Abstract
The extended (E) isoform unique to Fibrinogen420 (Fib420) is distinguished from the conventional chain of Fibrinogen340 by the presence of an additional 236-residue carboxyl terminus globular domain (EC). A recombinant form of EC (rEC), having a predicted mass of 27,653 Daltons, was expressed in yeast (Pichia pastoris) and purified by anion exchange column chromatography. Purified rEC appears to be predominantly intact, as judged by N-terminal sequence analysis, mass spectral analysis of the C-terminal cyanogen bromide (CNBr) fragment, and comparison of recognition by epitope-specific monoclonal antibodies. Carbohydrate determination, coupled with analysis of CNBr digestion fragments, confirms N-linked glycosylation at Asn667, the site at which sugar is attached in E. Analysis of CNBr digestion fragments confirms that two disulfide bridges exist at cysteine pairs E613/644 and E780/793. In the presence of 5 mmol/L EDTA, rEC is highly susceptible to plasmic degradation, but Ca2+ (5 mmol/L) renders rEC resistant. No protective effect from plasmic degradation was conferred to rEC by the peptides GPRPamide or GHRP, nor did rEC bind to a GPR peptide column. These results suggest that the EC domain contains a calcium-binding site, but lacks a polymerization pocket. By analogy with the site elucidated in the γC domain, we predict that the EC calcium binding site involves residues E772-778: DADQWEE.
TWO SUBCLASSES OF fibrinogen molecules can be distinguished in normal blood based on their α chain isoforms. Recognition of the existence of a second subclass evolved from the discovery that the complete sequence of the α gene contains an additional exon (exon VI of the human gene) encoding 236 amino acids (VI-domain) homologous to the fibrinogen β- and γ-chain carboxyl termini.1 The α gene, in addition to giving rise to the conventional form of the α subunit that lacks the VI-domain, also yields a transcript that, when alternatively spliced, encodes the extended α chain (αE isoform)2 containing the exon VI-encoded carboxy terminus (αEC).
Fibrinogen containing αE has subsequently been shown to be a symmetrical molecule of the structure (αEβγ)2,3 ie, the common fibrinogen α chains have both been replaced by αEchains against all stoichiometric odds, given the overwhelming 100:1 ratio of α mRNA:αE mRNA in the hepatocyte.2Based on mass prediction, (αEβγ)2 has been termed Fibrinogen420 (Fib420) to distinguish it from the common (αβγ)2 subclass, similarly referred to as Fibrinogen340(Fib340).
In the normal adult population, on average only 1 of every 100 molecules of fibrinogen is a molecule of Fib420.4 However, homologues of Fib420’s αE chains have been identified throughout the vertebrate kingdom and αEC itself represents the largest conserved segment of the α gene,1,5 6 a startling observation that suggests that the domain imparts a critical and distinctive functionality.
Clot formation and lysis, the primary functions of the major form of fibrinogen (Fib340), have been characterized in detail over a period of several decades.7 8 Briefly, thrombin cleavage of fibrinogen releases fibrinopeptides A and B from the amino termini of the α and β chains, and the resulting fibrin monomers spontaneously polymerize. Interactions between each newly exposed α-chain N-terminus and complementary polymerization pockets located in the carboxyl-terminal domains of the γ chains provide a main driving force for fibrin monomer association. This noncovalent association brings together each central amino terminal E-domain with the distal carboxyl terminal D-domains of two other fibrin molecules, forming a half-staggered, double array of protofibrils. This organization enables factor XIIIa-catalyzed covalent stabilization of the polymers by ε-amino-(γ-glutamyl) lysine cross-links. Calcium ions promote both the formation and cross-linking of fibrin polymers, and calcium binding limits fibrin(ogen)’s susceptibility to proteases such as plasmin.
Fib420 also participates in clot formation3(Applegate et al, manuscript in preparation); however, the influence of its two αEC domains on this process and on clot lysis is not known. In this context, the current study reports the generation of a soluble recombinant form of the human αEC (rαEC) and an evaluation of its physical and biochemical properties.
MATERIALS AND METHODS
Materials.
Human plasmin, thrombin, and fibrinogen fragment D19 were generous gifts of M. Mosesson (Sinai Samaritan Medical Center, Milwaukee, WI). Human factor XIII was graciously provided by P. Bishop (ZymoGenetics, Seattle, WA). A GPR peptide column was kindly provided by R. Doolittle (University of California at San Diego, La Jolla, CA).
rαEC expression.
A segment encoding the C-terminal domain of αE(Val610-Gln847) was generated by polymerase chain reaction (PCR), inserted in pPIC9 at the Avr II site, and verified by DNA sequencing. The recombinant αEC domain was expressed in the methylotrophic yeast strain Pichia pastoris (Mut−) according to the manufacturer’s protocol (Invitrogen, San Diego, CA). Throughout this report, residues in rαEC are numbered according to their corresponding position in the αE chain (GenBank accession no. M58569).
Large scale expression.
Production of rαEC was scaled up from flask to fermentor culture in a 12-L Microferm fermentor (New Brunswick Scientific, Edison, NJ), based on the Invitrogen fermentation protocol. Briefly, growth phase was initiated with inoculation of 6 L of medium with a P pastoris clone expressing rαEC and maintained as a glycerol batch culture for 24 hours, followed by continuous feeding of glycerol for another 24 hours. Feeding was interrupted for 0.5 hours (starvation) and the production phase was induced by the addition of methanol. The culture was stirred at 1,000 rpm and kept at 30°C throughout, with compressed air (5 psi) providing aeration. Dissolved oxygen, pH, and wet weight of cells were monitored. The antifoam agent Struktol KFO 673 (0.2%; Kabo Chemicals, Inc, Jackson Hole, WY) was present from the beginning. Because of fibrinogen’s sensitivity to low pH, a pH of 6 was maintained throughout the fermentation. Casamino acids (1%; Difco, Livonia, MI) were included during the production phase to suppress yeast proteases that may be active at this pH. Production of rαEC was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western analysis. After a total run of 100 hours (52-hour production phase), the yeast cells (wet weight, ∼200 g/L) were pelleted at 3,600 rpm. The recovered supernatant (5.4 L) was stored at 4°C overnight to dissolve the antifoam reagent. The conditioned medium was first filtered (0.45-μm Nalgene membrane, with a double layer of prefilters) and then concentrated approximately 10-fold with a Pellikon apparatus (Millipore, Bedford, MA; exclusion 10 kD). The crude fermentor culture supernatant was stored at −80°C.
Purification of rαEC.
Crude fermentor supernatant was dialyzed against 20 mmol/L Tris, pH 7.5, clarified by centrifugation at 5,000g, and subsequently applied to a Mono Q HR 5/5 anion exchange column (Pharmacia, Piscataway, NJ) pre-equilibrated with the same buffer. Bound protein was eluted with a 20-column volume linear salt gradient (20 mmol/L Tris, pH 7.5, to 1 mol/L NaCl, 20 mmol/L Tris, pH 7.5).
SDS-PAGE and Western blot analysis.
Samples were prepared for electrophoresis in Laemmli sample buffer in the absence or presence of 0.1 mol/L dithiothreitol10 and separated on SDS-PAGE using a Mini-Protean II Electrophoresis Cell (Bio-Rad, Hercules, CA). Protein was stained with Coomassie Blue R250. For Western blots, transfer onto 0.2-μm nitrocellulose membranes was performed with a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). Membranes were incubated with primary monoclonal antibodies followed by secondary antibody, horseradish peroxidase (HRPO)-labeled goat antimouse IgG (Pierce, Rockford, IL). To visualize enzyme activity, signals were developed by enhanced chemiluminescence (SuperSignal Substrate; Pierce) and filmed.
Isoelectric focusing.
Linear 4-6.5 gradient IEF PhastGels were run on the PhastSystem (Pharmacia). Recombinant αEC was diluted in 125 mmol/L NaCl, 25 mmol/L HEPES, pH 7.4, and compared with low isoelectric point calibration markers (Pharmacia Biotech, Piscatway, NJ) run under the same conditions.
N-terminal sequence determination.
The material was run on SDS-PAGE under reducing conditions, transferred to a polyvinylidene difluoride membrane, and stained with Coomassie Blue G250. The protein band was cut out and subjected to 20 cycles of microsequencing (Model 477A; Applied BioSystems, Inc, Foster City, CA).
Carbohydrate analysis.
N-linked glycosylation of rαEC was evaluated by Glyko, Inc (Novato, CA). In short, protein-bound oligosaccharides were released with PNGase F endoglycosidase, and the sugars were labeled with fluorophore, separated using PAGE, and visualized and analyzed using Glyko’s FACE Imager and software. The exact nature of the bands was determined by fingerprinting using several exoglycosidases alone or in combination: neuraminidase, β-galactosidase, β-N-acetylhexosaminidase, and α-mannosidase.
Cyanogen bromide (CNBr) cleavage.
rαEC was cleaved with CNBr in 70% formic acid for 18 hours as described by Blomback et al.11 The fragments were separated by high-performance liquid chromatography (HPLC) using a reverse-phase C18 column for analysis with a MALDI TOF mass spectrometer (Voyager DE; PE Biosystems, Framingham, MA).
Digestion with plasmin.
rαEC (0.4 mg/mL) was digested with human plasmin (0.5 U/mL) at 37°C in a buffer containing 125 mmol/L NaCl, 25 mmol/L HEPES, pH 7.4, in the presence of either 5 mmol/L EDTA or 5 mmol/L CaCl2. For plasmin protection experiments with peptides (5 mmol/L), the samples contained 5 mmol/L EDTA and either GPRPamide or GHRP (Sigma, St Louis, MO). In all cases, reactions were stopped by the addition of aprotinin (60 KIU/mL), followed by the addition of Laemmli sample buffer and boiling.
Affinity chromatography.
rαEC (0.2 mg; ∼33 nmol) in 135 μL of buffer containing 125 mmol/L NaCl, 25 mmol/L HEPES, pH 7.4, and 5 mmol/L CaCl2 was loaded on a 1.6-mL GPR peptide column pre-equilibrated with the same buffer. The column was then washed with 10 column volumes of loading buffer. Fractions (1 mL) were collected and absorbance at 280 nm was determined. After confirming that absorbance had returned to zero, bound protein was eluted with 3 column volumes of 1.0 mol/L NaBr, 0.05 mol/L sodium acetate, pH 5.3.12 13
Factor XIIIa-catalyzed cross-linking.
The reaction was performed in 5 mmol/L CaCl2, 100 mmol/L NaCl, 50 mmol/L HEPES, pH 7.4, at room temperature. It was initiated by the addition of human thrombin (1 U/mL) to a solution containing recombinant human factor XIII (40 μg/mL) plus substrate (4 mg/mL), either fibrinogen fragment D1 or rαEC. Aliquots were removed and the reaction was stopped by boiling in Laemmli sample buffer.
RESULTS AND DISCUSSION
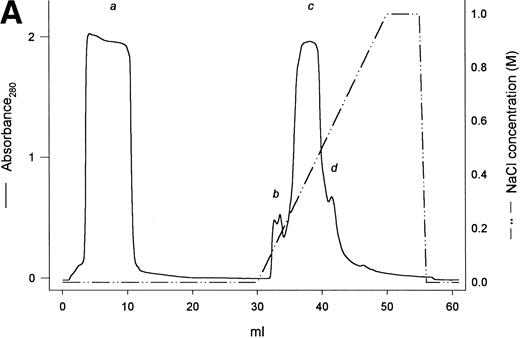
A protein of about 38 kD was the predominant protein secreted upon methanol induction of the yeast P pastoris transformed with a pPIC9 vector carrying the exon VI sequence of the human fibrinogen α gene (Fig 1B, lane 1). Recognition by a polyclonal antibody highly specific for the VI-domain of αE3 confirmed this band to be the recombinant αEC domain. In the first 52 hours of the methanol induction phase, rαEC accumulated to a level of about 200 μg/mL in the fermentation culture medium. After concentration, rαEC was purified from the medium supernatant by anion exchange chromatography (Fig 1A). Gel analysis showed that the majority of protein contaminants appeared in the flow-through (Fig 1B, lane 2). The rαEC domain eluted predominantly in five fractions between 350 and 400 mmol/L NaCl (peak c). These fractions were combined, dialyzed against 125 mmol/L NaCl, 25 mmol/L HEPES, pH 7.4, and concentrated for use in this study.
Purification of rEC by anion exchange chromatography. Crude fermentor supernatant was dialyzed and injected (10 mL) onto a 1-mL Mono Q HR 5/5 anion exchange column pre-equilibrated with the same buffer, as described in Materials and Methods. Bound protein was eluted with a 20-mL linear salt gradient (20 mmol/L Tris, pH 7.5, to 1 mol/L NaCl, 20 mmol/L Tris, pH 7.5) and fractions (1 mL) were collected. (A) Elution profile with absorbance plotted as a solid line (scale on the left) and the salt gradient (M NaCl) as the broken line (scale on the right). (B) Evaluation of fractions by SDS-PAGE. Fraction samples were run on 4% to 20% gradient SDS-PAGE under reducing conditions. S denotes crude fermentor supernatant. Lanes corresponding to sequentially collected fractions under the following peaks are indicated with lowercase letters: peak a, pooled flow-through; peak b, two fractions; peak c, four fractions; peak d, one fraction. Eluted material was deliberatedly overloaded on the gel to enhance detection of minor contaminants.
Purification of rEC by anion exchange chromatography. Crude fermentor supernatant was dialyzed and injected (10 mL) onto a 1-mL Mono Q HR 5/5 anion exchange column pre-equilibrated with the same buffer, as described in Materials and Methods. Bound protein was eluted with a 20-mL linear salt gradient (20 mmol/L Tris, pH 7.5, to 1 mol/L NaCl, 20 mmol/L Tris, pH 7.5) and fractions (1 mL) were collected. (A) Elution profile with absorbance plotted as a solid line (scale on the left) and the salt gradient (M NaCl) as the broken line (scale on the right). (B) Evaluation of fractions by SDS-PAGE. Fraction samples were run on 4% to 20% gradient SDS-PAGE under reducing conditions. S denotes crude fermentor supernatant. Lanes corresponding to sequentially collected fractions under the following peaks are indicated with lowercase letters: peak a, pooled flow-through; peak b, two fractions; peak c, four fractions; peak d, one fraction. Eluted material was deliberatedly overloaded on the gel to enhance detection of minor contaminants.
Characterization of rαEC.
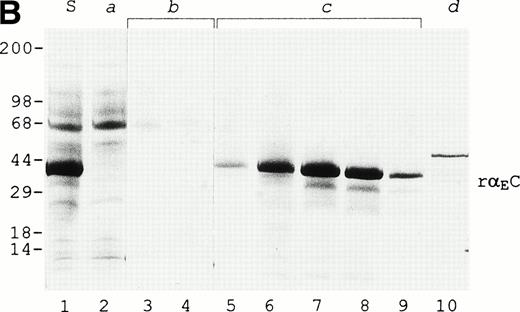
The purified rαEC appears to be essentially intact based on N-terminal sequence analysis, on C-terminal epitope recognition, and on mass spectrometry of CNBr fragments. Microsequencing established the 20 N-terminal residues of rαEC to be[YVEFPR]VRDXDDVLQTXPXG, representing an initial set of six residues generated by the multiple cloning region of the vector (in italics) and continuing with valine as the first bona fide fibrinogen residue (Val610), the final amino acid of the conventional α chain as it is found in circulating Fib340. In rαEC, this valine is followed, as in αE, by Arg611 and then the 236 residues of the VI-domain proper (Asp612-Gln847). A product beginning at Ile631 of the αE chain was also detected. Accounting for less than 10% of the purified rαEC, it is evident as the fainter band migrating just ahead of intact rαEC on SDS-PAGE (Fig 1B) and presumably represents partially degraded rαEC.
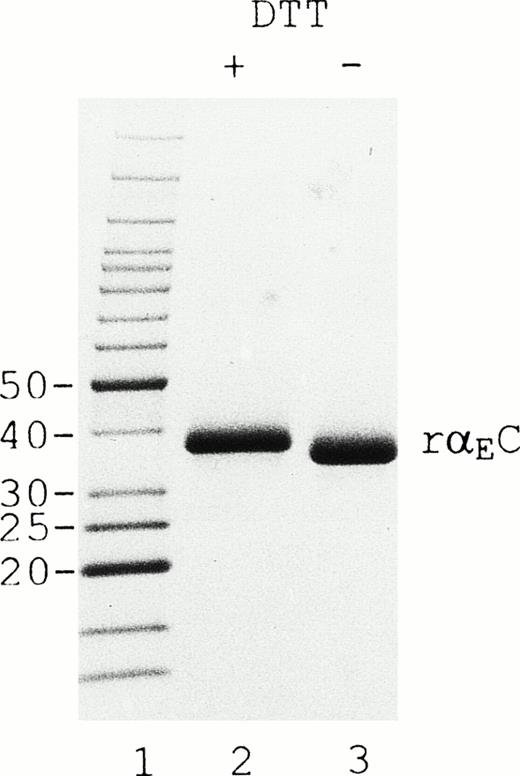
On SDS-PAGE (Fig 2), reduction of rαEC produces an upward mobility shift from an apparent size of 34 to 38 kD, indicating the presence of internal disulfide bridges. The αEC domain has 4 cysteine residues available for such bonds at positions 613, 644, 780, and 793. Their positions, invariant among αE homologues throughout vertebrate evolution, align precisely with cysteines in the βC and γC domains.2,5 6 By analogy with βC and γC, as well as from the results of mutational analysis of αE,13a we expected disulfide bonding in rαEC to be restricted to the cysteine pairs αE613/644 and αE780/793.
rEC contains internal disulfide bridges. Recombinant EC was run under reducing (lane 2) and nonreducing (lane 3) conditions on 4% to 20% gradient SDS-PAGE. A GIBCO/BRL (Grand Island, NY) Benchmark protein ladder was used for molecular mass estimation (lane 1).
rEC contains internal disulfide bridges. Recombinant EC was run under reducing (lane 2) and nonreducing (lane 3) conditions on 4% to 20% gradient SDS-PAGE. A GIBCO/BRL (Grand Island, NY) Benchmark protein ladder was used for molecular mass estimation (lane 1).
To test the prediction, rαEC was treated with CNBr. The three methionines in the sequence of the recombinant protein are distributed such that cleavage after these positions should generate four unequal fragments, with the predicted disulfide loops internally situated in two separate fragments (Table 1). Indeed, four major CNBr-generated fragments of rαEC were observed by SDS-PAGE under both reducing and nonreducing conditions. Moreover, upward mobility shifts upon reduction provided positive, albeit indirect, evidence for internal disulfide bonds within fragments 1 and 3, consistent with the predicted assignments. In contrast, fragment 2, with no cysteines, migrated as a broad band with the same mobility whether reduced or not. The smallest fragment, containing the domain’s C-terminal amino acid residues, was barely detectable by SDS-PAGE, but was identified by mass spectrometry as having the predicted molecular mass of 954 Daltons.
Analysis of CNBr Fragments of rEC
| Fragment . | Sequence . | Mass (kD) . | ||
|---|---|---|---|---|
| Observed . | Calculated . | |||
| −DTT . | +DTT . | |||
| 1 | YVEFPR-V610 to M660 | 5 | 6 | 6.462 |
| 2 | D661 to M764 | 18 | 18 | 11.649 |
| 3 | Q765 to M839 | 7 | 8.5 | 8.642 |
| 4 | K840 to Q847 | 1 | 1 | 0.954 |
| Fragment . | Sequence . | Mass (kD) . | ||
|---|---|---|---|---|
| Observed . | Calculated . | |||
| −DTT . | +DTT . | |||
| 1 | YVEFPR-V610 to M660 | 5 | 6 | 6.462 |
| 2 | D661 to M764 | 18 | 18 | 11.649 |
| 3 | Q765 to M839 | 7 | 8.5 | 8.642 |
| 4 | K840 to Q847 | 1 | 1 | 0.954 |
CNBr cleavage of rαEC was performed as described in Materials and Methods. The fragments were analyzed on 16.5% Tris-Tricine SDS-PAGE (Bio-Rad) in the presence and absence of 0.1 mol/L DTT. Fragment sizes were determined by comparison with low-range SDS-PAGE standards from Bio-Rad. Sequence numbering is according to the full αE sequence (GenBank accession no. M58569).
The calculated mass of the 244 amino acids of the full recombinant fusion protein, TyrValGluPheProArgVal610-Gln847, is 27,653 Daltons. The sizable discrepancy between this number and 34 kD, its apparent molecular mass on SDS-PAGE under nonreducing conditions (Fig2), can be attributed to glycosylation. Whereas the conventional α chain of Fib340 has no carbohydrate, the αEC domains of Fib420 are decorated, attachment occurring only at Asn66713a even though a second potential site exists at Asn812.2 Carbohydrate analysis of the yeast-expressed rαEC showed that roughly two thirds of the molecules contained high mannose N-linked oligosaccharides (Man6-13GlcNAc2) common to other glycoproteins expressed in P pastoris14; phosphorylated low mannose sugars (Man2P GlcNAc2) were deduced to be present on the remainder. This heterogeneity permits assignment of the attachment site in rαEC to CNBr fragment 2, which migrated on SDS-PAGE not only as the broadest band, but also displayed an apparent mass considerably greater than that calculated from the amino acid sequence alone (Table 1). This finding indicates sole use of the site at Asn667 by yeast, consistent with glycosylation of the αE chains of human Fib420. The structure of the carbohydrate attached to the native αE remains to be determined, although it is likely to be of the low mannose type ending predominantly in sialic acid, as in the β and γ chains,15 rather than the high mannose type found in the yeast-derived recombinant domain.
The recombinant domain’s predicted pI is 4.3, based on its amino acid sequence. Isoelectrofocusing of rαEC yielded two closely migrating bands located between the pI markers at 4.15 and 4.55 (not shown), consistent with the prediction. It may be that these doublet bands reflect the heterogeneity of the carbo- hydrate side chain described above.
Functional properties.
The primary structure of Fib420 is identical to that of Fib340 but for the αEC domains.2 3 We used rαEC to investigate whether these domains might contribute to Fib420 additional cross-linking sites, calcium-binding sites, and/or polymerization pockets. This yeast recombinant closely approximates the conformation of the native domain as indicated by its use of the same carbohydrate attachment site and the deduced congruence of its internal disulfide bridges described above.
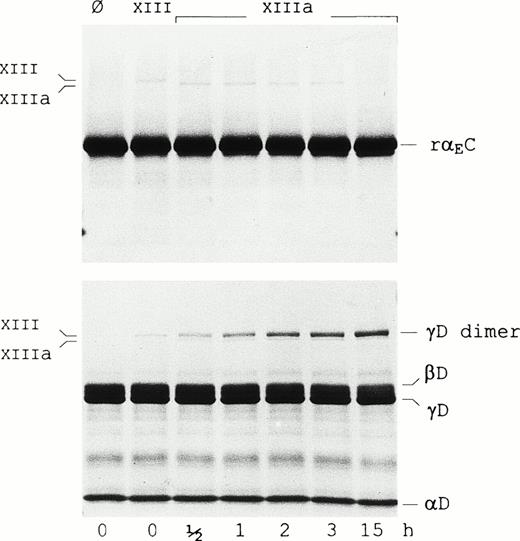
It has been suggested that the αEC domains participate in factor XIIIa-catalyzed cross-linking based on studies in lamprey.13 To evaluate cross-linking between the human αEC domains, rαEC was incubated with factor XIIIa (Fig 3). Although the reaction was performed at high substrate and enzyme concentrations to increase its efficiency, no formation of rαEC-dimers was observed, even after prolonged incubation. Under similar conditions, significant cross-linking of the γ-chain fragment of fragment D1(γD) occurred, as evidenced by the steady accumulation of γD-dimer over the 15-hour period. In addition, reactions conducted with Fib340 as substrate in a 10-fold molar excess of rαEC yielded no evidence of the domain cross-linking either to itself or to fibrinogen (not shown). These data suggest that Fibrin(ogen)420’s αEC domains may not cross-link. Taken together with recent mutagenesis studies finding no evidence of disulfide bonding between αEC domains and the rest of the Fib420 molecule,13a the findings support the notion that, in a clot, the αEC domains may be tethered only via their upstream “αC” protein sequence.
Factor XIIIa does not cross-link rEC. Factor XIII-catalyzed cross-linking reactions were performed as described in Materials and Methods with either the rEC domain (upper panel) or fragment D1 (lower panel) as substrate. Lanes Ø contain substrate alone. Samples in the adjacent lane in each panel were further supplemented with nonactivated factor XIII (FXIII), whereas those in the remaining lanes were treated with thrombin-activated factor XIII (FXIIIa) and incubated for the lengths of time indicated. Samples were run on 4% to 20% gradient SDS-PAGE under reducing conditions. Positions of factor XIII, factor XIIIa, the fragment D1 subunits (D, βD, and γD) as well as the γD-dimer are indicated in the margins.
Factor XIIIa does not cross-link rEC. Factor XIII-catalyzed cross-linking reactions were performed as described in Materials and Methods with either the rEC domain (upper panel) or fragment D1 (lower panel) as substrate. Lanes Ø contain substrate alone. Samples in the adjacent lane in each panel were further supplemented with nonactivated factor XIII (FXIII), whereas those in the remaining lanes were treated with thrombin-activated factor XIII (FXIIIa) and incubated for the lengths of time indicated. Samples were run on 4% to 20% gradient SDS-PAGE under reducing conditions. Positions of factor XIII, factor XIIIa, the fragment D1 subunits (D, βD, and γD) as well as the γD-dimer are indicated in the margins.
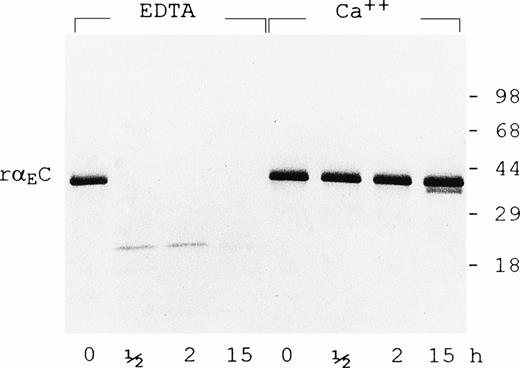
In the presence of 5 mmol/L EDTA, rαEC is highly susceptible to plasmin (0.5 U/mL), being degraded to species no larger than 18 kD within 30 minutes (Fig 4). However, calcium ions (5 mmol/L) largely protect rαEC from plasmin for at least 15 hours. The calcium protection is reminiscent of that observed for the carboxyl terminal domain of γ chains (γC)16,17 and argues strongly for an active calcium binding site within αEC. By analogy with the site elucidated in the γC domain,18-20 we predict that the αEC calcium binding site involves residues 772-778: DADQWEE. Interestingly, the corresponding region of the βC domain has a five-residue insert that, according to a recently reported structural study, diminishes but does not obliterate its affinity for calcium.21
Calcium protects EC domain from digestion by plasmin. Recombinant EC was digested with plasmin (0.5 U/mL) for the times indicated in the presence of either EDTA (5 mmol/L) or CaCl2 (5 mmol/L) as described in Materials and Methods. Samples were run on 4% to 20% gradient SDS-PAGE under reducing conditions.
Calcium protects EC domain from digestion by plasmin. Recombinant EC was digested with plasmin (0.5 U/mL) for the times indicated in the presence of either EDTA (5 mmol/L) or CaCl2 (5 mmol/L) as described in Materials and Methods. Samples were run on 4% to 20% gradient SDS-PAGE under reducing conditions.
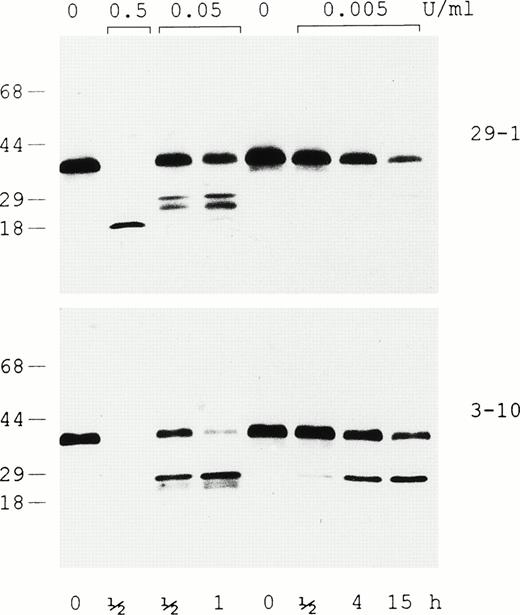
Plasmin digestion of rαEC in the presence of 5 mmol/L EDTA was evaluated by Western analysis with two different monoclonal antibodies, one that recognizes an epitope at the C-terminus (29-1) and another (3-10) that recogizes an epitope downstream in the N-terminal third of the VI-domain (Fig 5). The immunoblot shows generation of a relatively stable 18-kD degradation fragment containing the C-terminal epitope at the highest plasmin concentration, whereas lower levels of the enzyme produce a variety of fragments in the 20- to 25-kD range. The relative amounts of the latter, which bear either the N- or C-terminal epitope, are dependent on the time of exposure and the concentration of plasmin. In this context, it is noteworthy that both antibodies recognize the starting material’s major 38-kD band as well as the minor species migrating immediately below it (presumably the species beginning at αE631), providing further evidence that the yeast-expressed rαEC domain has a structurally intact C-terminus.
Evaluation of intact rEC and its plasmic degradation products by immunoblotting. In the presence of 5 mmol/L EDTA, rEC was digested with plasmin at either 0.5, 0.05, or 0.005 U/mL for the lengths of time indicated and run on 4% to 20% gradient SDS-PAGE under reducing conditions. Western analysis was performed using two monoclonal antibodies: 29-1, which recognizes an epitope at the domain’s C-terminus, and 3-10, which recognizes an epitope downstream in the N-terminal third of EC.
Evaluation of intact rEC and its plasmic degradation products by immunoblotting. In the presence of 5 mmol/L EDTA, rEC was digested with plasmin at either 0.5, 0.05, or 0.005 U/mL for the lengths of time indicated and run on 4% to 20% gradient SDS-PAGE under reducing conditions. Western analysis was performed using two monoclonal antibodies: 29-1, which recognizes an epitope at the domain’s C-terminus, and 3-10, which recognizes an epitope downstream in the N-terminal third of EC.
The synthetic peptide GPRPamide, resembling the amino terminus of the fibrin α chain, limits the plasmic digestion of fragment D and the γC domain.16 22 Analysis of rαEC incubated with plasmin showed that the synthetic peptides GPRPamide or GHRP (the latter mimicking the amino terminus of the fibrin β chain) rendered no protective effect (not shown). To test whether analogues of the fibrin α chain amino terminus bind rαEC without affecting its sensitivity to plasmin, we performed affinity chromatography by passing purified rαEC over a GPR peptide column as described in Materials and Methods. Under conditions in which intact fibrinogen bound completely, no rαEC bound to the column, all of it appearing in the flow-through. These results suggest that the αEC domain lacks a polymerization pocket.
In conclusion, this study strongly suggests that the conformation of recombinant αEC expressed in yeast closely approximates that of the native domain. Experiments with the recombinant domain indicate that Fib420 is endowed with one extra calcium-binding site per αEC but no additional polymerization pockets or cross-linking sites. The usefulness of the rαEC generated in this study is underscored by our recent observation that, when Fibrin(ogen)420 is treated with plasmin, the αEC domains are readily clipped off and, under physiologic conditions, remain intact (Applegate et al, manuscript in preparation). Intriguingly, the freed domain is detectable in human plasma, particularly under pathophysiologic conditions. Thus, rαEC may serve as an excellent model substance for testing the potential biological function(s) of this unique and stable degradation product generated from Fibrin(ogen)420.
ACKNOWLEDGMENT
The authors thank Dr James Farmar of the Microchemistry Laboratory for performing the mass spectral analyses and Dr Bohdan Kudryk for many helpful discussions. Our gratitude goes to Dr Jack Goldstein and members of his laboratory, particularly Drs Alex Zhu, Zhanfan Zhang, and Lin Leng, for help with the fermentation process.
Supported in part by the National Institutes of Health through grants to G.G. (HL 51050) and to C.M.R. (HL 37457) as well as by the American Heart Association and the Hugoton Foundation through grants to G.G.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
NOTE ADDED IN PROOF
As this report was being processed for publication, the x-ray structure of the αEC domain of Fib420 was solved using rαEC crystals, providing confirmation of the predictions of this biochemical study.23
Author notes
Address reprint requests to Gerd Grieninger, PhD, New York Blood Center, 310 E 67th St, New York, NY 10021; e-mail:ggrien@server.nybc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal