand GAPDH and 8 to 24 hours for hematopoietic genes. Ao, 5-week CD34+CD45+ aortic cells; 5L, 5-week CD34+CD45+ liver cells; 6.5L, 6.5-week CD34+CD38− liver cells.
Construction and differential screening of a library of embryonic aorta-associated HSC 3′cDNAs.
cDNAs amplified from FACS-sorted CD34+CD45+aorta-associated cells were ligated into phage arms to construct a representative cDNA library. This cDNA as well as cDNAs amplified from 5-week CD34+CD45+ and 6.5-week CD34+CD38− embryonic liver cells were32P-labeled and used as probes for differential library screening. Two differential screenings were performed in parallel. A first set of filters was hybridized with either CD34+CD45+ aortic HSCs cDNA probe or with cDNA probe originating from CD34+CD45+ same-stage liver HSCs. A second screening was performed with cDNA probes from CD34+CD45+ aortic cells and from 6.5-week CD34+CD38− hepatic HSCs. From the screening of 6,000 recombinant phages, 100 clones were isolated that showed specific hybridization to cDNA probe originating from aortic HSCs. To further assess the specificity of these clones, the corresponding inserts were amplified by PCR and hybridized on Southern blots with the three cDNA probes used for the differential screening according to Dulac and Axel.40 Thanks to this sensitive screen, the specificity of 10 clones was confirmed. The sequence of one of these clones appeared identical to a kinase-related sequence previously isolated from the KG-1 cell line and referred to as KIAA0096.42 The expression of this transcript, hereafter designated as KG-1 kinase, was next studied by in situ hybridization on 5-week embryo cross-sections.
Expression of KG-1-kinase mRNA in the human embryo.
Upon in situ hybridization of a radiolabeled KG-1 kinase probe, a faint specific signal was observed within aortic CD34+ blood cell foci in a 5-week embryo (Fig6C). Unexpectedly, the whole endothelial network of the embryo in veins, arteries, and capillaries also specifically expressed the KG-1 kinase messenger (Fig 6B), thus displaying an expression pattern similar to that of the CD34 antigen (Fig 6A). This observation prompted us to investigate more closely the ontogeny of KG-1 kinase expression in the human embryo.
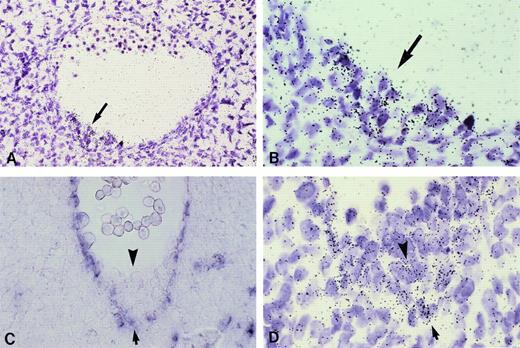
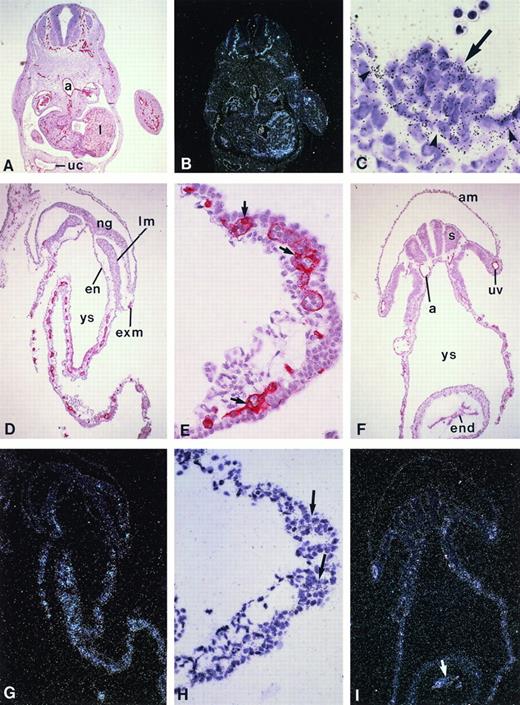
KG-1 kinase expression pattern in the early human embryo and yolk sac. (A through C) Cross-sections through a 5-week human embryo. (A) CD34 immunostaining (red color) is present in the whole endothelial network of the embryo. (B) Dark field illumination of an adjacent section hybridized with the KG-1 kinase riboprobe shows identical expression pattern in blood vessels and capillaries (original magnification × 26). (C) High magnification of (B) in the region of the aorta that shows a hematopoietic cell cluster. Note that endothelial cells (arrowheads) and associated hematopoietic cells (arrow) are labeled, whereas no significant hybridization signal is detected on subaortic mesodermal cells (original magnification × 650). (D through F) CD34 expression on transverse sections of a 5-somite (D and E) and a 15-somite human embryo (F). (D) Section through the postsomitic region of a 5-somite embryo shows CD34+ cells in the yolk sac and extraembryonic mesoderm (original magnification × 65). (E) Detail of (D) in the yolk sac: flattened endothelial and round hematopoietic CD34+ cells are clearly visible in the yolk sac blood islands (arrows; original magnification × 260). (F) Section through the heart region of a 15-somite embryo. CD34 staining is obvious in the yolk sac, as well as in the endocardium and developing dorsal aortae and umbilical veins (original magnification × 65). (G, H, and I) Dark field illumination of sections adjacent to those shown, respectively, in (D), (E), and (F), hybridized with the KG-1 kinaseriboprobe: labeling parallels that of the CD34 antigen in both the yolk sac and embryo proper. (H) Higher magnification (original magnification × 260) of (G; original magnification × 65) showing compact aggregates of labeled cells in yolk sac blood islands. The arrow in (I) points to the labeling in the endocardium (original magnification × 65). No signal was observed upon hybridization of a sense riboprobe (not shown). a, dorsal aorta; am, amnios; en, endoderm; end, endocardium; ex.m, extraembryonic mesoderm; lm, lateral mesoderm; ng, neural grove; s, somite; uc, umbilical cord; uv, umbilical vein; ys, yolk sac.
KG-1 kinase expression pattern in the early human embryo and yolk sac. (A through C) Cross-sections through a 5-week human embryo. (A) CD34 immunostaining (red color) is present in the whole endothelial network of the embryo. (B) Dark field illumination of an adjacent section hybridized with the KG-1 kinase riboprobe shows identical expression pattern in blood vessels and capillaries (original magnification × 26). (C) High magnification of (B) in the region of the aorta that shows a hematopoietic cell cluster. Note that endothelial cells (arrowheads) and associated hematopoietic cells (arrow) are labeled, whereas no significant hybridization signal is detected on subaortic mesodermal cells (original magnification × 650). (D through F) CD34 expression on transverse sections of a 5-somite (D and E) and a 15-somite human embryo (F). (D) Section through the postsomitic region of a 5-somite embryo shows CD34+ cells in the yolk sac and extraembryonic mesoderm (original magnification × 65). (E) Detail of (D) in the yolk sac: flattened endothelial and round hematopoietic CD34+ cells are clearly visible in the yolk sac blood islands (arrows; original magnification × 260). (F) Section through the heart region of a 15-somite embryo. CD34 staining is obvious in the yolk sac, as well as in the endocardium and developing dorsal aortae and umbilical veins (original magnification × 65). (G, H, and I) Dark field illumination of sections adjacent to those shown, respectively, in (D), (E), and (F), hybridized with the KG-1 kinaseriboprobe: labeling parallels that of the CD34 antigen in both the yolk sac and embryo proper. (H) Higher magnification (original magnification × 260) of (G; original magnification × 65) showing compact aggregates of labeled cells in yolk sac blood islands. The arrow in (I) points to the labeling in the endocardium (original magnification × 65). No signal was observed upon hybridization of a sense riboprobe (not shown). a, dorsal aorta; am, amnios; en, endoderm; end, endocardium; ex.m, extraembryonic mesoderm; lm, lateral mesoderm; ng, neural grove; s, somite; uc, umbilical cord; uv, umbilical vein; ys, yolk sac.
Primitive erythropoiesis occurs in the extraembryonic tissues of vertebrates in structures known as blood islands, in which endothelial and hematopoietic cells emerge synchronously.43 44 As judged by CD34 immunostaining on 5-somite embryos (end of week 3 of gestation), equivalent structures where flattened CD34+endothelial cells surround round hematopoietic CD34+/-cells are present in the human yolk sac at that stage (Fig 6D and E). As shown in Fig 6G and H, KG-1 kinase mRNA is also detected in these emerging yolk sac blood islands, whereas blood vessels have not formed yet in the embryo. At 15 somites, KG-1 kinase expression (Fig 6I) still parallels that of CD34 (Fig 6F) in the yolk sac, as well as in the endothelium of the main blood vessels now developing within the embryo proper and in the endocardium, ie, the inner endothelial lining of the heart. No KG-1 kinase expression was ever detected in any other cell type but hematopoietic and endothelial throughout embryonic tissues (Fig 6 and not shown). KG-1 kinase thus appears as an early specific marker of both hematopoietic and endothelial cell lineages in the human embryo.
DISCUSSION
The ontogeny of the hematopoietic system is not limited to prenatal stages, since most blood cells are permanently renewed during the whole life of the organism. It is therefore likely that the identification of novel factors governing the emergence and amplification of HSCs in the embryo could permit critical improvements in the experimental and clinical manipulation of adult HSCs. We first reasoned that blood-forming tissue rudiments should drive the active expansion of ingressing stem cells at the stage of hematopoiesis incipience. Therefore, these could be candidate sites for the identification of stromal cells stimulating early progenitors. Although that approach allowed us to describe the cellular environment of emerging hematopoiesis in the human early fetal bone marrow, no significant expansion of progenitors could be detected at these early stages in the medullary cavities, where blood cells even appeared to develop in the absence of phenotypically identifiable stem cells.45Unexpectedly, the highest local concentration of blood cell progenitors was detected in the ventral wall of the aorta, under the form of several hundred endothelium-adherent CD34+Lin− hematopoietic cells.33 This finding, together with converging results obtained in animal models, suggested that aorta-associated CD34+ hematopoietic cells represent the p-SP–derived stem of the human definitive blood system.
The present study was then undertaken to analyze the molecular events that determine the primary emergence and expansion of these ancestral hematopoietic cells.
Expression of early hematopoiesis-regulating factors.
We show here that CD34+ aorta-associated cells coexpress the tal-1/SCL, c-myb, GATA-2, and GATA-3 mRNAs. These transcription factors were previously identified in animal models as key players in the onset of definitive hematopoiesis. Although their expression within intravascular clumps of hematopoietic cells present at the equivalent stage of mouse development has not been yet reported, the tal-1/SCL and GATA-2 (but not GATA-3) transcripts were found in lymphohematopoietic progenitors generated in mouse day-7 embryo cultures.46tal-1, GATA-2, and c-myb are also expressed by hematopoietic progenitors emerging in vitro from ES cells in the course of their differentiation into embryoid bodies.47 Moreover, tal-1, GATA-2, GATA-3, and c-myb were shown to be continuously transcribed in Xenopus from the stage of early neurula in both the ventral blood islands and dorsal lateral plate, which are equivalent to the murine yolk sac and p-SP, respectively.48 Altogether these data suggest that hematopoietic commitment from either extraembryonic or intraembryonic mesoderm involves a similar early transcriptional program from lower vertebrates to humans.
We also show that HSCs associated with the 5-week human embryo aorta express two early receptor tyrosine kinase genes, c-kit and flk-1/KDR, as does the mouse p-SP/AGM,15,16,49 in which c-Kit+ cells are solely responsible for LTR activity.13,50 As expected, endothelial cells bordering the aortic clusters were strongly labeled with the flk-1/KDR probe, but interestingly, we also found a lower level of KDR mRNA in associated hematopoietic cells. This observation supports the hypothesis, first raised from the results of knock-out experiments in mice, that vascular endothelial growth factor (VEGF) receptor also plays a role in hematopoietic development.16,27 However, our observation that only CD34+ cells closest to the vessel wall express detectable KDR messengers, whereas the most peripheral CD34+ cells are negative is striking. This expression gradient may reflect the commitment to hematopoiesis of bipotential hemangioblasts, ie, common progenitors for both endothelial and hematopoietic cell lineages.43,51 The long-assumed existence of hemangioblasts was supported by recent experiments performed in the quail embryo, in which VEGF-R-2+ cells sorted at the primitive streak stage could be induced to differentiate along either hematopoietic or endothelial cell lineages according to culture conditions.52 In mammals, the existence of a bipotent hemangioblast is suggested by the identification, within early differentiating embryoid bodies, of blast colony-forming cells that develop in response to VEGF and generate mixed cultures of endothelial and hematopoietic cells.53 54
The hematopoietic molecular markers expressed by aortic human HSCs were also present in embryonic and fetal HSCs throughout development. Except for the flk-1/KDR growth factor receptor, these markers were previously detected in adult HSCs as well.55-59 The apparent identical expression of these factors in both quiescent CD34+CD38− and cycling CD34+CD38+ cells may be explained by the persistence of the tal-1/SCL transcription factor in the differentiating erythroid lineage60 and by the expression of GATA-2, c-myb, and c-Kit receptor by committed hematopoietic progenitors.59,61,62 However, of note, there was a significantly reduced amount of flk-1/KDR mRNA in fetal liver and bone marrow HSCs, which is consistent with previous findings of a dramatic reduction in mice in the number of Flk-1+ HSCs along development of the hematopoietic system.63
Cloning of a novel early marker of hematopoietic and endothelial development.
A cDNA library differential screening method permitted us to clone a sequence already isolated from undifferentiated KG-1 cells in a systematic attempt to clone unidentified human genes.42 The sequence of this gene includes a serine/threonine kinase consensus motif together with a presumptive prenyl group-binding site42 similar to that encountered in the Ras family of protein kinases. KG-1 kinase shows a striking expression in vascular endothelial cells in the whole embryo. In the late 3-week yolk sac, the earliest stage tested so far, the KG-1 kinase mRNA is already detected in the blood islands. This novel hematopoietic factor thus adds to the list of markers shared by cells of these two lineages from the early stages of development, such as MB1 in birds,64 CD34 in mouse and human,14,33 and, to a lesser extent, flk-1/KDR.16 The shared expression of the KG-1 kinase by endothelial and hematopoietic cells in both intraembryonic and extraembryonic territories of active primary hematopoiesis may be related to the derivation of these two cell lineages from common hemangioblastic precursors. Interestingly, the closely related expression of the KDR tyrosine and KG-1 serine/threonine kinase messengers in the early developing human embryo may imply that both act in concert. On the other hand, the expression pattern of the KG-1 kinase-encoding gene is reminiscent of that of cloche, whose mutation in zebrafish dramatically perturbs both hematopoietic and endothelial differentiation.65 Because cloche acts upstream of flk-1 and GATA-2,66 it would be of interest to compare the expression patterns of KG-1 kinase, KDR, and GATA-2 mRNAs in the early human embryo. The identification of the gene encoding the mouse KG-1 kinase gene will help to determine its expression from the stage of gastrulation and to analyze its role in the establishment of the endothelial and hematopoietic cell lineages through targeted mutation in ES cells.
ACKNOWLEDGMENT
The authors are indebted to P. Vaigot for expert cell sorting by flow cytometry and to C. Debacker for excellent technical assistance throughout. We are grateful to H. San Clemente, F. Viala, F. Beaujean, and S. Gournet for their help in the design and preparation of figures and to M. Scaglia who expertly typed the manuscript. We also thank C. Carrière and Prof E. Aubeny for procurement of first trimester embryos and Drs M. Catala, F. Narcy, A.-L. Delezoide, and Prof C. Nessmann for providing fetal tissues. We are thankful for the generous hospitality of R. Axel that was essential for the further construction of the PCR-based cDNA library from embryonic HSCs.
Supported in part by grants from Association pour la Recherche sur le Cancer. F.C. was the recipient of a fellowship from the European Commission.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Marie-Claude Labastie, PhD, Institut d’Embryologie Cellulaire et Moléculaire, CNRS UPR 9064, 49bis, avenue de la Belle Gabrielle, 94736 Nogent-sur-Marne Cedex, France; e-mail: labastie@infobiogen.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal