Abstract
Acute myeloid leukemia (AML) occurs as the result of malignant transformation in a hematopoietic progenitor cell, which proliferates to form an accumulation of AML blasts. Only a minority of these AML cells are capable of proliferation in vitro, suggesting that AML cells may be organized in a hierarchy, with only the most primitive of these cells capable of maintaining the leukemic clone. To further investigate this hypothesis, we have evaluated a strategy for purifying these primitive cells based on surface antigen expression. As an in vitro endpoint, we have determined the phenotype of AML progenitor cells which are capable of producing AML colony-forming cells (CFU) for up to 8 weeks in suspension culture (SC) and compared the phenotype with that of cells which reproduce AML in nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice. AML cells were fluorescence-activated cell sorted (FACS) for coexpression of CD34 and CD71, CD38, and/or HLA-DR and the subfractions were assayed in vitro and in vivo at various cell doses to estimate purification. While the majority of primary AML CFU lacked expression of CD34, most cells capable of producing CFU after 2 to 8 weeks in SC were CD34+/CD71−. HLA-DR expression was heterogeneous on cells producing CFU after 2 to 4 weeks. However, after 6 to 8 weeks in SC, the majority of CFU were derived from CD34+/HLA-DR− cells. Similarly, the majority of cells capable of long-term CFU production from SC were CD34+/CD38−. Most cells that were capable of engrafting NOD/SCID mice were also CD34+/CD71− and CD34+/HLA-DR−. Engraftment was not achieved with CD34+/CD71+ or HLA-DR+subfractions, however, in two patients, both the CD34+and CD34− subfractions were capable of engrafting the NOD/SCID mice. A three-color sorting strategy combining these antigens allowed approximately a 2-log purification of these NOD/SCID leukemia initiating cells, with engraftment achieved using as few as 400 cells in one experiment. Phenotyping studies suggest even higher purification could be achieved by combining lack of CD38 expression with the CD34+/CD71− or CD34+/HLA DR− phenotype. These results suggest that most AML cells capable of long-term proliferation in vitro and in vivo share the CD34+/CD71−/HLA-DR− phenotype with normal stem cells. Our data suggests that in this group of patients the leukemic transformation has occurred in a primitive progenitor, as defined by phenotype, with some degree of subsequent differentiation as defined by functional assays.
HUMAN ACUTE MYELOID leukemia (AML) is a clonal disorder characterized by the accumulation of aberrant myeloid blasts, defective in their maturation and function, in the blood and bone marrow of affected individuals. The cell of origin of AML has been the subject of much investigation, as AML patients with both multilineage and single lineage involvement within the leukemic clone have been identified.1-4 It has been proposed in the past that the morphologic appearance and phenotype of the bulk of the AML blasts would provide evidence for the cell of origin, with French-American-British (FAB) M1 being more primitive, while FAB M4, which express monocytic surface antigens showing evidence of differentiation.2 Other studies have demonstrated considerable heterogeneity in both the proliferative and self-renewal abilities of the transformed cells within patients,5 6suggesting that AML cell populations within individual patients are organized in a hierarchy. Thus, it is possible that only the most primitive of these AML cells is responsible for maintenance of the disease.
In recent years, it has been accepted that normal primitive long-term culture initiating cells (LTC-IC)7 can be distinguished and separated from more mature cells on the basis of their expression/lack of expression of specific cell surface markers. LTC-IC represent a rare population in the bone marrow, they have low forward light scatter, express CD34 and Thy-1, most lack HLA-DR, CD71, CD38, and CD45RA expression and they are Rhodamine123dull.7-13CD71 antibody is specific for the human transferrin receptor.14 It is thought to be essential for transporting iron into proliferating cells and is expressed on erythroid progenitors. HLA-DR is a human class II major histocompatibility complex (MHC) antigen, which controls responsiveness to soluble antigens.15 It is expressed on B lymphocytes, activated T lymphocytes, and natural killer cells, monocytes and macrophages,16,17 as well as multipotential clonogenic progenitor cells (colony-forming unit-granulocyte, erythroid, monocyte, megakaryocyte [CFU-GEMM]).18 The phenotype of AML cells, which are capable of forming colonies in semisolid medium has been reported.2,3,19 However, these assay systems, at least in normal hematopoiesis, detect progenitors that have limited self-renewal and proliferative capacities. We have previously described a suspension culture assay,20,21 which detects cells with long-term proliferative ability in vitro. AML progenitor cells, which are capable of long-term proliferation in vitro in this system, are less frequent than primary CFU20 and differ from the majority of AML cells in that they are enriched within the CD34+/CD38−/Thy-1−subfraction.20,21 More recently, work by our group and that of others has shown that AML cells, which are capable of engrafting nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice are CD34+/Thy-1−21 and CD34+/CD38−.22
To further phenotypically characterize and purify the cells that have long-term proliferative ability and may be responsible for maintenance of the disease, we have evaluated the coexpression of CD34 and CD71, HLA-DR, or CD38 on cells, which are capable of producing colonies for up to 8 weeks in suspension culture and on cells which are capable of engrafting NOD/SCID mice. We have also evaluated the ability of a multiparameter sorting strategy using CD34, CD71, and HLA-DR to enrich primitive AML cells with NOD/SCID engrafting capacity. Finally, a phenotyping study using a variety of monoclonal antibodies was performed to provide further information on which antibody combinations would be most useful to enrich for primitive AML cells.
MATERIALS AND METHODS
Patient cells.
Peripheral blood cells were obtained from patients at diagnosis of AML after informed consent and with approval of the Clinical Research Ethics Board of the University of British Columbia. Blood cells were Ficoll separated to obtain a mononuclear cell population and then frozen in Dulbecco’s modified Eagle’s medium (DMEM; StemCell Technologies, Inc, Vancouver, BC, Canada) with 30% fetal calf serum (FCS; StemCell Technologies, Inc) and 10% dimethyl sulfoxide (DMSO) and stored at −135°C.
AML cell phenotyping and sorting.
Before sorting, thawed AML cells were suspended in HFN (Hanks’ medium + 2% FCS and 0.1% sodium azide) at 107 cells/mL. Cells were stained for 30 minutes on ice with monoclonal antibodies directly coupled to the fluorochromes fluorescein isothiocyanate (FITC), phycoerythrin (PE), or cyanine5-succinimidylester (Cy5). CD34-Cy5/PE, CD71-PE/FITC were kind gifts from Dr Peter Lansdorp (Terry Fox Laboratory, Vancouver, Canada); CD38-PE, HLA-DR-FITC were obtained from Becton Dickinson (San Jose, CA); and CD33-FITC was obtained from Immunotech (Westbrook, ME). Antibodies were used at 1 μg/mL with the exception of CD34 and CD33, which were used at 4 μg/mL. Cells were then washed twice in HFN at 4°C, propridium iodide (PI) at 2 μg/mL was added to the cells before the second wash, and the cells were maintained on ice before sorting.
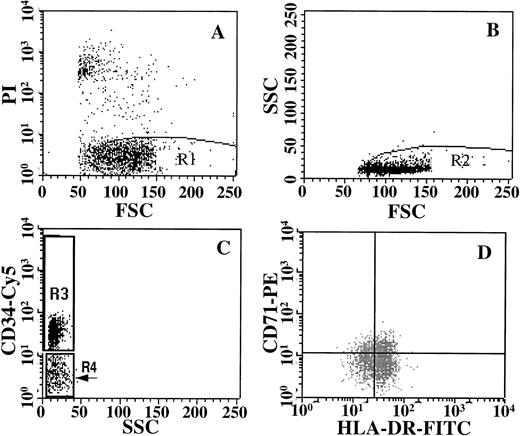
Cells were analyzed and sorted on a dual laser FACStarplus(Becton Dickinson) on the basis of fluorescence intensity after gating out nonviable (PI+) cells and gating on low side scatter as shown in Fig 1. Sort gates were set up to exclude 100% of the cells in an irrelevant isotype control. A 10 channel separation was used to discriminate positive from negative fractions to ensure purity. Fractions were sorted into Iscove’s modified Dulbecco’s medium (StemCell Technologies, Inc) with 50% FCS in microcentrifuge tubes at 4°C. Sort windows used to isolate the specific subfractions are shown in Fig 1. Sorted fractions were washed, resuspended at known cell concentrations, and used to initiate primary CFU and suspension culture assays or used in the NOD/SCID assay.
Purification strategy for isolating cells stained with CD34/CD71 and HLA-DR. Live cells were gated on the basis of PI staining (A), and these cells were then gated on the basis of low side scatter (B). Subsequently, cells were gated for expression of CD34-Cy5, into CD34+ (R3) or CD34− (R4) windows (C). The CD34+ cells in R3 were then sorted on the basis of their expression of CD71-PE and HLA-DR-FITC (D). Sort gates for C and D were set up to exclude 100% of the cells in an isotype control.
Purification strategy for isolating cells stained with CD34/CD71 and HLA-DR. Live cells were gated on the basis of PI staining (A), and these cells were then gated on the basis of low side scatter (B). Subsequently, cells were gated for expression of CD34-Cy5, into CD34+ (R3) or CD34− (R4) windows (C). The CD34+ cells in R3 were then sorted on the basis of their expression of CD71-PE and HLA-DR-FITC (D). Sort gates for C and D were set up to exclude 100% of the cells in an isotype control.
AML colony assays (CFU).
Unsorted AML mononuclear blood cells or FACS-sorted cells immediately following sorting or cells removed during media changes after at least 2 weeks in suspension culture were plated in α-methylcellulose culture medium (Methocult M3230, StemCell Technologies, Inc), containing 3 U/mL recombinant human (rh) erythropoietin (StemCell Technologies, Inc), 20 ng/mL rh interleukin-3 (IL-3) (Sandoz, Basel, Switzerland), 20 ng/mL rhIL-6 (Terry Fox Laboratory), 20 ng/mL rhG-CSF (Amgen Canada Inc, Mississauga, Ontario), 20 ng/mL rh granulocyte-macrophage colony-stimulating factor (GM-CSF) (Sandoz), and 50 ng/mL rhSteel Factor (SF) (Terry Fox Laboratory). After 14 days of incubation at 37°C in a 5% CO2 humidified incubator, AML blast clusters (10 to 20 cells) or colonies (>20 cells) were counted and the numbers were pooled to obtain CFU counts. Most colonies appeared abnormal (blastlike or monocytic). These abnormal appearing colonies comprised 55% of the CFU counts from the primary CFU assays in the 19 patients used for the in vitro studies, while clusters represented 33%, and the remainder were of erythroid origin. Secondary CFU assays using cells derived from SC or from NOD/SCID mice contained similar colony types to those seen in the primary assays from the individual patients.
Suspension culture assays (SC).
Suspension cultures were initiated and maintained as previously described.21 Briefly, unsorted cells or FACS-sorted AML cells were suspended at up to 106 cells/mL in serum-free medium (SFM) consisting of Iscove’s modified Dulbecco’s medium containing 10 μg/mL insulin, 200 μg/mL transferrin, 2% bovine serum albumin, 0.9% NaHCO3 (StemCell Technologies, Inc) and the recombinant growth factors IL-3, IL-6, G-CSF, GM-CSF, and SF in the concentrations described above for the CFU assay. Cultures were maintained at 37°C in a 5% CO2 humidified incubator and were demi-depopulated weekly by removal of half the cells plus media and replacing it with fresh media. Every second week the cells, which were removed, were cultured in α-methylcellulose to determine the CFU content of the suspension culture. Suspension cultures were maintained for 8 weeks and the entire contents of the wells were procured and assessed for CFU content. To allow comparisons between experiments, the proportion of progenitors derived from each sorted fraction at each time point was determined by comparison to a progenitor recovery of 100% from all fractions at that time point.
Transplantation of leukemic cells into NOD/SCID mice.
NOD/SCID mice (Jackson Laboratory, Bar Harbor, ME) were bred and maintained in sterile microisolator cages. Twenty-four hours before transplantation, mice were irradiated with 3.5 Gy γ irradiation from a 137Cs source at a dose rate of 1.25 cGy/minute. Unsorted AML cells and sorted subfractions were suspended in 0.3 mL α minimal essential medium (StemCell Technologies, Inc) with 5% FCS and injected intravenously into the lateral tail vein of 6- to 8-week-old NOD/SCID mice. Whenever possible, various cell concentrations were injected as a semiquantitative assessment of the number of cells required for engraftment. Normal human bone marrow (NBM) cells were given 15 Gy irradiation using 250 kVp x-rays (Philips RT-250, HVL 1.5 mm Cu) at a dose rate of 5.1 Gy/min. In each case, 106 irradiated NBM cells were coinjected with the unsorted cells and sorted AML subfractions to enhance engraftment potential. Animals were given subcutaneous injections of 6 μg human IL-3 and 10 μg human Steel Factor twice per week and were maintained until 8 weeks postinjection. After killing, the gross anatomy of each mouse was inspected, and femoral bone marrow samples were removed for flow cytometry, fluorescent in situ hybridization (FISH), and histologic analysis.
Flow cytometry analysis of murine tissues.
Cell suspensions from femoral bone marrow were lysed in ammonium chloride for 20 minutes and then washed in HFN with 5% human serum. Cells were stained with a human pan leukocyte antibody, 9.4 (anti-CD45)-FITC (Dr P. Lansdorp) for 30 minutes on ice. Separate aliquots were stained with an irrelevant IgG1-FITC antibody (Becton Dickinson) as an isotype control. Aliquots of normal human peripheral blood cells and bone marrow cells from a noninjected NOD/SCID mouse were also stained with CD45-FITC and the isotype control to serve as positive and negative controls for antibody specificity. After staining, the cells were washed in HFN containing PI (2 μg/mL) and resuspended in 300 μL HFN. Samples were analyzed using a FACScan (Becton Dickinson), nonviable cells were gated out based on PI uptake. Isotype controls were run for each tissue sample and used to set the gates. Cells were defined as positive using gate settings, which excluded ≥99.9% of the cells in the matched isotype control, any positive value for the isotype control was then subtracted from the percentage positive in samples stained with CD45-FITC. The expression of CD45-FITC is very specific, in 55 control (noninjected) mice, expression of CD45 was 0.002% ± 0.002% and levels of expression above 0.06% have never been observed in these control animals. We defined human engraftment as expression of ≥ 0.1% CD45+cells in a sample. Whenever possible, the CD45+ cells derived from murine tissue were sorted and set up in suspension culture or plated in α-methylcellulose with recombinant growth factors. The derived colonies were plucked onto slides and cells from the suspension cultures were transferred directly onto slides for subsequent cytogenetic analysis.
Cytogenetic analysis.
Colonies from primary CFU or from CFU, which were derived from SC were evaluated for the leukemia-specific cytogenetic change by either standard cytogenetics or by FISH whenever possible. Likewise, sorted CD45+ cells from bone marrow removed from NOD/SCID mice and CFU derived from these cells were evaluated for the leukemic transformation. For analysis, a random selection of single colonies or clusters from methylcellulose dishes were plucked and plated onto slides after colony synchronization with colcemid.23Colonies derived from these assays had a uniform morphology and appeared abnormal. Analysis was not restricted to colonies with a blastlike appearance or to only colonies and clusters of the myeloid lineage, those of erythroid origin were also evaluated. In one case only (patient 5), pools of 3 to 5 colonies were analyzed, rather than individual colonies. Whole chromosome 9 painting probe directly labeled with spectrum green (BRL Life Technologies, Gaithersburg, MD) was used to detect the t(9;11) in one patient. The inversion (inv) 16 probe directly labeled with digoxigenin (Oncor, Gaithersburg, MD) was used to detect inv16 in two other patients. The centromere-specific chromosome 8 probe directly labeled to digoxigenin (Oncor) was used to highlight at least one additional chromosome 8 in four other patients. The +8 probe and inv16 probes were hybridized in Hybrisol VI (Oncor) with amplification of the signal by rabbit antisheep FITC (Jackson Immuno Research Laboratories, West Grove, PA) and detection by sheep antidigoxigenin-FITC (Boehringer Mannheim, Quebec, Canada). Counterstaining was in PI at 200 ng/mL in Vectashield (Vector Laboratories, Inc, Burlingome, CA). The chromosome 9 probe was hybridized by the manufacturer’s instructions with counter-staining as above. Due to the technical difficulty in obtaining only cells derived from the small colonies, while excluding cells contained in surrounding methylcellulose by manual plucking, colonies were defined as positive whenever ≥60% of cells contained the respective leukemic karyotype of that patient. Colonies containing only 40% to 60% of cells with the leukemic change were classed as undetermined and those with <40% leukemic cells were defined as negative. In the three patients where both metaphase and interphase cells could be scored, due to the presence of an additional chromosome 8, approximately 200 cells (mean ± standard error [SE] 210 ± 3 cells) were counted per colony.
Statistics.
Statistical analysis was performed using analysis of variance (ANOVA) with Tukey’s posthoc analysis of means.
RESULTS
Expression of CD34 and CD71 or HLA-DR or CD38 on AML cells with long term proliferative ability in vitro.
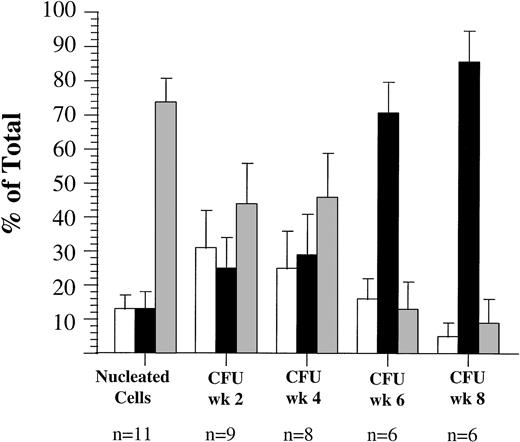
Ficoll-separated blood mononuclear cells from 19 AML patients at diagnosis were used for the present investigation, the clinical characteristics of these patients are shown in Table 1. Because of limitations in available cell numbers, only subsets of this group, as indicated, could be analyzed with each sorting strategy. Peripheral blood cells from 14 of these patients were sorted for coexpression of CD34 and CD71 (Fig 2). Eight of these patient samples had a high number of CD34+ nucleated cells (range, 10% to 87% CD34+), the remaining six samples had only a very small proportion of CD34+ cells (0.11% to 4%). Overall, the CD34+/CD71+ subfraction represented a small fraction of nucleated cells in the blood in these patients (mean ± SEM 12% ± 4%), the CD34+/CD71−subfraction represented 20% ± 7% and the majority of nucleated cells overall were CD34− (68% ± 9%). The majority of primary CFU were also derived from the CD34− fraction (54% ± 13%). However, after 2 weeks in suspension culture, the majority of detectable CFU (56% ± 10%) were derived from the CD34+/CD71−subfraction. Only 22% ± 7% and 22% ± 8% of CFU at week 2 were derived from the CD34+/CD71+ and CD34− subfractions, respectively. After 4 weeks in suspension culture (SC), 62% ± 11% of CFU were derived from the CD34+/CD71−subfraction. The proportion of CFU derived from the CD34+/CD71−subfraction increased significantly with length of time in SC to 66% ± 10% at week 6 and 83% ± 8% at week 8 (F = 5.77**, degrees of freedom [df] = 5, P < .0002). Of the six patients with very low numbers of CD34+ cells in their blast population (mean ± SEM 1.7% ± 0.7%), in four patient samples, all of the detectable CFU after at least 4 weeks in suspension culture were derived from the CD34+/CD71−subfraction. In one of these four samples, cells with this phenotype represented only 0.1% of nucleated cells, but produced 100% of the CFU in SC at week 4. In the two remaining patient samples, the majority of primary CFU (93% ± 6%) and CFU derived from SC at week 2 (99%) were derived from the CD34−subfraction. In one of these two samples, CFU could not be detected from any subfraction after 6 weeks in culture. In the other sample, the majority of CFU detected at week 4 (70%), at week 6 (64%), and week 8 (52%) were derived from the CD34− subfraction.
Clinical Characteristics of AML Patients and Growth of Cells in Control Cultures
| Patient No. . | FAB . | WBC × 109/L . | % Blasts . | Cytogenetic Change . | % CD34 . | CFU*/105 . | CFU in SC wk 4†/105 . | CFU in SC wk 8/105 . |
|---|---|---|---|---|---|---|---|---|
| 1 | M1 | 100 | 90 | Normal | 0.1 | 12 | 15 | 57 |
| 2 | M5 | 155 | 39 | t(9;11) | 1.1 | 88 | 80 | 806 |
| 3 | M5 | 47 | 23 | del 11q23 | 48 | 870 | 6 | 13 |
| 4 | M4 | 370 | 46 | +13 | 28 | 300 | 17,728 | 51 |
| 5 | M4 | 71 | 76 | +8 | 76 | 910 | 10 | 57 |
| 6 | M5 | 430 | 98 | ND | 2 | 5 | 5 | 0 |
| 7 | M4 | 103 | 42 | del 16q | 3 | 13 | 2 | 0 |
| 8 | M5 | 50 | 92 | +8,+8, t(9;11) | 0.4 | 1,060 | 3,413 | 0 |
| 9 | M4e | 130 | 70 | inv 16 | 10 | 3 | 178 | 0 |
| 10 | M4e | 65 | 80 | inv 16 | 87 | 6 | 133 | 7 |
| 11 | M4e | 19 | 34 | inv 16 | 48 | 10 | 3 | 6 |
| 12 | M4e | 18 | 52 | inv 16 | 78 | 18 | 11 | 96 |
| 13 | M2 | 57 | 51 | +8, t(12;22), del 20q | 11 | 182 | 928 | 256 |
| 14 | M2 | 29 | 38 | t(6;9) | 1.6 | 800 | 995 | 2,161 |
| 15 | M5 | 94 | 73 | Normal | 0.1 | 148 | 204 | 882 |
| 16 | M4 | 87 | 73 | del 16q22 | 4 | 116 | 4 | 14 |
| 17 | M1 | 27 | 80 | t(8;20) | 7 | ND | ND | ND |
| 18 | M5 | 94 | 73 | +8 | 0.5 | 2 | 2 | 14 |
| 19 | M5b | 12 | 23 | Normal | 3 | 9 | 2 | 21 |
| Patient No. . | FAB . | WBC × 109/L . | % Blasts . | Cytogenetic Change . | % CD34 . | CFU*/105 . | CFU in SC wk 4†/105 . | CFU in SC wk 8/105 . |
|---|---|---|---|---|---|---|---|---|
| 1 | M1 | 100 | 90 | Normal | 0.1 | 12 | 15 | 57 |
| 2 | M5 | 155 | 39 | t(9;11) | 1.1 | 88 | 80 | 806 |
| 3 | M5 | 47 | 23 | del 11q23 | 48 | 870 | 6 | 13 |
| 4 | M4 | 370 | 46 | +13 | 28 | 300 | 17,728 | 51 |
| 5 | M4 | 71 | 76 | +8 | 76 | 910 | 10 | 57 |
| 6 | M5 | 430 | 98 | ND | 2 | 5 | 5 | 0 |
| 7 | M4 | 103 | 42 | del 16q | 3 | 13 | 2 | 0 |
| 8 | M5 | 50 | 92 | +8,+8, t(9;11) | 0.4 | 1,060 | 3,413 | 0 |
| 9 | M4e | 130 | 70 | inv 16 | 10 | 3 | 178 | 0 |
| 10 | M4e | 65 | 80 | inv 16 | 87 | 6 | 133 | 7 |
| 11 | M4e | 19 | 34 | inv 16 | 48 | 10 | 3 | 6 |
| 12 | M4e | 18 | 52 | inv 16 | 78 | 18 | 11 | 96 |
| 13 | M2 | 57 | 51 | +8, t(12;22), del 20q | 11 | 182 | 928 | 256 |
| 14 | M2 | 29 | 38 | t(6;9) | 1.6 | 800 | 995 | 2,161 |
| 15 | M5 | 94 | 73 | Normal | 0.1 | 148 | 204 | 882 |
| 16 | M4 | 87 | 73 | del 16q22 | 4 | 116 | 4 | 14 |
| 17 | M1 | 27 | 80 | t(8;20) | 7 | ND | ND | ND |
| 18 | M5 | 94 | 73 | +8 | 0.5 | 2 | 2 | 14 |
| 19 | M5b | 12 | 23 | Normal | 3 | 9 | 2 | 21 |
Abbreviations: ND, not determined; wbc, white blood cells.
CFU, leukemic clusters (10 to 20 cells) plus colonies (>20 cells) scored after 14 days in methylcellulose culture per 105cells plated.
CFU in SC wk4, CFU in 4-week-old suspension cultures initiated with 105 cells. CFU numbers were corrected for weekly depopulation.
Proportion of AML peripheral blood cells and progenitors expressing CD34 and CD71 antigens in 14 patients (patients 2 to 5, 8 to 16, and 18). (□), CD34+/CD71+; (▪), CD34+/CD71−; (▩), CD34−.
Proportion of AML peripheral blood cells and progenitors expressing CD34 and CD71 antigens in 14 patients (patients 2 to 5, 8 to 16, and 18). (□), CD34+/CD71+; (▪), CD34+/CD71−; (▩), CD34−.
Peripheral blood cells from 11 patients were sorted for coexpression of CD34 and HLA-DR (Fig 3). The majority of nucleated cells at diagnosis were CD34− (74% ± 7%), while 13% ± 4% and 13% ± 5% of cells were CD34+/HLA-DR+ and CD34+/HLA-DR−, respectively. Cells, which were capable of producing CFU after 2 weeks and 4 weeks in SC, were heterogeneous in their HLA-DR expression and some were in the CD34− subfraction. However, after 6 weeks and 8 weeks in SC, significantly higher numbers of CFU (71% ± 9% and 86% ± 9%, respectively) were derived from the CD34+/HLA-DR− subfraction, compared with earlier time points (F = 10.82**, df = 4, P < .00003).
Proportion of AML peripheral blood cells and progenitors expressing CD34 and HLA-DR antigens in 11 patients (patients 1, 2, 4, 5, 7, 8, 10, 11, 13, 15, and 19). □), CD34+/HLA-DR+; (▪), CD34+/HLA-DR−; (▩), CD34−.
Proportion of AML peripheral blood cells and progenitors expressing CD34 and HLA-DR antigens in 11 patients (patients 1, 2, 4, 5, 7, 8, 10, 11, 13, 15, and 19). □), CD34+/HLA-DR+; (▪), CD34+/HLA-DR−; (▩), CD34−.
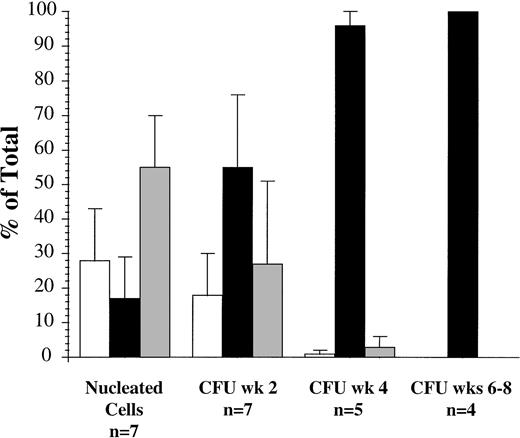
Cells from seven patients were sorted for coexpression of CD34 and CD38 and evaluated in the suspension culture assay (Fig 4). The CD34+/CD38+ subfraction represented 28% ± 15% of nucleated cells at diagnosis, 17% ± 12% (range, 0.03 to 45%) were CD34+/CD38− and the majority were CD34− (55% ± 15%). However, the majority of CFU detected after 2 weeks (55% ± 12%) and 4 weeks (96% ± 4%) in suspension culture were derived from the CD34+/CD38− subfraction. Subsequently, at weeks 6 to 8, in the four patient samples where CFU were still detectable, all were derived from the CD34+/CD38− subfraction.
Proportion of AML peripheral blood cells and progenitors expressing CD34 and CD38 antigens in seven patients (patients 2, 3, 5, 6, 8, 10, and 13). (□), CD34+/CD38+; (▪), CD34+/CD38−; (▩), CD34−.
Proportion of AML peripheral blood cells and progenitors expressing CD34 and CD38 antigens in seven patients (patients 2, 3, 5, 6, 8, 10, and 13). (□), CD34+/CD38+; (▪), CD34+/CD38−; (▩), CD34−.
Cytogenetic analysis of in vitro studies.
Standard cytogenetic or FISH analysis was performed on primary CFU derived from unsorted and sorted subfractions and on CFU derived from suspension cultures initiated with these subfractions (Table 2). In experiments with seven patient samples, all primary CFU colonies analyzed had the respective leukemic change. The majority (40 of 41) of CFU derived from SC of unsorted cells at weeks 2 to 8 were found to be derived from the leukemic clone, with only one undetermined colony. All primary CFU derived from sorted subfractions were found to have the expected chromosomal abnormality. CFU derived from SC of CD34+/CD71−, CD34+/HLA-DR+, and CD34+/HLA-DR− subfractions were analyzed in 18 cases (eight patients). The majority of colonies analyzed were found to be positive for the leukemic karyotype (77 of 84), while five colonies were undetermined and two were negative.
No. of Colonies Detected With the Leukemic Change by FISH or Cytogenetics After In Vitro Assay
| Patient No. . | Positive Colonies From Primary CFU . | Colonies from CFU in SC wk 2-8 . | ||
|---|---|---|---|---|
| Positive . | Undetermined . | Negative . | ||
| Unsorted | ||||
| 2 | 5/5 | 4/5 | 1/5 | |
| 3 | ND | 7/7 | ||
| 5 | 5/5 | 12/12 | ||
| 8 | 5/5 | 5/5 | ||
| 10 | ND | 2/2 | ||
| 11 | 2/2 | 2/2 | ||
| 13 | 5/5 | 8/8 | ||
| CD34+/CD71+ | ||||
| 5 | ND | 4/4 | ||
| 10 | 2/2 | ND | ND | ND |
| 13 | ND | 8/8 | ||
| CD34+/CD71− | ||||
| 2 | ND | 7/7 | ||
| 3 | ND | 3/3 | ||
| 5 | ND | 6/6 | ||
| 8 | ND | 10/10 | ||
| 10 | ND | 2/2 | ||
| 11 | 2/2 | 3/3 | ||
| 13 | 10/10 | 9/9 | ||
| CD34+/HLA-DR+ | ||||
| 2 | ND | 3/3 | ||
| 4 | ND | 1/2 | 1/2 | |
| 5 | ND | 2/3 | 1/3 | |
| 13 | 3/3 | 2/2 | ||
| CD34+/HLA-DR− | ||||
| 2 | ND | 2/2 | ||
| 4 | 3/3 | 4/5 | 1/5 | |
| 5 | ND | 3/3 | ||
| 10 | ND | 4/4 | ||
| 13 | 5/5 | 8/8 | ||
| CD34− | ||||
| 2 | 5/5 | 3/5 | 1/5 | 1/5 |
| 5 | ND | 2/5 | 3/5 | |
| 8 | ND | 10/10 | ||
| Patient No. . | Positive Colonies From Primary CFU . | Colonies from CFU in SC wk 2-8 . | ||
|---|---|---|---|---|
| Positive . | Undetermined . | Negative . | ||
| Unsorted | ||||
| 2 | 5/5 | 4/5 | 1/5 | |
| 3 | ND | 7/7 | ||
| 5 | 5/5 | 12/12 | ||
| 8 | 5/5 | 5/5 | ||
| 10 | ND | 2/2 | ||
| 11 | 2/2 | 2/2 | ||
| 13 | 5/5 | 8/8 | ||
| CD34+/CD71+ | ||||
| 5 | ND | 4/4 | ||
| 10 | 2/2 | ND | ND | ND |
| 13 | ND | 8/8 | ||
| CD34+/CD71− | ||||
| 2 | ND | 7/7 | ||
| 3 | ND | 3/3 | ||
| 5 | ND | 6/6 | ||
| 8 | ND | 10/10 | ||
| 10 | ND | 2/2 | ||
| 11 | 2/2 | 3/3 | ||
| 13 | 10/10 | 9/9 | ||
| CD34+/HLA-DR+ | ||||
| 2 | ND | 3/3 | ||
| 4 | ND | 1/2 | 1/2 | |
| 5 | ND | 2/3 | 1/3 | |
| 13 | 3/3 | 2/2 | ||
| CD34+/HLA-DR− | ||||
| 2 | ND | 2/2 | ||
| 4 | 3/3 | 4/5 | 1/5 | |
| 5 | ND | 3/3 | ||
| 10 | ND | 4/4 | ||
| 13 | 5/5 | 8/8 | ||
| CD34− | ||||
| 2 | 5/5 | 3/5 | 1/5 | 1/5 |
| 5 | ND | 2/5 | 3/5 | |
| 8 | ND | 10/10 | ||
Colonies were defined according to the number of cells with leukemic karyotype, positive (≥60%), undetermined (40% to 59%), negative (<40%).
Abbreviation: SC, suspension culture.
In vivo NOD/SCID assay.
Unsorted and sorted AML cells from 14 patients were evaluated for their ability to engraft NOD/SCID mice after sorting for coexpression of CD34 and CD71 and/or HLA-DR. However, engraftment with any sorted or unsorted fraction was achieved using cells from only 12 of these patients, with injections of up to 5 × 106 unsorted cells per mouse. Unsorted cells from 11 of the 12 patients engrafted in the NOD/SCIDs (Table 3). Engraftment was achieved using 106 cells from nine of 11 patients evaluated (mean ± SEM, 27% ± 11% CD45+) using 5 × 105 unsorted cells from six of nine patients evaluated (26% ± 15% CD45+) and using 105 unsorted cells from four of eight patients evaluated (20% ± 15% CD45+). One patient, of eight tested, engrafted with 5 × 104 cells (26% CD45+) and no engraftment was achieved using ≤104 unsorted cells.
Percentage of CD45+ Cells Detected in the BM of NOD/SCID Mice Injected With Unsorted AML Peripheral Blood Cells
| Patient No. . | 5 × 106 . | No. of Cells Injected3-150 . | No. of Positive Colonies by FISH . | |||
|---|---|---|---|---|---|---|
| 106 . | 5 × 105 . | 105 . | ≤5 × 104 . | |||
| 2 | 92 | 57 ± 3 | 2 | 0 | 0 | 7/8 |
| 3 | 21 | 15 | ND | ND | ND | 6/6 |
| 4 | 30 ± 10 | 42 | 5 | 0 | 4/123-151 | |
| 5 | 3 | 0 | ND | ND | ND | ND |
| 8 | 95 | 97 ± 0.6 | 92 | 66 | 26 | 26/26 |
| 9 | 2 | 0.2 | ND | ND | ND | 10/10 |
| 10 | 0.1 | 0 | 0 | 0 | ND | |
| 12 | ND | 0.2 | 0 | 0 | ND | |
| 13 | 0.14 | 0 | 0 | 0 | ND | |
| 15 | 30 ± 6 | 9 | 1 | 0 | ND | |
| 17 | 13 | 11 | 8 | 0 | ND | |
| 18 | 0 | 0 | ND | ND | ND | |
| Patient No. . | 5 × 106 . | No. of Cells Injected3-150 . | No. of Positive Colonies by FISH . | |||
|---|---|---|---|---|---|---|
| 106 . | 5 × 105 . | 105 . | ≤5 × 104 . | |||
| 2 | 92 | 57 ± 3 | 2 | 0 | 0 | 7/8 |
| 3 | 21 | 15 | ND | ND | ND | 6/6 |
| 4 | 30 ± 10 | 42 | 5 | 0 | 4/123-151 | |
| 5 | 3 | 0 | ND | ND | ND | ND |
| 8 | 95 | 97 ± 0.6 | 92 | 66 | 26 | 26/26 |
| 9 | 2 | 0.2 | ND | ND | ND | 10/10 |
| 10 | 0.1 | 0 | 0 | 0 | ND | |
| 12 | ND | 0.2 | 0 | 0 | ND | |
| 13 | 0.14 | 0 | 0 | 0 | ND | |
| 15 | 30 ± 6 | 9 | 1 | 0 | ND | |
| 17 | 13 | 11 | 8 | 0 | ND | |
| 18 | 0 | 0 | ND | ND | ND | |
Abbreviation: ND, not determined.
In four of the cases shown, the values represent the mean ± SEM % of CD45+ cells in femoral marrow of at least 3 NOD/SCID mice. All other values represent the result from a single mouse injected with the number of cells indicated.
Of the remaining 8 colonies, 5 were undetermined and 3 were negative.
Cells from nine AML patients were sorted for CD34 and CD71 expression and the subfractions were evaluated in NOD/SCID mice. In five patients who had high numbers of CD34+ nucleated cells (28% to 75%) (Table 4), engraftment was achieved using the CD34+/CD71− sorted subfraction from all patients, although there was considerable heterogeneity in the degree of engraftment achieved (range, 0.16% to 74% CD45+ using 104 to 106 cells). Engraftment was not obtained when CD34+/CD71+or CD34− subfractions from those patients were injected. Four patients who had very low numbers (0.11% to 1.3%) of CD34+ cells (Table5) were also evaluated using this strategy. Engraftment was achieved using small numbers of CD34+/CD71− cells from all of these patients (range, 0.13% to 27% CD45+ with 103to 104 cells). Interestingly, engraftment was achieved using the CD34− fraction from patient 2 (37% CD45+ with 4 ×105 cells) and patient 8 (11% CD45+) with 105 cells, (54%) with 5 × 105 and (31%) with 106 cells. The majority of CFU in suspension culture at 2 to 8 weeks was derived from the CD34− subfraction in these two patients. However, considerably higher numbers of CD34− cells (≥105) had to be injected to achieve engraftment equivalent to that achieved with ≤4 × 103CD34+/CD71− cells. No engraftment was achieved with ≤106 CD34− cells from the other two patients. Engraftment was not achieved with CD34+/CD71+ cells from any patient.
Expression of Human CD45 Antigen in NOD/SCID Recipients of AML Cells Sorted for Coexpression of CD34 and CD71 (CD34+ patients)
| Patient No. . | Phenotype . | % Nucleated Cells . | No. Cells Injected . | % CD45+ BM . | FISH Analysis on Colonies . | ||
|---|---|---|---|---|---|---|---|
| Positive . | Undetermined . | Negative . | |||||
| 3 | Unsorted | 106 | 15 | 3/4 | 1/4 | ||
| CD34+/CD71+ | 1.8 | 1.5 × 105 | 0 | ||||
| CD34+/CD71− | 45.8 | 106 | 74 | 2/4 | 1/4 | 1/4 | |
| CD34− | 52.4 | 106 | 0 | ||||
| 4 | Unsorted | 106 | 24 | ||||
| Unsorted | 5 × 105 | 43 | 5/6 | 1/6 | |||
| Unsorted | 105 | 5 | |||||
| Unsorted | ≤5 × 104 | 0 | |||||
| CD34+/CD71+ | 24.5 | ≤5 × 105 | 0 | ||||
| CD34+/CD71− | 3.9 | 2.5 × 105 | 5 | 1/5 | 2/5 | 2/5 | |
| CD34+/CD71− | ≤5 × 104 | 0 | |||||
| CD34− | 71.6 | ≤5 × 105 | 0 | ||||
| 5 | Unsorted | 106 | 0 | ||||
| CD34+/CD71+ | 62.6 | ≤106 | 0 | ||||
| CD34+/CD71− | 13.2 | 105 | 0.2 | ||||
| CD34+/CD71− | 5 × 104 | 0.2 | |||||
| CD34+/CD71− | 104 | 0.3 | |||||
| CD34− | 24.2 | 1.4 × 106 | 0 | ||||
| 11 | Unsorted | 106 | 0.1 | ||||
| CD34+/CD71+ | 8.8 | 105 | 0 | ||||
| CD34+/CD71+ | 5.4 × 104 | 0 | |||||
| CD34+/CD71− | 39.1 | 2 × 105 | 0.2 | ||||
| CD34+/CD71− | 6 × 104 | 0.2 | |||||
| CD34+/CD71− | 5 × 104 | 0 | |||||
| CD34− | 52.1 | ≤3 × 105 | 0 | ||||
| 12 | Unsorted | 5 × 105 | 0.2 | ||||
| Unsorted | ≤105 | 0 | |||||
| CD34+/CD71+ | 5.1 | 106 | 0 | ||||
| CD34+/CD71− | 67.2 | 105 | 3 | ||||
| CD34+/CD71− | ≤5 × 104 | 0 | |||||
| CD34− | 27.7 | ≤5 × 105 | 0 | ||||
| Patient No. . | Phenotype . | % Nucleated Cells . | No. Cells Injected . | % CD45+ BM . | FISH Analysis on Colonies . | ||
|---|---|---|---|---|---|---|---|
| Positive . | Undetermined . | Negative . | |||||
| 3 | Unsorted | 106 | 15 | 3/4 | 1/4 | ||
| CD34+/CD71+ | 1.8 | 1.5 × 105 | 0 | ||||
| CD34+/CD71− | 45.8 | 106 | 74 | 2/4 | 1/4 | 1/4 | |
| CD34− | 52.4 | 106 | 0 | ||||
| 4 | Unsorted | 106 | 24 | ||||
| Unsorted | 5 × 105 | 43 | 5/6 | 1/6 | |||
| Unsorted | 105 | 5 | |||||
| Unsorted | ≤5 × 104 | 0 | |||||
| CD34+/CD71+ | 24.5 | ≤5 × 105 | 0 | ||||
| CD34+/CD71− | 3.9 | 2.5 × 105 | 5 | 1/5 | 2/5 | 2/5 | |
| CD34+/CD71− | ≤5 × 104 | 0 | |||||
| CD34− | 71.6 | ≤5 × 105 | 0 | ||||
| 5 | Unsorted | 106 | 0 | ||||
| CD34+/CD71+ | 62.6 | ≤106 | 0 | ||||
| CD34+/CD71− | 13.2 | 105 | 0.2 | ||||
| CD34+/CD71− | 5 × 104 | 0.2 | |||||
| CD34+/CD71− | 104 | 0.3 | |||||
| CD34− | 24.2 | 1.4 × 106 | 0 | ||||
| 11 | Unsorted | 106 | 0.1 | ||||
| CD34+/CD71+ | 8.8 | 105 | 0 | ||||
| CD34+/CD71+ | 5.4 × 104 | 0 | |||||
| CD34+/CD71− | 39.1 | 2 × 105 | 0.2 | ||||
| CD34+/CD71− | 6 × 104 | 0.2 | |||||
| CD34+/CD71− | 5 × 104 | 0 | |||||
| CD34− | 52.1 | ≤3 × 105 | 0 | ||||
| 12 | Unsorted | 5 × 105 | 0.2 | ||||
| Unsorted | ≤105 | 0 | |||||
| CD34+/CD71+ | 5.1 | 106 | 0 | ||||
| CD34+/CD71− | 67.2 | 105 | 3 | ||||
| CD34+/CD71− | ≤5 × 104 | 0 | |||||
| CD34− | 27.7 | ≤5 × 105 | 0 | ||||
The remainder of cells to 100% were CD45−.
Expression of Human CD45 Antigen in NOD/SCID Recipients of AML Cells Sorted for Coexpression of CD34 and CD71 (CD34− patients)
| Patient No. . | Phenotype . | % Nucleated Cells . | No. Cells Injected . | % CD45+ BM . | FISH Analysis on Colonies . | ||
|---|---|---|---|---|---|---|---|
| Positive . | Undetermined . | Negative . | |||||
| 2 | Unsorted | 106 | 55 | 3/4 | 1/4 | ||
| Unsorted | 5 × 105 | 2 | |||||
| Unsorted | ≤105 | 0 | |||||
| CD34+/CD71+ | 0.02 | 7 × 103 | 0 | ||||
| CD34+/CD71− | 1.1 | 2 × 104 | 0 | ||||
| CD34+/CD71− | 103 | 1 | |||||
| CD34− | 98.88 | 4 × 105 | 37 | ||||
| CD34− | ≤104 | 0 | |||||
| 8 | Unsorted | 106 | 96 | 4/4 | |||
| Unsorted | 5 × 105 | 92 | 3/3 | ||||
| Unsorted | 105 | 66 | 4/4 | ||||
| Unsorted | 5 × 104 | 26 | |||||
| ≤104 | 0 | ||||||
| CD34+/CD71+ | 0.4 | 7 × 103 | 0 | ||||
| CD34+/CD71− | 0.01 | 4 × 103 | 11 | 4/4 | |||
| CD34− | 99.59 | 106 | 31 | 3/3 | |||
| CD34− | 5 × 105 | 54 | 5/5 | ||||
| CD34− | 105 | 11 | 3/3 | ||||
| CD34− | ≤5 × 104 | 0 | |||||
| 15 | Unsorted | 106 | 24 | ||||
| Unsorted | 5 × 105 | 9 | |||||
| Unsorted | 105 | 1 | |||||
| Unsorted | ≤104 | 0 | |||||
| CD34+/CD71+ | 0.01 | 103 | 0 | ||||
| CD34+/CD71− | 0.1 | 5 × 103 | 27 | ||||
| CD34− | 99.89 | ≤106 | 0 | ||||
| 18 | Unsorted | ≤106 | 0 | ||||
| CD34+/CD71+ | 0.3 | ≤5 × 104 | 0 | ||||
| CD34+/CD71− | 0.2 | 5 × 104 | 0.1 | ||||
| CD34+/CD71− | 104 | 0.2 | |||||
| CD34+/CD71− | 104 | 0.1 | |||||
| CD34− | 99.5 | ≤106 | 0 | ||||
| Patient No. . | Phenotype . | % Nucleated Cells . | No. Cells Injected . | % CD45+ BM . | FISH Analysis on Colonies . | ||
|---|---|---|---|---|---|---|---|
| Positive . | Undetermined . | Negative . | |||||
| 2 | Unsorted | 106 | 55 | 3/4 | 1/4 | ||
| Unsorted | 5 × 105 | 2 | |||||
| Unsorted | ≤105 | 0 | |||||
| CD34+/CD71+ | 0.02 | 7 × 103 | 0 | ||||
| CD34+/CD71− | 1.1 | 2 × 104 | 0 | ||||
| CD34+/CD71− | 103 | 1 | |||||
| CD34− | 98.88 | 4 × 105 | 37 | ||||
| CD34− | ≤104 | 0 | |||||
| 8 | Unsorted | 106 | 96 | 4/4 | |||
| Unsorted | 5 × 105 | 92 | 3/3 | ||||
| Unsorted | 105 | 66 | 4/4 | ||||
| Unsorted | 5 × 104 | 26 | |||||
| ≤104 | 0 | ||||||
| CD34+/CD71+ | 0.4 | 7 × 103 | 0 | ||||
| CD34+/CD71− | 0.01 | 4 × 103 | 11 | 4/4 | |||
| CD34− | 99.59 | 106 | 31 | 3/3 | |||
| CD34− | 5 × 105 | 54 | 5/5 | ||||
| CD34− | 105 | 11 | 3/3 | ||||
| CD34− | ≤5 × 104 | 0 | |||||
| 15 | Unsorted | 106 | 24 | ||||
| Unsorted | 5 × 105 | 9 | |||||
| Unsorted | 105 | 1 | |||||
| Unsorted | ≤104 | 0 | |||||
| CD34+/CD71+ | 0.01 | 103 | 0 | ||||
| CD34+/CD71− | 0.1 | 5 × 103 | 27 | ||||
| CD34− | 99.89 | ≤106 | 0 | ||||
| 18 | Unsorted | ≤106 | 0 | ||||
| CD34+/CD71+ | 0.3 | ≤5 × 104 | 0 | ||||
| CD34+/CD71− | 0.2 | 5 × 104 | 0.1 | ||||
| CD34+/CD71− | 104 | 0.2 | |||||
| CD34+/CD71− | 104 | 0.1 | |||||
| CD34− | 99.5 | ≤106 | 0 | ||||
The remainder of cells to 100% were CD45−.
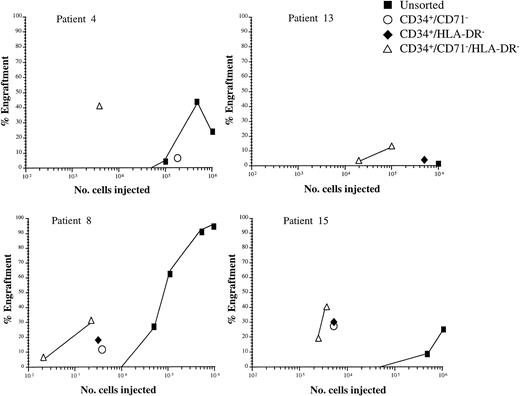
To determine the HLA-DR phenotype of NOD/SCID engrafting cells and to evaluate the possible enrichment of these cells with three-color FACS sorting, cells from six patients, which included three evaluated in Table 4 or 5, were sorted for CD34, CD71, and HLA-DR combinations (Table 6). The number of CD34+ nucleated cells was low (0.11% to 28%) in these patients. In five patients, both CD34+/HLA-DR+and CD34+/HLA-DR− fractions were evaluated and engraftment was achieved with CD34+/HLA-DR− cells from four of these patients (4% to 38% CD45+). However, engraftment was not achieved with CD34+/HLA-DR+ subfraction, where approximately equivalent numbers of cells were transplanted, or with the CD34− subfraction, with the exception of patient 8 (98% CD45+ with 106 cells). Cells from four of these five patients, plus one other, were sorted for expression of CD34, lack of CD71, and expression/lack of HLA-DR to further define HLA-DR phenotype and determine enrichment. In all cases, engraftment was achieved using very low numbers of CD34+/CD71−/HLA-DR−cells (1% to 40% CD45+ using 4 × 102 to 3.7 × 104 cells), but was not seen with equivalent or greater numbers of CD34+/CD71−/HLA-DR+ cells. This triple sorting strategy achieved ≈2 log enrichment of NOD/SCID engrafting cells in the four patients where this could be evaluated and improved on the enrichment observed with CD34+/CD71− or CD34+/HLA-DR− sorting alone (Fig 5).
Expression of Human CD45 Antigen in NOD/SCID Recipients of AML Cells Sorted for Coexpression of CD34, HLA-DR, and CD71
| Patient No. . | Phenotype . | % Nucleated Cells . | No. Cells Injected . | % CD45+ BM . | FISH Analysis on Colonies . | ||
|---|---|---|---|---|---|---|---|
| Positive . | Undetermined . | Negative . | |||||
| 4 | Unsorted | 106 | 16 | ||||
| Unsorted | 5 × 105 | 41 | 3/5 | 2/5 | |||
| CD34+/CD71+/DR+ | 21.95 | NA | |||||
| CD34+/CD71+/DR− | 4.36 | 104 | 0 | ||||
| CD34+/CD71−/DR+ | 1.14 | 4 × 104 | 0 | ||||
| 9 × 103 | 0 | ||||||
| CD34+/CD71−/DR− | 0.56 | 5 × 103 | 39 | 1/4 | 3/4 | ||
| 500 | 0 | ||||||
| CD34− | 71.99 | ≤106 | 0 | ||||
| 8 | Unsorted | 106 | 97 | 5/5 | |||
| CD34+/DR+ | 0.1 | 8 × 103 | 0 | ||||
| 5 × 103 | 0 | ||||||
| CD34+/DR− | 0.01 | 3 × 103 | 18 | 5/5 | |||
| CD34+/CD71+/DR+ | 0.06 | NA | |||||
| CD34+/CD71+/DR− | 0.008 | NA | |||||
| CD34+/CD71−/DR+ | 0.04 | 5 × 103 | 0 | ||||
| CD34+/CD71−/DR− | 0.002 | 2272 | 32 | 5/5 | |||
| 400 | 4 | 3/3 | |||||
| CD34− | 99.89 | 106 | 98 | 4/4 | |||
| 105 | 29 | 4/4 | |||||
| 9 | Unsorted | 106 | 0.2 | ||||
| CD34+/DR+ | 4.6 | 7 × 104 | 0 | ||||
| CD34+/DR− | 4.5 | 105 | 0 | ||||
| 8 × 104 | 0 | ||||||
| 7 × 104 | 0 | ||||||
| CD34+/CD71+/DR+ | 3.7 | NA | |||||
| CD34+/CD71+/DR− | 2.9 | NA | |||||
| CD34+/CD71−/DR+ | 0.9 | 8 × 104 | 0 | ||||
| 4 × 104 | 0 | ||||||
| CD34+/CD71−/DR− | 1.6 | 3.7 × 104 | 1 | ||||
| CD34− | 90.9 | ≤106 | 0 | ||||
| 13 | Unsorted | 106 | 0.1 | ||||
| CD34+/DR+ | 10.3 | 5 × 105 | 0 | ||||
| 3.5 × 105 | 0 | ||||||
| CD34+/DR− | 0.07 | 5 × 105 | 4 | 3/3 | |||
| 105 | 0 | ||||||
| CD34+/CD71+/DR+ | 9.6 | NA | |||||
| CD34+/CD71+/DR− | 0.05 | NA | |||||
| CD34+/CD71−/DR+ | 0.7 | 2.5 × 104 | 0 | ||||
| CD34+/CD71−/DR− | 0.02 | 105 | 13 | 3/3 | |||
| 2 × 104 | 3 | 3/3 | |||||
| CD34− | 89.63 | 5 × 106 | 0 | ||||
| 15 | Unsorted | 106 | 36 | ||||
| CD34+/DR+ | 0.11 | 1.7 × 104 | 0 | ||||
| CD34+/DR− | 0.09 | 5.6 × 103 | 29 | ||||
| CD34+/CD71+/DR+ | 0.02 | NA | |||||
| CD34+/CD71+/DR− | 0.01 | NA | |||||
| CD34+/CD71−/DR+ | 0.09 | 5 × 103 | 0 | ||||
| CD34+/CD71−/DR− | 0.08 | 3.7 × 103 | 40 | ||||
| 2.5 × 103 | 19 | ||||||
| CD34− | 99.8 | 106 | 0 | ||||
| 17 | Unsorted | 106 | 13 | ||||
| 5 × 105 | 11 | ||||||
| 105 | 8 | ||||||
| CD34+/DR+ | 1.3 | 2 × 105 | 0 | ||||
| CD34+/DR− | 5.3 | 3 × 105 | 38 | ||||
| CD34− | 93.4 | 106 | 0 | ||||
| 6 × 105 | 0 | ||||||
| Patient No. . | Phenotype . | % Nucleated Cells . | No. Cells Injected . | % CD45+ BM . | FISH Analysis on Colonies . | ||
|---|---|---|---|---|---|---|---|
| Positive . | Undetermined . | Negative . | |||||
| 4 | Unsorted | 106 | 16 | ||||
| Unsorted | 5 × 105 | 41 | 3/5 | 2/5 | |||
| CD34+/CD71+/DR+ | 21.95 | NA | |||||
| CD34+/CD71+/DR− | 4.36 | 104 | 0 | ||||
| CD34+/CD71−/DR+ | 1.14 | 4 × 104 | 0 | ||||
| 9 × 103 | 0 | ||||||
| CD34+/CD71−/DR− | 0.56 | 5 × 103 | 39 | 1/4 | 3/4 | ||
| 500 | 0 | ||||||
| CD34− | 71.99 | ≤106 | 0 | ||||
| 8 | Unsorted | 106 | 97 | 5/5 | |||
| CD34+/DR+ | 0.1 | 8 × 103 | 0 | ||||
| 5 × 103 | 0 | ||||||
| CD34+/DR− | 0.01 | 3 × 103 | 18 | 5/5 | |||
| CD34+/CD71+/DR+ | 0.06 | NA | |||||
| CD34+/CD71+/DR− | 0.008 | NA | |||||
| CD34+/CD71−/DR+ | 0.04 | 5 × 103 | 0 | ||||
| CD34+/CD71−/DR− | 0.002 | 2272 | 32 | 5/5 | |||
| 400 | 4 | 3/3 | |||||
| CD34− | 99.89 | 106 | 98 | 4/4 | |||
| 105 | 29 | 4/4 | |||||
| 9 | Unsorted | 106 | 0.2 | ||||
| CD34+/DR+ | 4.6 | 7 × 104 | 0 | ||||
| CD34+/DR− | 4.5 | 105 | 0 | ||||
| 8 × 104 | 0 | ||||||
| 7 × 104 | 0 | ||||||
| CD34+/CD71+/DR+ | 3.7 | NA | |||||
| CD34+/CD71+/DR− | 2.9 | NA | |||||
| CD34+/CD71−/DR+ | 0.9 | 8 × 104 | 0 | ||||
| 4 × 104 | 0 | ||||||
| CD34+/CD71−/DR− | 1.6 | 3.7 × 104 | 1 | ||||
| CD34− | 90.9 | ≤106 | 0 | ||||
| 13 | Unsorted | 106 | 0.1 | ||||
| CD34+/DR+ | 10.3 | 5 × 105 | 0 | ||||
| 3.5 × 105 | 0 | ||||||
| CD34+/DR− | 0.07 | 5 × 105 | 4 | 3/3 | |||
| 105 | 0 | ||||||
| CD34+/CD71+/DR+ | 9.6 | NA | |||||
| CD34+/CD71+/DR− | 0.05 | NA | |||||
| CD34+/CD71−/DR+ | 0.7 | 2.5 × 104 | 0 | ||||
| CD34+/CD71−/DR− | 0.02 | 105 | 13 | 3/3 | |||
| 2 × 104 | 3 | 3/3 | |||||
| CD34− | 89.63 | 5 × 106 | 0 | ||||
| 15 | Unsorted | 106 | 36 | ||||
| CD34+/DR+ | 0.11 | 1.7 × 104 | 0 | ||||
| CD34+/DR− | 0.09 | 5.6 × 103 | 29 | ||||
| CD34+/CD71+/DR+ | 0.02 | NA | |||||
| CD34+/CD71+/DR− | 0.01 | NA | |||||
| CD34+/CD71−/DR+ | 0.09 | 5 × 103 | 0 | ||||
| CD34+/CD71−/DR− | 0.08 | 3.7 × 103 | 40 | ||||
| 2.5 × 103 | 19 | ||||||
| CD34− | 99.8 | 106 | 0 | ||||
| 17 | Unsorted | 106 | 13 | ||||
| 5 × 105 | 11 | ||||||
| 105 | 8 | ||||||
| CD34+/DR+ | 1.3 | 2 × 105 | 0 | ||||
| CD34+/DR− | 5.3 | 3 × 105 | 38 | ||||
| CD34− | 93.4 | 106 | 0 | ||||
| 6 × 105 | 0 | ||||||
The remainder of cells to 100% were CD45−.
Abbreviation: NA, data is not available.
Enrichment of NOD/SCID leukemia initiating cells. Graphs depict the engraftment achieved with variable numbers of number of unsorted and sorted cells from four patients (patients 4, 8, 13, and 15). All points represent at least 0.1% engraftment in a single NOD/SCID mouse femoral bone marrow.
Enrichment of NOD/SCID leukemia initiating cells. Graphs depict the engraftment achieved with variable numbers of number of unsorted and sorted cells from four patients (patients 4, 8, 13, and 15). All points represent at least 0.1% engraftment in a single NOD/SCID mouse femoral bone marrow.
Cytogenetic analysis of transplanted cells.
It was possible to analyze colonies derived from sorted CD45+ cells from NOD/SCID recipients injected with AML cells in seven experiments (five patients). In six cases, the majority of CFU derived from recipients of unsorted cells contained ≥ 60% leukemic cells (25 of 29) and four were negative. Plucked colonies from patient 4 were heterogeneous in detection of the +13 change by FISH from both sorted and unsorted cells. However, the cytogenetic analysis of this patient at presentation reported that only half of the metaphases analyzed had trisomy 13. Our results are therefore consistent with this finding. In all other cases, the majority of CFU derived from recipients of either the CD34+/71− (6 of 8), CD34+/HLA-DR− (8 of 8), or CD34+/CD71−/HLA-DR− (14 of 14) subfractions were defined as being positive for the specific karyotype of that patient. CFU derived from recipients of the CD34− subfraction were analyzed in one patient and all were found to be leukemic (19 of 19 positive colonies).
Cell phenotyping study.
To determine which combinations of antibodies might be optimal to obtain purification of primitive AML progenitors, peripheral blood mononuclear cells from 13 AML patients at diagnosis were stained with combinations of three monoclonal antibodies (Table 7). These AML patients were FAB M0 (n = 1), M1 (n = 2), M2 (n = 2), M4 (n = 4), M5 (n = 3), and one patient had an undetermined subtype. Expression of CD34 was high in most patients of this set (44% ± 10% CD34+). The CD34+/CD38−/HLA-DR−subfraction represented only 0.3% ± 0.1% of the total nucleated cell population, thus a sorting strategy using this combination of monoclonal antibodies could potentially enrich for primitive cells with this phenotype over 300-fold. Using CD34 in combination with CD38/CD71 or CD71/HLA-DR could produce enrichments of >100-fold and 67-fold, respectively, of primitive cells, which expressed CD34 and lacked expression of the other markers. The other antibody combinations that were evaluated would also enrich for primitive cells, although to a lesser extent, and may be less useful in a purification strategy.
Summary of Triple Phenotyping Study in 13 AML Patients at Diagnosis
| Phenotype . | % NC . | Potential Fold Enrichment7-150 . |
|---|---|---|
| CD34+/CD38−/HLA-DR− | 0.3 ± 0.1 | >300 |
| CD34+/CD38−/CD71− | 0.7 ± 0.4 | >100 |
| CD34+/CD71−/HLA-DR− | 1.5 ± 0.9 | 67 |
| CD34+/CD38−/CD33− | 2 ± 1 | 50 |
| CD34+/HLA-DR−/CD33− | 5 ± 3 | 20 |
| CD34+/CD71−/CD33− | 6.4 ± 3 | 15 |
| Phenotype . | % NC . | Potential Fold Enrichment7-150 . |
|---|---|---|
| CD34+/CD38−/HLA-DR− | 0.3 ± 0.1 | >300 |
| CD34+/CD38−/CD71− | 0.7 ± 0.4 | >100 |
| CD34+/CD71−/HLA-DR− | 1.5 ± 0.9 | 67 |
| CD34+/CD38−/CD33− | 2 ± 1 | 50 |
| CD34+/HLA-DR−/CD33− | 5 ± 3 | 20 |
| CD34+/CD71−/CD33− | 6.4 ± 3 | 15 |
Abbreviation: NC, nucleated cells.
Potential fold enrichment, calculated from the % of NC. This is equal to the degree of enrichment of primitive cells, which could be achieved by using a sorting strategy to select cells with this phenotype, providing all cells of interest were contained within this subfraction.
DISCUSSION
The finding that AML cells with different phenotypes differ in their proliferative ability in functional assays suggests that AML cells may exist as a hierarchy in most patients. We have previously shown that only a minority of AML blasts have long-term proliferative ability, with cells that have longer term proliferative ability being less frequent than those with shorter term proliferative potential.20 In this study, we have compared the phenotype of cells that grow long-term in vitro with those that engraft NOD/SCID mice. AML cells capable of producing human AML in immunodeficient NOD/SCID mice may be similar to the AML sustaining cells in the patients. The possibility that only a small minority of AML tumor cells have the ability to act as stem cells in vivo to maintain the malignant population has important therapeutic significance, as these cells may be the only relevant cells to target with treatment regimens. The main objectives in our study were to further phenotypically characterize and purify the cells which are responsible for maintenance of AML. Here we investigated the coexpression of CD34 and CD71, HLA-DR, or CD38 antigens on AML cells with long-term proliferative ability in vitro. Subsequently, we evaluated whether the phenotype of AML cells capable of long-term proliferation in vitro matched that of cells, which were capable of engrafting sublethally-irradiated NOD/SCID mice.
Our findings here indicate that among AML cells the CD34+/CD71− subfraction contained a relatively small proportion of nucleated cells (20% ± 7%) and primary CFU (34% ± 12%), but the majority of cells capable of producing colonies after 2 to 8 weeks in suspension culture. The CD34+/HLA-DR− phenotype selected 13% ± 5% of leukemic cells at diagnosis. However, after 8 weeks in suspension culture, the majority of CFU (86% ± 9%) were derived from cells with the CD34+/HLA-DR−phenotype. To further confirm the CD34+, CD71−, HLA-DR− phenotype of primitive AML cells, we evaluated the ability of the sorted subfractions to engraft NOD/SCID mice. Engraftment (0.13% to 74% CD45+) was achieved using 103 to 106 CD34+/CD71− cells from nine patient samples. However, there was no engraftment (<0.1% CD45+) with as many as 106 cells when the CD34+/CD71+ subfraction was used. Among five patients, a mean of 2% of the leukemic cell population was CD34+/HLA-DR−. Engraftment was achieved using this subfraction (range, 4% to 38% CD45+ with 3 × 103 to 5 × 105 cells) from four of these samples. In contrast, engraftment could not be attained using equal or greater numbers of CD34+/HLA-DR+ cells from the same patients. These findings show that the majority of AML cells, which are capable of engrafting NOD/SCID mice, as well as the majority of those generating CFU in long-term suspension culture, have the phenotype CD34+/CD71− and HLA-DR−.
In addition, AML progenitors detected after 4 to 8 weeks in the SC were also shown to be CD34+/CD38−, although only 17% ± 12% of the starting cell population expressed this phenotype. The CD34+/CD38− phenotype of NOD/SCID leukemia initiating cells has been previously described by other investigators22 and in one experiment, we confirmed this result with 48% CD45+ cells in the mouse transplanted with 106 CD34+/CD38− cells and no engraftment in the mouse injected with 106CD34+/CD38+ cells. Thus, AML cells capable of long-term proliferation in vitro and in vivo repopulation share a phenotype, also seen on primitive normal cells (LTC-IC), which express CD34 and lack expression of CD71, HLA-DR, and CD38.
The majority of nucleated cells in six patients were CD34− (<4% CD34+). In two of these patients, the majority of detectable CFU were always derived from the CD34− subfraction. CFU produced by the CD34− cells in these two patients were subsequently confirmed to have the leukemic karyotype by FISH. Engraftment in NOD/SCID mice was also achieved using CD34− cells in these two patients. This finding raises the possibility that in these two patients, the leukemic transformation may have occurred in a CD34− cell. However, in both cases, engraftment could also be achieved using considerably smaller numbers of CD34+/CD71− cells (103 and 4 × 103, respectively) and in patient 8, an equivalent degree of engraftment was attained with as few as 4 × 102CD34+/CD71−/HLA-DR−cells. These results suggest that in these patients, the leukemic transformation may have occurred in a primitive cell, which expressed CD34, with subsequent clonal evolution so that cells which no longer express the antigen, nevertheless retain their ability to proliferate and engraft NOD/SCID mice. These findings are consistent with our previous report21 and that of Terpstra et al,24demonstrating the presence of cells with high proliferative ability in the CD34− subfraction of some AML patients. The CD34− cells from the remainder of the patients analyzed in the present study were not capable of long-term proliferation in vitro and had no engraftment potential, suggesting that loss of expression of CD34 in these cases was associated with loss of proliferative ability. This finding provides evidence that differentiation, as defined by this phenotypic change, is usually, but not always, associated with loss of long-term proliferative ability in AML.
As a semiquantitative assessment of the number of AML cells, which are required to engraft the mice, a number of cell doses were evaluated whenever possible. The frequency of NOD/SCID leukemia initiating cells has previously estimated to be 0.2 to 100/106 AML cells,22 while other groups have reported a minimum of 1.1 × 106 unsorted cells were required for engraftment of SCID mice.25 Our data using the NOD/SCID model suggests that in these patients, the frequency of the NOD/SCID leukemic initiating cells ranges from <1 to 20/106 unsorted cells and further confirms the considerable heterogeneity between individual patients.
The CD34+/CD71−/HLA-DR−subfraction from five patients, representing on average 0.35% of nucleated cells, was capable of engrafting NOD/SCID mice (1% to 40% CD45+) using very small numbers of cells (4 × 102 to 105), while engraftment could not be achieved using equivalent numbers of CD34+/CD71− or CD34+/HLA-DR− cells. As shown in Fig 5, the combined use of all three antigens in a sorting strategy lead to approximately a 2 log enrichment of NOD/SCID leukemia initiating cells. Furthermore, as illustrated in Table 7, using combinations of CD34 and CD38 plus CD71 or HLA-DR may make it possible to achieve even greater (up to 300-fold) enrichment of primitive AML cells. The ability to enrich for NOD/SCID engrafting activity in some subfractions and correspondingly deplete this activity from other subfractions provides additional evidence for the stem cell model in AML.
Our results support the notion that the leukemic transformation occurs in a primitive cell, which has a CD34+/CD71−/HLA-DR−/CD38−phenotype similar to primitive normal hematopoietic cells. Other groups have also found the primitive AML phenotype to be CD34+/CD38−22,26,27 or CD34+/CD38−/CD33−.28We have previously shown these primitive AML cells differ from normal primitive progenitor cells in their lack of expression of Thy-1.21 Understanding of the phenotypic similarities and differences between normal and leukemic stem cells may enable the development of new purging strategies for use in autologous transplantation. The ability to isolate purified populations of primitive AML progenitors and to study their functional characteristics in biological assays will facilitate studies designed to investigate the molecular basis of human leukemogenesis.
ACKNOWLEDGMENT
The authors thank R. Zapf, B. Gerhard, G. Thornbury, and G. Shaw for excellent technical assistance.
Supported by a grant from the National Cancer Institute of Canada. A.B. is a fellow of the Leukaemia Research Fund of Canada.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to H.J. Sutherland, MD, PhD, Terry Fox Laboratory, 601 W 10th Ave, Vancouver, BC, Canada V5Z 1L3.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal