Abstract
Three novel monoclonal antibodies (MoAbs) have been established that recognize distinct epitopes of a human pre–B-cell receptor (pre-BCR) composed of a μ heavy (μH) chain and a λ5/VpreB surrogate light (SL) chain. HSL11 reacts with λ5 whereas HSL96 reacts with VpreB. Intriguingly, HSL2 does not bind to each component of the pre-BCR but does bind to the completely assembled pre-BCR complex. Flow cytometric analyses with cytoplasmic staining of a panel of human cell lines showed that HSL11 and HSL96 specifically stained cell lines derived from the pro–B and pre–B-cell stages of B-cell development. In contrast, HSL2 stained exclusively cell lines derived from the pre–B-cell stage. These results prompted us to explore the possibility of clinical application of these MoAbs for the determination of the cell lineage and developmental stage of acute lymphoblastic leukemia (ALL). Whereas none of mature B-lineage ALLs (B-ALLs), T-lineage ALLs (T-ALLs), and acute myeloid leukemias analyzed were stained in the cytoplasm with these three MoAbs, the vast majority of non–B- and non–T-ALLs (53 out of 56 cases) were found positive for either λ5, Vpre-B, or both in their cytoplasm. Among these 53 cytoplasmic SL chain-positive ALLs, 19 cases were also positive for cytoplasmic μH chain, indicative of pre–B-cell origin. Interestingly, 6 out of these 19 pre–B-ALL cases were found negative for cytoplasmic staining with HSL2. From these results, we propose a novel classification of B-ALL in which five subtypes are defined on the basis of the differential expression of SL chain, μH chain, pre-BCR, and light chain along the B-cell development.
ACUTE LYMPHOBLASTIC leukemia (ALL) is the most common malignancy in children and accounts for approximately 80% of childhood leukemia.1,2 ALL seems to be a heterogeneous group of disorders with subgroups that have distinct clinical and prognostic features. Several attempts have been made to establish the classification of ALL cells that can be correlated with clinical characteristics and the behavior of the disease such as response to therapy.1 2
The French-American-British (FAB) classification is based on morphology of leukemic cells and identifies three subgroups of ALL, namely L1, L2, and L3.3 However, this classification does not tell the cell lineage and maturational stage from which leukemia arises. Indeed, the vast majority of childhood ALLs are classified as L1. On the other hand, the immunophenotypic classification is based on expression of certain antigens on the surface of leukemic cells and seems more clinically relevant in case of ALL.4,5 Because normal lymphocytes express specific antigens in an orderly fashion through their different stages of development, the expression of certain antigens on the surface of ALL cells can be considered to indicate the specific stage of lymphocyte development where the malignant transformation occurred. Based on reactivity with a panel of antibodies, ALL is broadly classified as having a T- or B-cell origin. B-lineage ALL (B-ALL) is further subdivided into three distinct subtypes: early pre-B cell (or B precursor), pre-B cell, and B cell2,4,5; or into four subtypes: pro-B (B-I), common (B-II), pre-B (B-III), and mature-B (B-IV).6 Approximately 85% to 90% of childhood ALL and 75% to 80% of adult ALL is assigned to be of B-cell lineage, of which nearly 70% falls into the early pre–B-cell subtype and less than 5% into the mature B-cell subtype. Markers such as CD10, CD19, CD20, CD21, CD22, CD23, CD24, and HLA-DR are commonly used to characterize B-ALL. However, one must be cautious in use of these markers, because no marker is absolutely lineage specific and stage specific. For example, CD19 can be found in at least 50% of acute myeloid leukemia (AML) with t(8;21),7 and CD10 in T-lineage ALL (T-ALL) as well.8 Moreover, approximately 5% to 10% of childhood ALL and 30% of adult ALL cases express myeloid markers.9,10 The rearrangement patterns of immunoglobulin (Ig) and T-cell receptor (TCR) genes have also been used as markers for clonality and origin of leukemic cells. However, the molecular technique to detect these patterns is not as easy and fast as immunophenotyping, and also has a limitation in the determination of the cellular origin. TCR rearrangements are often observed in B-ALL and also in AML, but at a lower frequency.11,12 IgH gene rearrangements occur in 10% to 15% of T-ALL and in about 20% of AML cases.12 13 Thus, there is a clear need to discover more reliable markers to define the cellular origin of leukemic cells.
Recent studies have shown that pre–B-cell receptor (pre-BCR) plays critical roles in early B-cell development,14-17 and the lack of its expression resulted in severe impairment of B-cell production.18-21 The pre-BCR is composed of a μ heavy (μH) chain and surrogate light (SL) chain,22,23 and its expression is restricted to the pre–B-cell stage of B-cell development.24,25 The SL chain is composed of λ5 and VpreB,26-30 both of which are already produced before the μH chain is expressed, that is, before the formation of pre-BCR. Thus, the expression of the SL chain is restricted to both pro–B and pre–B-cell stages of B-cell development.24 25 These lineage- and stage-specific expression patterns of the SL chain and pre-BCR make them very attractive as potential markers to determine the cell lineage and developmental stage of ALL cells.
In the present study, we have established three monoclonal antibodies (MoAbs) with distinct specificity to the human SL chain and pre-BCR. Analysis of a panel of human leukemia samples has shown that the flow cytometry with those MoAbs is indeed an easy, fast, and reliable way to diagnose the cell lineage and developmental stage of ALL. Based on this analysis, we will propose a novel classification of B-ALL.
Several different nomenclatures are used by researchers to distinguish different stages of B-cell development and subtypes of B-ALL.2,4-6,25 31 To avoid possible confusion, in this study we use the following nomenclature for the stages of B-cell development: pro-B cells, pre-B cells, and B cells as H chain–L chain–-, H chain+L chain–-, and H chain+L chain+-B-lineage cells, respectively.
MATERIALS AND METHODS
Cell lines.
Human pro–B-cell lines include RS4;11,32REH,33 NALM16,34 NALM19,35NALM20,36 and NALM27.37 Human pre–B-cell lines include NALM6,38 NALM17,37 697,39HPB-NULL,37 and SMS-SB.40 LBW-4,41Daudi,42 and GM60743 are human B-cell lines whereas Jurkat44 and MOLT-445 are human T-cell lines. All the cell lines were maintained in RPMI 1640 supplemented with 10% fetal calf serum (Equitech-Bio, Inc, Ingram, TX), 100 U/mL of penicillin-streptomycin (GIBCO-BRL, Grand Island, NY), 2 mmol/L L-glutamine (GIBCO-BRL), and 5 × 10–5 mol/L 2-mercaptoethanol.
Leukemia samples.
Sixty-three childhood leukemia samples which had been diagnosed by conventional phenotypic analyses and stocked in liquid nitrogen were analyzed in this study. They included 56 samples of non–B- and non–T-ALL, 3 of mature B-ALL (L3 of FAB classification), 2 of T-ALL, and 2 of AML. All the non–B- and non–T-ALL samples had been categorized into L1 of FAB classification and assigned to be of B-lineage origin based on the expression of CD19 on their surface, but further subtyping had not been performed. Sources of samples were mainly bone marrow or peripheral blood, but in 1 case of T-ALL the sample was obtained from pleural effusion, and in all 3 cases of mature B-ALL the samples were taken from lymph nodes. Immediately before immunofluorescence staining, frozen samples were thawed, washed, and resuspended in staining buffer. The research followed the tenets of the Declaration of Helsinki.
Antibodies.
Mouse MoAbs specific to human CD10, CD19, CD22, CD79b, κL chain and λL chain were purchased from Pharmingen (San Diego, CA). A mouse MoAb specific to human μH chain was purified from culture supernatants of a hybridoma clone M-2E6 (American Type Culture Collection, HB138).46 Phycoerythrin (PE)-conjugated goat anti-mouse κ antibody was purchased from Southern Biotechnology Associates Inc (Birmingham, AL).
Establishment of transfectants expressing a human SL chain and a human/mouse-chimeric pre-BCR.
cDNAs encoding human λ5 and VpreB were cloned from a human pre–B-cell line NALM6 by reverse transcription polymerase chain reaction (RT-PCR) based on the published nucleotide sequences.28-30 Ig-nonproducing X63.Ag8-653 myeloma cells47 were transfected with the expression vector BCMGSHyg22 carrying either the λ5 or VpreB cDNA as described previously.48 Ltk–μC37 fibroblast cells expressing mouse μH chain22 were transfected with BCMGSHyg carrying both human λ5 and VpreB cDNAs in the same way as described previously22 to establish Ltk–pre-BCR transfectants that secrete a soluble form of chimeric pre-BCR (mouse μH chain/human SL chain complex).
Establishment of MoAbs specific to human λ5, VpreB, and pre-BCR.
The chimeric pre-BCR complexes were purified by absorption to M4149 (anti-mouse μH MoAb)-conjugated Sepharose beads (Pharmacia Biotech, Uppsala, Sweden) from culture supernatants of the Ltk–pre-BCR transfectants. BALB/c mice (6-week-old female; Japan SLC, Hamamatsu, Japan) were immunized intraperitoneally seven times at 1-week intervals, each time with approximately 1 μg of the complex absorbed on beads, and then administered a final boost with intravenous injection of 30 μg of the complex eluted from the M41-conjugated beads. Three days after final injection, spleen cells were fused with PAI myeloma cells50 and cultured in medium containing hypoxanthine, aminopterin, thymidine (GIBCO-BRL) and recombinant interleukin-6.51Culture supernatants of resulting hybridomas were initially screened by enzyme-linked immunosorbent assay for reactivity to the chimeric pre-BCR but not to human IgM (μκ and μλ). Supernatants of selected cultures were further characterized by cell staining as well as immunoprecipitation and cloned twice by limiting dilution. All the MoAbs established and described here were of the IgG1 subclass and had κL chains as determined by the isotyping kit (Pharmingen).
Immunoprecipitation.
Cells were biosynthetically labeled with [35S]-methionine (Dupont, Wilmington, DE) for 4 hours and lysed with 1% NP-40 lysis buffer as described previously.48 Cell lysates were precleared with protein G-Sepharose beads (Pharmacia Biotech AB, Uppsala, Sweden) and then reacted for 2 hours with protein G-Sepharose beads preincubated with hybridoma culture supernatants or purified antibodies. Immunoprecipitates were electrophoresed on 13% sodium dodecyl sulfate (SDS) polyacrylamide gel, the gel was dried, and the radioactive bands were visualized by the phosphoimager Fuji BAS2000 (Fuji Photo Film Co Ltd, Tokyo, Japan).
Cell-surface and cytoplasmic staining.
For cell-surface staining, cells suspended in staining buffer (phosphate-buffered saline with 0.1% bovine serum albumin, 0.05%NaN3) were incubated with 1 μg/mL of mouse MoAbs for 30 minutes on ice and then with 2 μg/mL of PE-conjugated goat anti-mouse κ antibodies. For cytoplasmic staining, cells were suspended in staining buffer containing 0.05% saponin (Sigma Chemical Co, St Louis, MO) and 10 mmol/L HEPES, pH 7.3, and stained as for cell-surface staining. Throughout staining and washing procedures, 0.05% of saponin was kept in buffer. Stained cells were applied to the flow cytometer FACSCalibur (Becton Dickinson, Mountain View, CA). Cells present in the lymphocyte gate defined by light scatter were analyzed.
RESULTS
Establishment of MoAbs specific to λ5, VpreB, or a conformational epitope of the human pre-BCR complex.
Three MoAbs, designated HSL2, HSL11, and HSL96, were established as described in Materials and Methods. All the MoAbs precipitated from NALM6 pre-B cells three proteins of 70 kD, 22 kD, and 18 kD corresponding to μH chain, λ5, and VpreB, respectively, as did an anti-μH chain MoAb (Fig 1A). Intriguingly, HSL11 and HSL96 precipitated the λ5/VpreB SL chain complex from μH chain-negative RS4;11 pro-B cells whereas HSL2 did not (Fig 1B). To identify the specificity of each MoAb further, X63.Ag8-653 cells transfected with either the λ5 or VpreB gene were used for immunoprecipitation. HSL96 precipitated an 18-kD protein from the VpreB transfectant but not from the λ5 transfectant, indicating its specificity to VpreB (Fig 1C). On the other hand, HSL11 precipitated 22-kD and 16-kD proteins from the λ5 transfectant but not from the VpreB transfectant, indicating its specificity to λ5 (Fig 1C). Though the exact nature of the 16-kD protein coprecipitated with λ5 remains to be elucidated, it has been reported that a similar 16-kD protein was detected in the immunoprecipitates of human pre-BCR complexes.25 52 It should be related to λ5 because it could be detected only in the λ5-expressing cells (NALM6, RS4;11, the λ5-transfectant) but not in the VpreB transfectant. In contrast to HSL11 and HSL96, HSL2 precipitated neither λ5 nor VpreB from the λ5 and VpreB transfectants (Fig 1C). It also did not precipitate IgM (μκ and μλ) from human B-cell lines Daudi and LBW-4 (data not shown). Thus, HSL2 reacted with the complete pre-BCR complex but not with each component of the pre-BCR complex or the λ5/VpreB SL chain. This indicates that HSL2 is a unique MoAb that recognizes a conformational epitope formed only when the μH chain and λ5/VpreB SL chain associate with each other to make the pre-BCR complex. On the other hand, HSL11 and HSL96 reacted with λ5 and VpreB, respectively, in free form, in the form of the λ5/VpreB SL chain, or in the form of the complete pre-BCR complex.
Specificity of the newly established MoAbs HSL11, HSL96, and HSL2 for human λ5, VpreB, and pre-BCR, respectively. A pre–B-cell line NALM6 (A), a pro–B-cell line RS4;11 (B), and X63.Ag8-653 cells transfected with either the λ5 or VpreB gene (C) were biosynthetically labeled with [35S]-methionine for 4 hours and lysed with 1% NP-40 lysis buffer. Detergent-soluble lysates were incubated with indicated MoAbs, and immunoprecipitates were analyzed by 13% SDS polyacrylamide gel electrophoresis under reducing conditions. The positions of μH, λ5, and VpreB are indicated by arrows, and a 16-kD band precipitated by HSL11 together with λ5 is indicated by an arrow head.
Specificity of the newly established MoAbs HSL11, HSL96, and HSL2 for human λ5, VpreB, and pre-BCR, respectively. A pre–B-cell line NALM6 (A), a pro–B-cell line RS4;11 (B), and X63.Ag8-653 cells transfected with either the λ5 or VpreB gene (C) were biosynthetically labeled with [35S]-methionine for 4 hours and lysed with 1% NP-40 lysis buffer. Detergent-soluble lysates were incubated with indicated MoAbs, and immunoprecipitates were analyzed by 13% SDS polyacrylamide gel electrophoresis under reducing conditions. The positions of μH, λ5, and VpreB are indicated by arrows, and a 16-kD band precipitated by HSL11 together with λ5 is indicated by an arrow head.
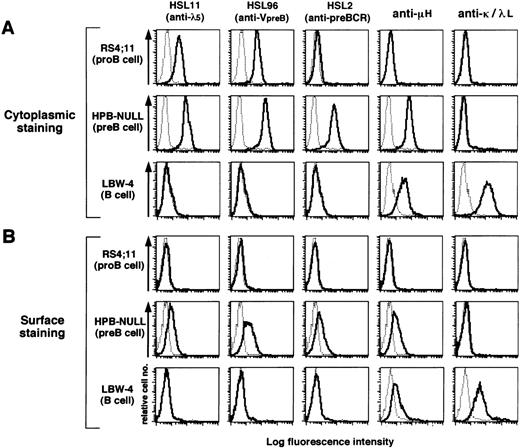
Differential reactivity of the MoAbs to human pro–B and pre–B-cell lines.
The reactivity of the three MoAbs to a panel of human cell lines was examined by immunofluorescence flow cytometry on the cell surface as well as in the cytoplasm (Fig 2 and Table 1). For cytoplasmic staining, the cell membrane was permealized with 0.05% of saponin added in the staining buffer. HSL11, HSL96, and HSL2 were all found to stain the cytoplasm and cell surface of all five μH+ pre–B-cell lines analyzed (HPB-NULL in Fig 2 and Table 1; 697, SMS-SB, NALM6, and NALM17 in Table 1). The intensity of the surface staining was 7 to 10 times less than that of the cytoplasmic staining, indicating that the majority of pre-BCR was retained in the cytoplasm. None of the MoAbs stained mature B-and T-cell lines (LBW-4 in Fig 2 and Table 1; Daudi, GM607, Jurkat, and MOLT-4 in Table 1), confirming no cross-reactivity to Ig or TCR.
Flow cytometric analysis of human B-lineage cell lines with the MoAbs. A pro–B-cell line RS4;11, a pre–B-cell line HPB-NULL, and a B-cell line LBW-4 were stained by the indicated MoAbs in the cytoplasm (A) and on the cell surface (B). For cytoplasmic staining, 0.05% of saponin was added to the staining buffer. Cells were first incubated with the MoAbs and then with PE–anti-mouse κL chain antibody. The fluorescence intensity of the stained cells was analyzed by a flow cytometer. The thin dotted-line histograms indicate control staining with an isotype-matched control MoAb whereas the thick solid-line histograms indicate staining with MoAbs specific to λ5, VpreB, pre-BCR, μH chain, κL chain, or λL chain. All data including other cell lines are summarized in Table 1.
Flow cytometric analysis of human B-lineage cell lines with the MoAbs. A pro–B-cell line RS4;11, a pre–B-cell line HPB-NULL, and a B-cell line LBW-4 were stained by the indicated MoAbs in the cytoplasm (A) and on the cell surface (B). For cytoplasmic staining, 0.05% of saponin was added to the staining buffer. Cells were first incubated with the MoAbs and then with PE–anti-mouse κL chain antibody. The fluorescence intensity of the stained cells was analyzed by a flow cytometer. The thin dotted-line histograms indicate control staining with an isotype-matched control MoAb whereas the thick solid-line histograms indicate staining with MoAbs specific to λ5, VpreB, pre-BCR, μH chain, κL chain, or λL chain. All data including other cell lines are summarized in Table 1.
The Lineage- and Differentiation Stage–Specific Expression of SL Chain and Pre-BCR Detected by the MoAbs in a Panel of Human Cell Lines
| Cell Type . | Cell Line . | Cytoplasmic Staining . | Surface Staining . | ||||
|---|---|---|---|---|---|---|---|
| HSL 11 (anti-λ5) . | HSL 96 (anti-VpreB) . | HSL 2 (anti-pre-BCR) . | HSL 11 (anti-λ5) . | HSL 96 (anti-VpreB) . | HSL 2 (anti-pre-BCR) . | ||
| Pro B (μ−, L−) | RS4; 11 | + | + | − | − | − | − |
| REH | + | − | − | − | − | − | |
| NALM 16 | + | + | − | − | − | − | |
| NALM 19 | + | + | − | − | − | − | |
| NALM 20 | + | + | − | − | − | − | |
| NALM 27 | + | + | − | − | − | − | |
| Pre B (μ+, L−) | HPB-NULL | + | + | + | + | + | + |
| 6 9 7 | + | + | + | + | + | + | |
| SMS-SB | + | + | + | + | + | + | |
| NALM 6 | + | + | + | + | + | + | |
| NALM 17 | + | + | + | + | + | + | |
| B (μ+, L+) | LBW-4 | − | − | − | − | − | − |
| Daudi | − | − | − | − | − | − | |
| GM607 | − | − | − | − | − | − | |
| T (CD3+) | Jurkat | − | − | − | − | − | − |
| MOLT-4 | − | − | − | − | − | − | |
| Cell Type . | Cell Line . | Cytoplasmic Staining . | Surface Staining . | ||||
|---|---|---|---|---|---|---|---|
| HSL 11 (anti-λ5) . | HSL 96 (anti-VpreB) . | HSL 2 (anti-pre-BCR) . | HSL 11 (anti-λ5) . | HSL 96 (anti-VpreB) . | HSL 2 (anti-pre-BCR) . | ||
| Pro B (μ−, L−) | RS4; 11 | + | + | − | − | − | − |
| REH | + | − | − | − | − | − | |
| NALM 16 | + | + | − | − | − | − | |
| NALM 19 | + | + | − | − | − | − | |
| NALM 20 | + | + | − | − | − | − | |
| NALM 27 | + | + | − | − | − | − | |
| Pre B (μ+, L−) | HPB-NULL | + | + | + | + | + | + |
| 6 9 7 | + | + | + | + | + | + | |
| SMS-SB | + | + | + | + | + | + | |
| NALM 6 | + | + | + | + | + | + | |
| NALM 17 | + | + | + | + | + | + | |
| B (μ+, L+) | LBW-4 | − | − | − | − | − | − |
| Daudi | − | − | − | − | − | − | |
| GM607 | − | − | − | − | − | − | |
| T (CD3+) | Jurkat | − | − | − | − | − | − |
| MOLT-4 | − | − | − | − | − | − | |
When μH-negative pro–B-cell lines were analyzed, the three MoAbs showed differential reactivity. HSL11 and HSL96 stained cytoplasm of RS4;11 pro–B-cell line whereas HSL2 did not (Fig 2). This was also the case for other pro–B-cell lines NALM16, NALM19, NALM20, and NALM27 (Table 1). One pro–B-cell line, REH, was found to be stained by HSL11 but not by HSL96 or HSL2 (Table 1), in agreement with the result of RT-PCR analysis that showed the presence of λ5 transcripts but not VpreB transcripts in REH (data not shown). The lack of reactivity of HSL2 with μH– pro–B-cell lines is in line with the conclusion drawn from the immunoprecipitation experiments that HSL2 reacts with the complete pre-BCR complex but not with each component of the complex or λ5/VpreB SL chain. Notably, none of the MoAbs stained the cell surface of the pro–B-cell lines even though λ5 and VpreB proteins were detected intracellularly by HSL11 and HSL96, respectively. This suggests that only completely assembled pre-BCR complexes can be expressed on the cell surface in humans, in agreement with a previous report,25 but in contrast to the observation in mouse pro-B cells.24 48
Collectively, HSL11 and HSL96 specifically reacted with cell lines derived from both pro–B- and pre–B-cell stages when analyzed by the cytoplasmic staining. In contrast, HSL2 exclusively recognized cell lines derived from the pre–B-cell stage but not from the earlier or later stages of B-cell development. Thus, flow cytometric analysis with the combination of these MoAbs seems to be a convenient and reliable way to distinguish the developmental stage of B-lineage cells.
Flow cytometric analysis of human acute leukemia cells.
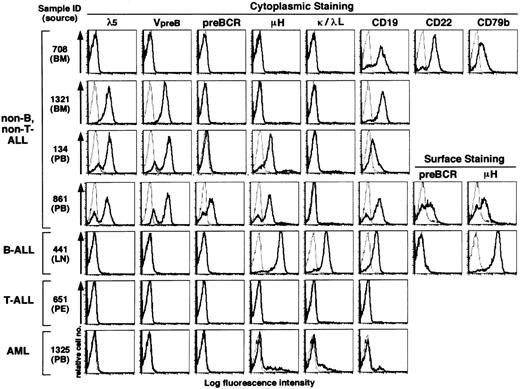
The analysis of human cell lines described above strongly suggested a possible clinical application of the MoAbs to determine the cellular origin of human leukemia cells. Therefore, we next performed flow cytometric analysis of 63 childhood leukemia samples that had been diagnosed by conventional phenotypic analyses and stocked in liquid nitrogen. These included 56 samples of non–B- and non–T-ALL, 3 of mature B-ALL, 2 of T-ALL, and 2 of AML (Fig3 and Table 2). All the non–B-, non–T-ALL samples had been assigned to be of B-lineage origin based on the expression of CD19 on their surface, but further subtyping had not been performed. Because the analysis of human cell lines showed that the surface expression of pre-BCR was extremely low as compared with its cytoplasmic expression, we focused on the cytoplasmic staining in the following experiments. The MoAbs used were HSL11 (anti-λ5), HSL96 (anti–VpreB), HSL2 (anti–pre-BCR), anti-μH chain, anti-κL chain, anti-λL chain, anti-CD19, anti-CD22, and anti-CD79b. Staining profiles of representative samples are shown in Fig 3, and all data are summarized in Tables 2 and 3.
Flow cytometric analysis of human acute leukemia samples by using the MoAbs HSL11, HSL96, and HSL2. Frozen samples of childhood acute leukemia were thawed and stained in the cytoplasm (in a few cases on the cell surface as well) by the MoAbs specific to λ5, VpreB, pre-BCR, μH chain, κL chain, λL chain, CD19, CD22, or CD79b in the same way as in Fig 2. Staining profiles with these MoAbs (thick solid line) and staining profiles with an isotype-matched control MoAb (thin dotted line) are overlaid in the histograms. Though data are not shown, the T-ALL sample was positive for CD3, CD4, and CD8 whereas the AML sample was positive for CD13 and CD33. All data including other leukemia samples are summarized in Tables 2 and 3. Sources of leukemia samples: BM, bone marrow; PB, peripheral blood; LN, lymph node; PE, pleural effusion.
Flow cytometric analysis of human acute leukemia samples by using the MoAbs HSL11, HSL96, and HSL2. Frozen samples of childhood acute leukemia were thawed and stained in the cytoplasm (in a few cases on the cell surface as well) by the MoAbs specific to λ5, VpreB, pre-BCR, μH chain, κL chain, λL chain, CD19, CD22, or CD79b in the same way as in Fig 2. Staining profiles with these MoAbs (thick solid line) and staining profiles with an isotype-matched control MoAb (thin dotted line) are overlaid in the histograms. Though data are not shown, the T-ALL sample was positive for CD3, CD4, and CD8 whereas the AML sample was positive for CD13 and CD33. All data including other leukemia samples are summarized in Tables 2 and 3. Sources of leukemia samples: BM, bone marrow; PB, peripheral blood; LN, lymph node; PE, pleural effusion.
Summary of Flow Cytometric Analysis of Human Leukemias
| Type of Leukemia . | No. of Samples Analyzed . | No. of Samples Positive for (%)* . | ||||
|---|---|---|---|---|---|---|
| λ5 . | VpreB . | pre-BCR . | μH Chain . | L Chain (κ or λ) . | ||
| Non–B-, non–T-ALL | 56 | 46 (82) | 49 (88) | 13 (23) | 19 (34) | 0 (0) |
| Mature B-ALL | 3 | 0 (0) | 0 (0) | 0 (0) | 3 (100) | 3 (100) |
| T-ALL | 2 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| AML | 2 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Type of Leukemia . | No. of Samples Analyzed . | No. of Samples Positive for (%)* . | ||||
|---|---|---|---|---|---|---|
| λ5 . | VpreB . | pre-BCR . | μH Chain . | L Chain (κ or λ) . | ||
| Non–B-, non–T-ALL | 56 | 46 (82) | 49 (88) | 13 (23) | 19 (34) | 0 (0) |
| Mature B-ALL | 3 | 0 (0) | 0 (0) | 0 (0) | 3 (100) | 3 (100) |
| T-ALL | 2 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| AML | 2 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
All samples were cytoplasmically stained with the specific MoAbs and analyzed by flow cytometer as described in Materials and Methods.
A Novel Classification of Non–B-, Non–T-ALL Based on the Expression of SL Chain, μH Chain, and Pre-BCR, and the Incidence of Each Group Observed in This Study
| Cytoplasmic Expression . | No. of Bone Marrow Samples [A] . | No. of Peripheral Blood Samples [B] . | [A] + [B] (%) . | Classification Proposed in This Study . | |||
|---|---|---|---|---|---|---|---|
| SL Chain (λ5 and/or VpreB) . | μH Chain . | Pre-BCR . | L Chain . | ||||
| − | − | − | − | 3 | 0 | 3 (5) | Pro–B-I |
| +3-150 | − | − | − | 17 | 17 | 34 (61) | Pro–B-II |
| +3-151 | + | − | − | 4 | 2 | 6 (11) | Pre-B (pre-BCR−) |
| +3-152 | + | + | − | 7 | 6 | 13 (23) | Pre-B (pre-BCR+) |
| Total | 31 | 25 | 56 (100) | ||||
| Cytoplasmic Expression . | No. of Bone Marrow Samples [A] . | No. of Peripheral Blood Samples [B] . | [A] + [B] (%) . | Classification Proposed in This Study . | |||
|---|---|---|---|---|---|---|---|
| SL Chain (λ5 and/or VpreB) . | μH Chain . | Pre-BCR . | L Chain . | ||||
| − | − | − | − | 3 | 0 | 3 (5) | Pro–B-I |
| +3-150 | − | − | − | 17 | 17 | 34 (61) | Pro–B-II |
| +3-151 | + | − | − | 4 | 2 | 6 (11) | Pre-B (pre-BCR−) |
| +3-152 | + | + | − | 7 | 6 | 13 (23) | Pre-B (pre-BCR+) |
| Total | 31 | 25 | 56 (100) | ||||
Among 34 samples in this group, 24 cases were positive for both λ5 and VpreB, 3 were positive for λ5 only, and 7 cases were positive for VpreB only.
Among 6 samples in this group, 5 cases were positive for both λ5 and VpreB, and one case was positive for λ5 only.
All 13 samples in this group were positive for both λ5 and VpreB.
None of the mature B-ALL, T-ALL, and AML samples were positive for λ5, VpreB, or pre-BCR as detected by HSL11, HSL96, and HSL2, respectively (three rows from the bottom in Fig 3 and Table 2). Among 56 samples of non–B-, non–T-CD19+ ALL analyzed, 19 were found positive for cytoplasmic μH chain, indicative of pre–B-cell origin, whereas the rest (37 samples) were negative, indicative of pro–B-cell origin (Table 2). Whereas none of these non–B-, non–T-ALL samples expressed L chains (κ or λ), 82%, 88%, and 23% of them expressed λ5, VpreB, and pre-BCR, respectively (Table 2). The vast majority (95%) of the non–B-, non–T-ALL samples were found to express at least one of the λ5 and VpreB components of the SL chain (Table 3). In the remaining 3 cases (5%), no components of λ5, VpreB, μH chain, and L chain were detected (for example, top row in Fig 3). Nevertheless, these three “null” samples were found to express not only CD19 but also CD22 and CD79b in the cytoplasm (for example, top row in Fig 3), indicative of B-lineage origin.
Thirty-four cases (61%) of the non–B-, non–T-ALL samples expressed SL chain (λ5 and/or VpreB) but not μH chain (Table 3): 24 cases were positive for both λ5 and VpreB (for example, second row in Fig 3), 3 were positive for λ5 only, and 7 were positive for VpreB only. All the 19 μH+ pre–B-ALL samples were found to express SL chain (λ5 and/or VpreB; Table 3): 18 were positive for both λ5 and VpreB (for example, third and fourth rows of Fig 3), and 1 was positive for λ5 only. When the expression of pre-BCR was analyzed by HSL2, 13 out of the 19 μH+ pre–B-ALL samples were positive for pre-BCR (for example, fourth row in Fig 3) whereas the remaining 6 samples were negative for pre-BCR (for example, third row in Fig 3) as summarized in Table 3. In a few cases in which enough cell numbers were available, cell-surface staining was also performed. The surface expression of pre-BCR in pre–B-ALL samples was found to be extremely low when compared with the surface expression of IgM in mature B-ALL samples (for example, compare the expression of surface μH chain in fourth and fifth rows of Fig 3).
DISCUSSION
The differentiation of B cells from hematopoietic stem cells progresses through a series of successive stages that are defined by sequential rearrangements of Ig loci, first in IgH locus and then in IgL locus, and expression of various stage-associated markers including Ig H and L chain proteins.31,53 Recent studies have clarified our understanding of mechanisms by which the Ig gene rearrangements are controlled and how Ig gene products participate in the regulation of the B-cell differentiation program.15 Pre-BCR composed of μH chain and SL chain has been shown to transduce signals for cell proliferation and differentiation as well as the allelic exclusion at IgH chain loci during the pre–B-cell stage of B-cell differentiation.14-17 The importance of pre-BCR in human B-cell development was highlighted by the discovery of B-cell–deficient patients in which the gene coding for μH chain or SL chain had a deletion or mutation.20 21
Besides the physiological importance of pre-BCR, its expression pattern restricted to an early stage of B-cell development makes it a hallmark of early B-cell precursors. Indeed, it has been shown that the expression of VpreB and λ5 genes coding for SL chain could be used as a specific marker for B-cell precursor leukemias.54,55However, the detection of the gene expression by Northern blotting as shown in previous studies would not be a routine laboratory work.54,55 Though the RT-PCR technique might be more sensitive to detect the expression and more convenient as a routine work,55 a possible drawback of this sensitive technique is to have false positive signals derived from normal B-cell precursor cells contaminated in leukemia samples.
In the present study, we showed that flow cytometric analysis with the combination of the SL chain– and pre-BCR–specific MoAbs was an easy, fast, and reliable way to diagnose leukemias derived from early B-cell precursors. Because the expression of SL chain and pre-BCR on the cell surface was found to be extremely low and often undetectable, cytoplasmic staining was needed for definite determination of their expression in leukemic cells. Fortunately, we figured out that cytoplasmic staining of leukemia samples could be performed as easily as cell-surface staining by just adding 0.05% of saponin in staining buffer without any other treatment of cells such as fixation with paraformaldehyde or ethanol. Though several laboratories have established MoAbs specific to either human λ5 or VpreB protein,25,56-58 no study has been reported that uses those MoAbs for analysis of leukemia specimens as far as we know. Because all those MoAbs are of IgM subclass in contrast to our MoAbs (IgG1 subclass), they might not be adequate for cytoplasmic staining. In accordance with the previous studies analyzing the gene expression using Northern blot and RT-PCR,54 55 the vast majority of non–B-, non–T-ALL (53 out of 56 cases) analyzed in the present study were found to express λ5 protein, VpreB protein, or both. On the contrary, none of mature B-ALL, T-ALL, and AML analyzed so far expressed these proteins, indicating the lineage and stage fidelity of SL chain expression even in leukemic cells.
The flow cytometric analysis using the MoAbs established in this study seems to be extremely useful for the precise classification of B-ALL according to the stages of B-cell development from which leukemic cells have arisen. In the course of B-cell development, the production of μH chains starts at the pre–B-cell stage and continues until the mature B-cell stage, whereas the production of SL chains already starts at the pro–B-cell stage and ceases before B-cell stage.24 25 Therefore, the B-cell differentiation process can be divided at least into three stages: the pro–B-cell stage expressing SL chains but not μH chains; the pre–B-cell stage expressing both SL and μH chains, hence pre-BCR; and the B-cell stage expressing μH chains and conventional L chains in place of SL chains. In the present study, we found that the expression patterns of λ5 and VpreB proteins were parallel in most of the cell lines and leukemic samples though some cases expressed only either of these proteins. To avoid complexity in the ALL classification and make it as simple as possible, we decided to score cells as positive for SL chain when λ5 or VpreB or both were detected in them by HSL11 and HSL96 MoAbs (Table3). By these criteria, among 56 non–B-, non–T-CD19+ ALL samples analyzed in the present study, 34 cases (61%) are assigned to pro-B ALL whereas 19 cases (34%) are assigned to pre-B ALL. The remaining 3 cases (5%) were found negative for both SL and μH chains. Because these 3 expressed not only CD19 but also CD22 and CD79b (B29, Igβ) in their cytoplasm, it is most likely that they were derived from very early progenitor cells that had been committed to B-cell lineage. Therefore, we would like to propose to designate this group as pro–B-I-ALL and the SL chain+μH chain– group as pro–B-II-ALL (Table 3).
Intriguingly, not all of the 19 μH chain+ pre–B-ALL cases were stained by the MoAb HSL2 which recognizes pre-BCR: 13 cases were positive for HSL2 staining whereas 6 cases were negative (Table3). In 1 case of the HSL2-negative pre–B-ALL group, the reason for nonreactivity with HSL2 was easily understandable because that case lacked VpreB expression. In contrast, the remainder (5 cases) of the HSL2-negative pre–B-ALL cases expressed all the components of pre-BCR detected by the specific MoAbs. Because HSL2 recognizes a completely assembled pre-BCR complex but not each component of the pre-BCR, one may assume that in these 5 cases both μH chain and λ5/VpreB SL chain were produced but not associated with each other as efficiently as to form pre-BCR. Though limited cell numbers in stocked leukemic specimens could not allow us to test this assumption by biochemical analyses, a recent study on the repertoire of variable region of IgH chains (VH) of mouse bone marrow precursor B cells suggests this assumption is plausible. The study showed that only half of μH chains expressed in early pre-B cells (c-kit+cytoplasmic μH+ cells) had the capacity to associate with SL chains to form a pre-BCR.59 Though such an analysis has not yet been performed for human counterparts, our observation in human pre–B-ALL samples strongly suggests that the same thing may happen in an early pre–B-cell stage of normal human B-cell development. The study on mouse pre-B cells predicted distinct fates of pre-BCR+ and pre-BCR– pre-B cells in B-cell development: the former can differentiate further toward B cells whereas the latter cannot unless another rearrangement or VH gene replacement takes place to replace μH chains.59 Based on these, we would like to propose to categorize pre–B-ALL into two groups, pre–B- (pre-BCR+) ALL and pre–B- (pre-BCR–) ALL, which can be easily distinguished by the reactivity with HSL2 (Table 3). We believe that transitional pre–B-ALL, which was recently reported to express μH chains on the surface but not with conventional L chains,60will likely fall into the pre–B- (pre-BCR+) ALL category of our classification when tested. As far as we analyzed, none of the leukemia samples expressed both SL chains and conventional L chains simultaneously (Tables 2 and 3), in accordance with the observation in normal human precursor B cells.25 61
In conclusion, the flow cytometric analysis with the combination of the MoAbs established in this study made it possible to categorize B-ALL into five subgroups: pro–B-I-ALL, pro–B-II-ALL, pre–B- (pre-BCR–) ALL, pre–B- (pre-BCR+) ALL, and B-ALL. This classification is based on the differential expression of SL chain, μH chain, pre-BCR, and L chain along normal B-cell development. Therefore, it would improve the precision of diagnosis of ALL by providing specific information regarding the lineage and stage of differentiation of the malignant cells. A large-scale survey of leukemia cases will be necessary to correlate each subtype of B-ALL defined by the novel classification with prognosis or response to treatment. Flow cytometric analysis with the MoAbs might also facilitate the detection of minimal residual leukemia. Furthermore, the MoAbs established in this study could be useful tools to study normal early B-cell development in human bone marrow as well.
ACKNOWLEDGMENT
We thank Mariko Yamagishi for excellent secretarial assistance.
Supported in part by grants-in-aid from the Ministry of Education, Science, Sports and Culture, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hajime Karasuyama, MD, PhD, Department of Immunology, The Tokyo Metropolitan Institute of Medical Science, 3-18-22 Honkomagome, Bunkyo-ku, Tokyo 113, Japan; e-mail:karasuyama@rinshoken.or.jp.

![Fig. 1. Specificity of the newly established MoAbs HSL11, HSL96, and HSL2 for human λ5, VpreB, and pre-BCR, respectively. A pre–B-cell line NALM6 (A), a pro–B-cell line RS4;11 (B), and X63.Ag8-653 cells transfected with either the λ5 or VpreB gene (C) were biosynthetically labeled with [35S]-methionine for 4 hours and lysed with 1% NP-40 lysis buffer. Detergent-soluble lysates were incubated with indicated MoAbs, and immunoprecipitates were analyzed by 13% SDS polyacrylamide gel electrophoresis under reducing conditions. The positions of μH, λ5, and VpreB are indicated by arrows, and a 16-kD band precipitated by HSL11 together with λ5 is indicated by an arrow head.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/11/10.1182_blood.v92.11.4317/5/m_blod42343001w.jpeg?Expires=1765885976&Signature=jROGNBwb9hA86Oa1IyEUUxCkOUAmFxz32GQIGivOoDU8BZPzCIo~Y77I55pxpPVAxf5mM9M3QD-Q0~2OBK4J5nz3D5GsZh4MdVxuhFvHMCyfPzzH95q35SeZ4wWBdlYwzBjL0w~AIzsG9l3oi-XelkybuRySPRWor5BzrqNIhgSnR6~HSMF30QyzbjMbwn8hFlCO8GDH7D3RKPunOWo5RN4RvLz4utLjG55goY5rpyeHfVfjDSaLsmJgq7hD2dNuQAj6e~MlyolVTdK92-c8t4RTen8SfBTx3Q2gFlNWv1MbYuqJUXeKQPCaTWuWuotFrWO5BhsgwJyo-iiYCYT-nQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal