Abstract
The differentiation of naive T-helper (Th) cells into cytokine-secreting effector Th cells requires exposure to multiple signals, including exogenous cytokines. Interleukin-4 (IL-4) plays a major role in this process by promoting the differentiation of IL-4–secreting Th2 cells. In Th2 cells, IL-4 gene expression is tightly controlled at the level of transcription by the coordinated binding of multiple transcription factors to regulatory elements in the proximal promoter region. Nuclear factor of activated T cell (NFAT) family members play a critical role in regulating IL-4 transcription and interact with up to five sequences (termed P0 through P4) in the IL-4 promoter. The molecular mechanisms by which IL-4 induces expression of the IL-4 gene are not known, although the IL-4–activated transcription factor signal transducer and activator of transcription 6 (Stat6) is required for this effect. We report here that Stat6 interacts with three binding sites in the human IL-4 promoter by electrophoretic mobility shift assays. These sites overlap the P1, P2, and P4 NFAT elements. To investigate the role of Stat6 in regulating IL-4 transcription, we used Stat6-deficient Jurkat T cells with different intact IL-4 promoter constructs in cotransfection assays. We show that, whereas a multimerized response element from the germline IgE promoter was highly induced by IL-4 in Stat6-expressing Jurkat cells, the intact human IL-4 promoter was repressed under similar conditions. We conclude that the function of Stat6 is highly dependent on promoter context and that this factor promotes IL-4 gene expression in an indirect manner.
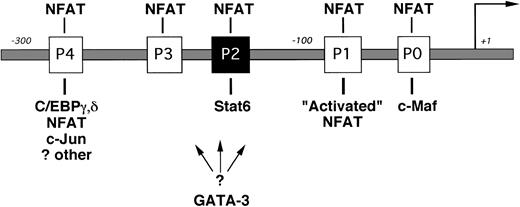
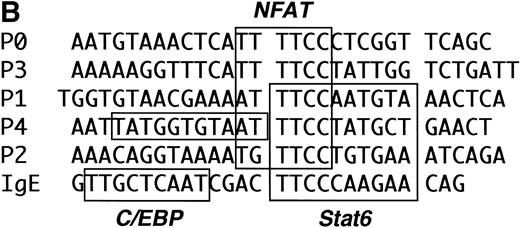
INTERLEUKIN-4 (IL-4) is a prototypic immunoregulatory cytokine.1 By virtue of its ability to induce IgE isotype switching in B cells, mast cell differentiation, and adhesion molecule expression, IL-4 plays a central role in many inflammatory responses.2-4 IL-4 is also the primary cytokine promoting the differentiation of naive T cells into cytokine-secreting T-helper 2 (Th2) cells.5Cytokine gene expression in Th2 cells is controlled primarily at the level of gene transcription,6 and dysregulation of this process is thought to contribute to the development of allergic diseases.7 Several transcription factors have recently been implicated in regulating Th2-restricted IL-4 gene expression (Fig 1).8-13 Nuclear factors of activated T cell (NFAT) are involved in this process by interacting with up to five sites in the IL-4 promoter (termed P0 through P4).14,15 The precise role of individual NFAT family members in regulating IL-4 transcription is currently unknown.16-18
The proximal IL-4 promoter (not drawn to scale). NFAT binding sites (the P elements) are indicated by open boxes, and the known Stat6 site is indicated by the solid box. Transcription factors implicated in Th2-specific IL-4 gene expression are indicated below their respective binding sites. A binding site for GATA-3 has not yet been reported.13 NFAT activity is higher in effector Th2 cells (“Activated” NFAT10) and is shown for simplicity binding only to the P1 NFAT site.
The proximal IL-4 promoter (not drawn to scale). NFAT binding sites (the P elements) are indicated by open boxes, and the known Stat6 site is indicated by the solid box. Transcription factors implicated in Th2-specific IL-4 gene expression are indicated below their respective binding sites. A binding site for GATA-3 has not yet been reported.13 NFAT activity is higher in effector Th2 cells (“Activated” NFAT10) and is shown for simplicity binding only to the P1 NFAT site.
Using T-cell lines derived from NFAT-reporter transgenic mice, Rincón and Flavell10 found that NFAT transcriptional activity is preferentially induced in Th2 cells but not in Th1 cells. Growing evidence suggests that Th2-specific NFAT cofactors may specifically enhance IL-4 transcription in these cells. For example, Ho et al9 found that the proto-oncogene c-maf was preferentially expressed in Th2 clones and that c-Maf acts synergistically with NFATp to induce IL-4 production in IL-4–negative cells. Additionally, Li-Weber et al12 detected a multiprotein complex containing C/EBP, NFAT, and AP-1 proteins forming on the P4 element using nuclear extracts from Th2 but not Th1 cells in electrophoretic mobility shift assays (EMSA).
Exogenous IL-4 is a critical stimulus for the effective differentiation of naive T cells into IL-4–secreting Th2 cells (for review, see O’Garra5). The mechanism by which IL-4 induces expression of the IL-4 gene is currently not known. IL-4 interacts with a multichain cell-surface receptor (IL-4R) that is expressed by several cell types, including B cells, T cells, and endothelial cells.4,19 IL-4 binding to the IL-4Rα chain induces different intracellular signals, including Jak-mediated phosphorylation of the transcription factor Stat6.20 Stat6 response elements share a consensus sequence5′TTCN3/4GAA3′ and are located in the promoter regions of many IL-4–responsive genes.21,22 The fundamental role of Stat6 in IL-4–driven responses was demonstrated by the phenotype of Stat6-deficient mice in which IgE synthesis and Th2 responses were abrogated.23-25Lederer et al11 discovered a Stat6 binding site in the mouse IL-4 promoter and found that Stat6 bound this sequence in EMSA using nuclear extracts from IL-4 induced Th2 cells but not Th1 cells. We and others identified a corresponding site in the human IL-4 promoter (−169TTCACAGGAA−160).26,27Because multimers of these elements were inducible by IL-4 when linked to heterologous promoters and transfected into Stat6-expressing B-cell lines,11 26 it seemed reasonable to conclude that Stat6 would directly enhance IL-4 transcription in T cells.
However, recent studies have suggested that activation of IL-4R signaling pathways is not required for IL-4 gene expression in effector T cells. For example, Huang et al28 found that IL-4 did not enhance transcription driven by a mouse IL-4 promoter construct in anti-CD3–activated Th2 cells, although the specific role of Stat6 in regulating the intact IL-4 promoter was not examined in that study. Additionally, Moriggl et al29 reported that a neutralizing anti–IL-4 monoclonal antibody (MoAb) actually enhanced anti-CD3–induced IL-4 production in committed Th2 cells. Using Stat6-deficient Jurkat T cells in cotransfection assays, we report here that, although cotransfected Stat6 strongly enhanced transcription driven by a multimerized response element, the human IL-4 promoter was significantly repressed under similar conditions. The repressive effects of Stat6 appeared to involve sequence-specific DNA binding, because a Stat6 DNA binding domain mutant failed to inhibit the IL-4 promoter. We describe two novel Stat6 binding sites within the proximal IL-4 promoter and show that Stat6 and NFAT bind competitively to overlapping nucleotides.
MATERIALS AND METHODS
Plasmid construction.
IL-4 promoter constructs were amplified from human genomic DNA using the polymerase chain reaction (PCR). A 25-bp 5′ primer annealing 265 bp upstream from the transcription start site (tss) according to Otsuka et al30 was used with a 25-bp 3′ primer ending at position +65. PCR products were sequenced using the dideoxy method and then ligated into the HindIII and Xba I sites of pCAT Basic (Promega, Madison, WI) to yield pCAT 265. The reporter C/EBP-N4 luc contains 4 copies of the composite C/EBP/Stat6 response element from the germline ε promoter fused to a thymidine kinase (TK) minimal promoter driving the firefly luciferase gene.21 The wild-type Stat6 expression vector (TPU 388) and the DNA-binding domain mutant vector (TPU522, in which the 3 amino acids VVI at positions 411 to 413 were replaced by EAA), both driven by the cytomegalovirus (CMV) promoter, have been described.21
Cell lines and transfections.
Jurkat T cells (a kind gift of Dr Jack Strominger, Harvard University, Cambridge, MA) were maintained in complete medium (RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum [FCS; Life Technologies, Gaithersburg, MD] and 50 μg/mL gentamycin [Life Technologies]). As previously reported, these cells constitutively express IL-4 mRNA.31 HepG2 cells were obtained from the ATCC (Rockville, MD) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS and 50 μg/mL gentamycin. In cotransfection experiments, 3 × 106 cells were transfected with 1 μg reporter, 2 μg expression or empty vector as a control, and 7 μL Superfect (Qiagen, Valencia, CA) in 5 mL complete medium and allowed to recover for 24 hours. Cells were then stimulated with combinations of the following agonists as indicated in the text for the last 18 hours: 1 μmol/L calcium ionophore (A23187; Calbiochem, San Diego, CA), 25 ng/mL phorbol-12-myristate-13-acetate (PMA; Calbiochem), and IL-4 (10 or 50 ng/mL; Peprotech, Rocky Hill, NJ). Cells were lysed by three freeze-thaw cycles and reporter gene expression was determined either by measuring CAT enzyme levels using a sensitive enzyme-linked immunosorbent assay (ELISA; Boehringer Mannheim, Indianapolis, IN) or by assaying for luciferase activity using standard techniques (Analytic Luminescence Laboratories, Sparks, MD). Cell extracts were normalized for protein content using the Bradford technique (Bio-Rad, Hercules, CA) before assays for reporter gene expression.
EMSA.
The following 30-bp oligonucleotides and their complements were synthesized (mutations in the P2 oligonucleotide are indicated as lowercase letters, and the Stat6 consensus sequence is underlined): 5′-ATTGCTGAAACCGAGGGAAAATGAGTTTACAT- TG-3′ (P0 −69 to −36); 5′-TGAGTTTACATTGGAAATTTTCGTTACACCAGATTG-3′ (P1 −92 to −60); 5′-TCTGATTTCACAGGAACATTTTACCTGTTT-3′ (P2 wt −175 to −146); 5′-gagac-TTTCACAGGAACATTTTACCTGTTT-3′ (P2 m1); 5′-TCTGAgggacCAGGAACATTTTACCTGTTT-3′ (P2 m2); 5′-TCTGATTTCAgctcgACATTTTACCTGTTT-3′ (P2 m3); 5′-TCTGATTTCACAGGActcggTTACCTGTTT-3′ (P2 m4); 5′-TCTGATTTCACAGGAACATTTgcgtt-TGTTT-3′ (P2 m5); 5′-TCTGATTTCACAGGAACATTTTTACCcaccg-3′ (P2 m6); 5′-AATCAGACCAATAGGAAAA- TGAAACCTTTTTAA-3′ (P3 −201 to −169); and 5′-AGTTTCAGCATAGGAAATTACACCATAATTTGC-3′ (P4 −248 to −216).
The Bcl-6 oligonucleotide (B6BS: 5′-GAAAATTCCTAGAAAGCATA-3′; donated by Dr Riccardo Dalla-Favera, Columbia University, New York, NY) has been described.32 Nuclear extracts were obtained from 5 × 106 Jurkat cells treated without or with IL-4 (20 ng/mL for 20 minutes; Peprotech) using the method of Schrieber et al.33 EMSAs were performed using 5 μg nuclear protein, 0.8 μg poly (dG-dC) (Amersham Pharmacia Biotech, Piscataway, NJ), and [γ32P] end-labeled probe in a final volume of 10 μL per reaction. Free probes and protein-DNA complexes were resolved by 5% polyacrylamide gel electrophoresis (PAGE) with 0.5× TBE. In antibody experiments, extracts were incubated at room temperature with 1 μL of the following antisera for 30 minutes after the addition of labeled probe: anti-Stat6 (Santa Cruz Biotech, Santa Cruz, CA), N70-6 (anti–Bcl-632; donated by Dr Riccardo Dalla-Favera), or isotype-matched control antisera.
Recombinant proteins.
A recombinant fragment of murine NFATp (including 298 amino acids of the DNA binding domain [DBD] that is highly conserved among different NFAT family members34) was expressed as a hexahistidine-tagged protein and extracted as described.35The NFATp expression vector was kindly donated by Dr Anjana Rao (Harvard University). Recombinant full-length, in vitro phosphorylated Stat6 has been described.21
Statistical analysis.
All transfections were performed in duplicate using cells of similar passage number. Average results of the indicated numbers of independent experiments were analyzed using the paired Student’s t-test (Statview II Software; SAS Institute, San Francisco, CA), and a P value less than .05 was considered to be statistically significant.
RESULTS
The IL-4R signaling pathway is intact in Jurkat T cells.
To investigate the role of Stat6 in regulating transcription of the intact IL-4 promoter, we studied Jurkat T cells transiently transfected with different IL-4 promoter reporter constructs. In preliminary experiments, we did not detect immunoreactive Stat6 in nuclear extracts from IL-4–activated Jurkat cells in EMSA, suggesting that the cells used in these experiments express negligible levels of this factor (not shown). Consistent with this result, IL-4 stimulation alone did not induce a full-length IL-4 promoter construct (not shown) or the reporter construct C/EBP-N4 luc (which contains 4 copies of the IgE C/EBP/Stat6 element driving the firefly luciferase gene; Fig 2A). However, as previously reported,21 C/EBP-N4 luc was strongly induced by IL-4 in Stat6-expressing HepG2 cells (Fig 2B).
The IL-4 receptor signaling pathway is intact in Jurkat T cells. (A) The luciferase reporter construct C/EBP-N4 luc (see text) was transiently transfected into Jurkat cells together with a control (Empty) or Stat6 expression vector. Cells were then stimulated without (□) or with (▪) IL-4 (50 ng/mL) for 18 hours before assays for reporter gene expression. In the absence of either cotransfected Stat6 or IL-4 stimulation, C/EBP-N4 luc is not active in Jurkat cells, but it is highly inducible by IL-4 in cells expressing Stat6. (B) Consistent with the known expression of Stat6 by Hep G2 cells,21C/EBP-N4 luc was induced by IL-4 in these cells, but its activity was further increased by overexpressing Stat6 (note the different scales). Results are the mean ± SEM of two (B) or three (A) independent experiments.
The IL-4 receptor signaling pathway is intact in Jurkat T cells. (A) The luciferase reporter construct C/EBP-N4 luc (see text) was transiently transfected into Jurkat cells together with a control (Empty) or Stat6 expression vector. Cells were then stimulated without (□) or with (▪) IL-4 (50 ng/mL) for 18 hours before assays for reporter gene expression. In the absence of either cotransfected Stat6 or IL-4 stimulation, C/EBP-N4 luc is not active in Jurkat cells, but it is highly inducible by IL-4 in cells expressing Stat6. (B) Consistent with the known expression of Stat6 by Hep G2 cells,21C/EBP-N4 luc was induced by IL-4 in these cells, but its activity was further increased by overexpressing Stat6 (note the different scales). Results are the mean ± SEM of two (B) or three (A) independent experiments.
To verify that the IL-4R signaling pathway was otherwise functional in Jurkat cells, we first analyzed the inducibility of C/EBP-N4 luc in cells cotransfected with a full-length Stat6 expression vector. Activity of C/EBP-N4 luc is strongly dependent on the coordinated binding of C/EBP and Stat6 to adjacent sites.21 Figure 2A shows that C/EBP-N4 luc was highly induced by IL-4 in Stat6-expressing Jurkat cells, but not in control cells cotransfected with the corresponding empty vector. This result confirms the known expression of C/EBP proteins by Jurkat cells.36 Consistent with previous findings,21 C/EBP-N4 luc was induced by IL-4 in HepG2 cells in the absence of exogenous Stat6 (Fig 2B). Thus, although Jurkat cells do not constitutively express functional Stat6 protein, cotransfected Stat6 is highly induced by IL-4 in these cells.
Stat6 represses transcription driven by the intact IL-4 promoter.
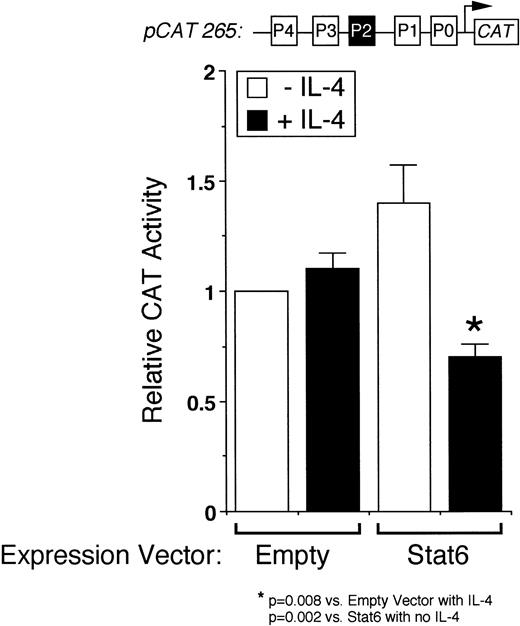
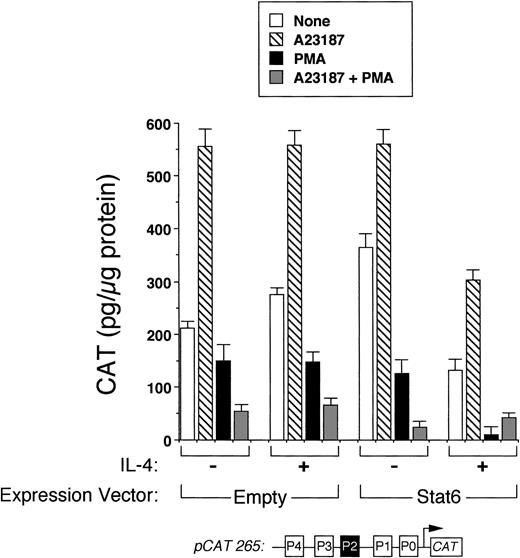
We next analyzed the ability of Stat6 to transactivate the intact IL-4 promoter. Figure 3 shows that a full-length human IL-4 promoter construct (pCAT 265) was consistently inhibited by IL-4 in Stat6-cotransfected Jurkat cells. To determine whether unstimulated Jurkat cells lacked an activation-induced Stat6 cofactor or whether Stat6 needed a further activation signal, which is necessary for IL-4 transcription, we next analyzed the effects of IL-4 in activated cells cotransfected with Stat6 and pCAT 265. As previously reported,37,38 a calcium-dependent signal alone was sufficient to maximally induce the IL-4 promoter (Fig 3). PMA downregulated IL-4 promoter activity, which we previously found was due to the displacement of NFATp from the human P1 sequence by induced nuclear NF-κB heterodimers.37 Interestingly, IL-4 consistently inhibited calcium-induced promoter activity in Stat6-cotransfected cells and almost completely repressed the promoter in combination with PMA (Fig 4). Thus, even in conjunction with activation of intracellular calcium and PKC-signaling pathways, Stat6 inhibited transcription driven by the full-length IL-4 promoter.
Stat6 inhibits transcription driven by the intact IL-4 promoter. pCAT 265, which contains all of the known NFAT P elements including the P2 Stat6 site (solid box), was transfected into unstimulated Jurkat cells together with a control (Empty) or a Stat6 expression vector, and the cells were incubated without (□) or with (▪) IL-4 (50 ng/mL) for 18 hours before assays for reporter gene expression by ELISA. Results are expressed relative to the constitutive activity of pCAT 265 without IL-4 and are the mean ± SEM of four independent experiments. IL-4 significantly downregulated promoter activity only in Stat6-cotransfected cells.
Stat6 inhibits transcription driven by the intact IL-4 promoter. pCAT 265, which contains all of the known NFAT P elements including the P2 Stat6 site (solid box), was transfected into unstimulated Jurkat cells together with a control (Empty) or a Stat6 expression vector, and the cells were incubated without (□) or with (▪) IL-4 (50 ng/mL) for 18 hours before assays for reporter gene expression by ELISA. Results are expressed relative to the constitutive activity of pCAT 265 without IL-4 and are the mean ± SEM of four independent experiments. IL-4 significantly downregulated promoter activity only in Stat6-cotransfected cells.
Stat6 does not synergize with other signals to activate the IL-4 promoter. Methods were similar to those described in Fig 3, except that cells were also stimulated with calcium ionophore (▧), PMA (▪), both (░), or no agonists (□) with or without IL-4 as indicated. In the presence of IL-4, reporter activity was consistently decreased in Stat6-cotransfected cells for each condition examined. Note that the combination of PMA and IL-4 almost completely repressed the promoter. Results are from one experiment performed in duplicate and are representative of three.
Stat6 does not synergize with other signals to activate the IL-4 promoter. Methods were similar to those described in Fig 3, except that cells were also stimulated with calcium ionophore (▧), PMA (▪), both (░), or no agonists (□) with or without IL-4 as indicated. In the presence of IL-4, reporter activity was consistently decreased in Stat6-cotransfected cells for each condition examined. Note that the combination of PMA and IL-4 almost completely repressed the promoter. Results are from one experiment performed in duplicate and are representative of three.
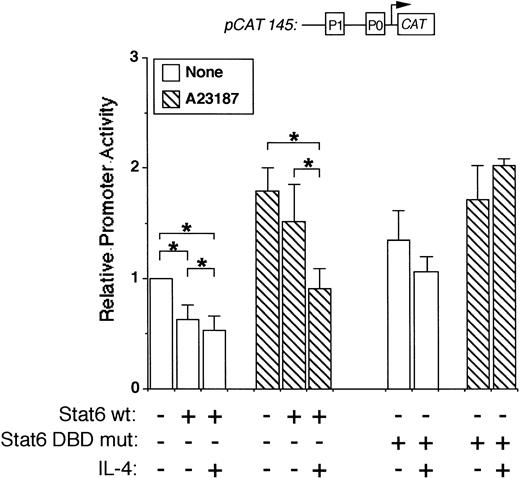
To map the IL-4 promoter element(s) necessary for Stat6-mediated transcriptional repression, we used a smaller promoter construct in additional cotransfection experiments. This construct (pCAT 145) contains 145 bp of the human promoter, including the P1 and P0 NFAT elements, but lacks the previously described Stat6 binding site (−169TTCACAGGAA−160). Interestingly, pCAT145 activity was significantly inhibited by IL-4 in both resting and stimulated Stat6 cotransfected cells (Fig 5). Importantly, an expression vector encoding a Stat6 DNA binding domain mutant (TPU522, see Materials and Methods) did not inhibit pCAT145 activity in either resting or activated cells (Fig 5, right side). These results suggested that Stat6-induced repression involved binding sites located downstream of the known Stat6 sequence and that the DNA binding ability of Stat6 was required for this effect.
Stat6 inhibits a minimal IL-4 promoter construct. Jurkat cells were cotransfected with a minimal promoter construct lacking the known P2 Stat6 element (pCAT 145) with or without a wild-type (wt) or DNA-binding domain mutant (DBD mut) Stat6 expression vector as indicated. Cells were stimulated for 18 hours with calcium ionophore (▧) with or without IL-4 as indicated, followed by cell lysis and analysis of reporter gene expression by ELISA. Results are expressed relative to CAT production in unstimulated cells and are the mean ± SEM of four (DBD mut) or seven (wt) independent experiments. *P< .05.
Stat6 inhibits a minimal IL-4 promoter construct. Jurkat cells were cotransfected with a minimal promoter construct lacking the known P2 Stat6 element (pCAT 145) with or without a wild-type (wt) or DNA-binding domain mutant (DBD mut) Stat6 expression vector as indicated. Cells were stimulated for 18 hours with calcium ionophore (▧) with or without IL-4 as indicated, followed by cell lysis and analysis of reporter gene expression by ELISA. Results are expressed relative to CAT production in unstimulated cells and are the mean ± SEM of four (DBD mut) or seven (wt) independent experiments. *P< .05.
The human IL-4 promoter contains multiple overlapping Stat6 and NFAT binding sites.
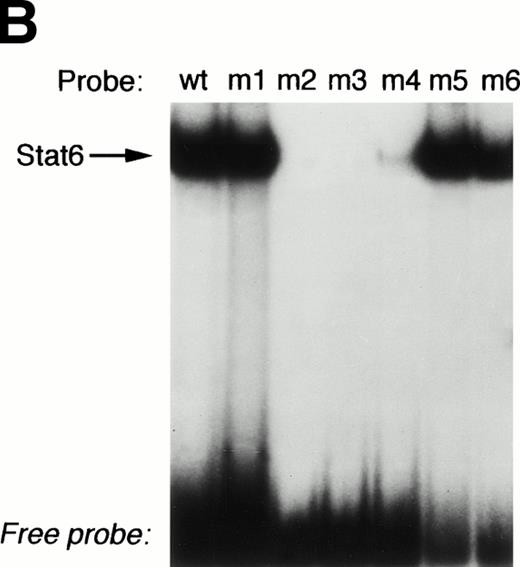
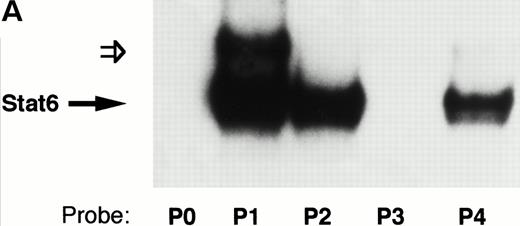
The previously described Stat6 binding site is contained within the P2 NFAT element (Fig 6). We and others have shown that this element can support the cooperative binding of NFAT and AP-1 proteins.39 40 As shown in Fig 6, the 3′-half of the Stat6 site (5′GAA3′) overlaps the 5′-end of the NFAT site (5′GGAA3′). In view of our functional data showing repression of a construct lacking the P2 element (Fig 5), we speculated that Stat6 could interact with additional P elements from the IL-4 promoter. To test this hypothesis, we analyzed the ability of recombinant Stat6 to bind similar length oligonucleotides including the five known IL-4 P elements by EMSA. Figure 7 shows that recombinant Stat6 bound 30-bp oligonucleotides containing the P1, P2, and P4 NFAT sites, but not the P0 or P3 sites.
The P2 element contains overlapping binding sites for Stat6 and NFATp. (A) Alignment of the human IL-4 P2 NFAT element with canonical binding sites for NFAT (overline) and Stat6 (underline). Note that the 3′-end of the Stat6 sequence overlaps the 5′-end of the NFAT site. (B) The ability of recombinant Stat6 to interact with wild-type (wt) and mutated probes (see Materials and Methods) was determined by EMSA. Stat6 no longer bound the m2-m4 oligonucleotides, confirming the overlapping nature of the Stat6 and NFAT binding sites.
The P2 element contains overlapping binding sites for Stat6 and NFATp. (A) Alignment of the human IL-4 P2 NFAT element with canonical binding sites for NFAT (overline) and Stat6 (underline). Note that the 3′-end of the Stat6 sequence overlaps the 5′-end of the NFAT site. (B) The ability of recombinant Stat6 to interact with wild-type (wt) and mutated probes (see Materials and Methods) was determined by EMSA. Stat6 no longer bound the m2-m4 oligonucleotides, confirming the overlapping nature of the Stat6 and NFAT binding sites.
The IL-4 promoter contains multiple Stat6 binding sites. (A) The ability of recombinant Stat6 to bind oligonucleotides containing the human P elements was determined by EMSA. Stat6 bound the P1, P2, and P4 probes (solid arrow). An additional slowly migrating complex (open arrow) formed on the P1 oligonucleotide (and occasionally on the P2 probe; see Fig 8). The relative migration of the free probes, which were of similar length (see Materials and Methods) and radiolabeled with similar specific activity, is not shown in this figure. (B) Alignment of the P elements that supported Stat6 binding with the composite C/EBP-Stat6 site from the germline IgE promoter. The P2 oligonucleotide is shown in opposite orientation than in Fig 2. Binding sites for NFAT, Stat6, and C/EBP are indicated by boxes. C/EBP proteins may interact with the P0, P1, and P4 NFAT elements, although the precise nucleotide binding sites have been reported only for the P4 sequence.36
The IL-4 promoter contains multiple Stat6 binding sites. (A) The ability of recombinant Stat6 to bind oligonucleotides containing the human P elements was determined by EMSA. Stat6 bound the P1, P2, and P4 probes (solid arrow). An additional slowly migrating complex (open arrow) formed on the P1 oligonucleotide (and occasionally on the P2 probe; see Fig 8). The relative migration of the free probes, which were of similar length (see Materials and Methods) and radiolabeled with similar specific activity, is not shown in this figure. (B) Alignment of the P elements that supported Stat6 binding with the composite C/EBP-Stat6 site from the germline IgE promoter. The P2 oligonucleotide is shown in opposite orientation than in Fig 2. Binding sites for NFAT, Stat6, and C/EBP are indicated by boxes. C/EBP proteins may interact with the P0, P1, and P4 NFAT elements, although the precise nucleotide binding sites have been reported only for the P4 sequence.36
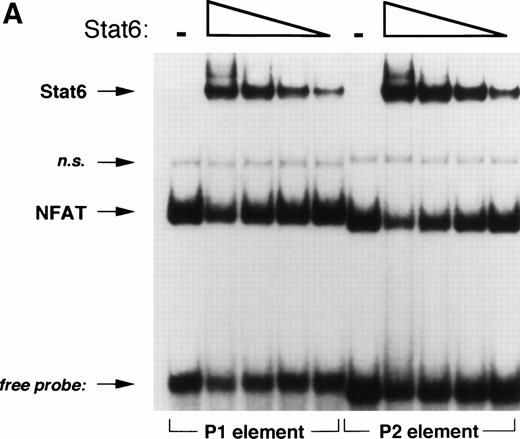
Based on sequence homology and mutational analysis (Fig 6), we predicted that Stat6 and NFAT would bind competitively to overlapping sites in the IL-4 promoter. To test this hypothesis and to exclude any combinatorial interactions of these factors on the IL-4 promoter, we analyzed the effect of Stat6 on the ability of the NFATp DBD (see Materials and Methods) to interact with oligonucleotides containing the P1 and P2 elements in EMSA. As expected, NFATp readily bound both probes (Fig 8). Interestingly, increasing amounts of Stat6 displaced NFATp from its cognate sites on both oligonucleotides. Displacement of NFAT from the P1 element by Stat6 may provide an explanation for the repressive effects of Stat6 on pCAT 145.
Stat6 binds competitively with NFAT to the P1 and P2 elements. (A) The ability of a recombinant fragment of NFATp (see Materials and Methods) to bind oligonucleotide probes containing the P1 and P2 elements in the presence of increasing concentrations of recombinant Stat6 was determined by EMSA. The relative mobilities of each factor are indicated. Serial twofold dilutions of Stat6 were examined against a constant amount of NFATp. n.s., nonspecific. (B) The relative intensities of observed bands were analyzed by densitometry and expressed relative to intensity of the NFAT complex for each oligonucleotide in the absence of Stat6 (which was defined as 1).
Stat6 binds competitively with NFAT to the P1 and P2 elements. (A) The ability of a recombinant fragment of NFATp (see Materials and Methods) to bind oligonucleotide probes containing the P1 and P2 elements in the presence of increasing concentrations of recombinant Stat6 was determined by EMSA. The relative mobilities of each factor are indicated. Serial twofold dilutions of Stat6 were examined against a constant amount of NFATp. n.s., nonspecific. (B) The relative intensities of observed bands were analyzed by densitometry and expressed relative to intensity of the NFAT complex for each oligonucleotide in the absence of Stat6 (which was defined as 1).
DISCUSSION
The molecular basis by which IL-4 induces the production of Th2 cytokines is currently not known. Experiments with Stat6-deficient mice have conclusively demonstrated a requirement for this IL-4–inducible transcription factor during the differentiation of naive T cells into Th2 cells.23-25 However, recent studies have found that activation of the IL-4R-signaling pathway is not required for the induction of IL-4 gene expression in committed Th2 cells.28,29,41 Additionally, Stat6 can repress IL-4 gene expression in Th1 cells by binding a cell-type specific silencer in the 3′ untranslated region (UTR).42 Thus, the precise role of Stat6 in regulating IL-4 expression is currently not clear.
Th2-restricted IL-4 gene expression is thought to be controlled at the level of transcription by the coordinated interactions of transcription factors binding to a proximal promoter region. Regulatory elements within the promoter have been shown not only to bind nuclear factors unique to Th2 cells, but also to be preferentially induced in Th2 cells.8-13 The observation that Stat6 can interact with a consensus sequence from both the mouse11 and human26 IL-4 promoters suggested that this factor might provide a direct link between IL-4R activation and IL-4 gene expression. However, a conclusion of our studies is that Stat6 might only facilitate IL-4 gene expression in T cells in an indirect fashion.
This report, showing for the first time the specific effect of Stat6 on the intact IL-4 promoter, contains several novel observations. First, we found that, although IL-4 receptor-mediated signals are faithfully transduced in Jurkat T cells (Fig 2), Stat6 was unable to transactivate the proximal human IL-4 promoter in these cells (Figs 3 through 5). Together with previous studies showing that IL-4 can induce multimers of the P2 Stat6 site linked to heterologous promoters,11,26,28 our findings suggest that the transactivation potential of Stat6 is dependent on promoter context. This conclusion is in keeping with recent analyses of Stat6 and the germline IgE43 and β-casein genes44 and with the study of Huang et al,28 who found that Stat6 differentially regulated multimers of the P2 Stat6 element in M12 B cells depending on the minimal promoter construct used.
Second, we found that transcription driven by two deletion constructs of the human IL-4 promoter was consistently inhibited by IL-4 in Stat6-expressing Jurkat T cells. Interestingly, recent studies suggest that IL-4 can inhibit IL-4 gene expression in a negative feedback fashion. For example, Moriggl et al29 found that committed Th2 cells secreted more IL-4 when restimulated with anti-CD3 in the presence of a neutralizing anti–IL-4 MoAb. Additionally, IL-4 seemed to inhibit anti-CD3–induced transcription driven by the full-length mouse IL-4 promoter in a Th2 clone.28 Taken together, these results suggest that activated Stat6 might downregulate IL-4 expression in effector Th cells and emphasize the need to distinguish between Th2 differentiation and IL-4 gene expression (see below).
Third, we have identified two novel Stat6 binding sites (within the P1 and P4 elements) in the IL-4 promoter. Although these sites do not contain the consensus Stat6 binding site (5′-TTCN4GAA-3′) defined by binding site selection assays, a significant fraction (10/42) of sequences selected by Stat6 in these assays contained single nucleotide substitutions within the dyad half-sites.45 Thus the ability of Stat6 to bind the P1 oligonucleotide (5′-TTCN4GTA-3′) is not surprising. The reason why Stat6 bound the P4 element but not the P0 element is less apparent, because they contain similar noncanonical dyad half sites, although they do differ in the spacer region.
Fourth, the demonstration that Stat6 and NFAT can bind competitively to the IL-4 promoter provides evidence of a previously unreported interaction between these two factors. Given the fundamental role of NFAT family members in regulating IL-4 gene expression, this observation provides a plausible explanation for the observed inhibitory effects of Stat6 on transcription driven by the IL-4 promoter. This would be similar to the recently described antagonism of NF-κB by Stat6 in the E-selectin promoter.46 Because the P1 element in particular plays a major role in activating IL-4 transcription,8 47 competition by Stat6 for NFAT binding to this element might have particularly repressive effects. Alternatively, we cannot exclude the possibility that Stat6 might actively repress the basal transcription complex or that it might also bind to and titrate away from the promoter a factor necessary for maximal IL-4 transcription.
Stat6 might require a coactivator not expressed by Jurkat cells to maximally transactivate the IL-4 promoter. For example, Stat6 cooperates with both C/EBP21 and NF-κB/Rel proteins48 to promote germline IgE transcription. The fact that C/EBP-N4 luc is highly induced (Fig 2) argues that the inability of Stat6 to activate IL-4 transcription in our experiments is not due to the lack of C/EBP proteins and confirms the known expression of C/EBP in Jurkat cells.36Additionally, we have shown that PMA-activated Jurkat cells contain abundant nuclear NF-κB.37 We cannot formally exclude the requirement for as yet unidentified Stat6 coactivators necessary for IL-4 transcription, although the studies by Huang et al28argue against the existence of such factors in a differentiated Th2 clone. Another possibility is that Stat6 might require additional posttranslational modification to achieve full transcriptional competency in Jurkat cells. However, the observations that (1) PMA enhanced the repressive effects of Stat6 (Fig 4) and (2) C/EBP-N4 luc was inducible by IL-4 in Stat6-cotransfected Jurkat cells (Fig 2) argue against this explanation.
Finally, it is possible that other factors inhibit the transactivation potential of Stat6 on the IL-4 promoter. For example, it has recently been suggested that Bcl-6, a transcriptional repressor deregulated in many lymphomas,32,49 can specifically inhibit the ability of Stat6 to transactivate gene expression. In fact, Bcl-6–deficient mice were found to have enhanced IL-4 production and Th2 responses.50 To exclude the possibility that Bcl-6 was inhibiting transactivation of the IL-4 promoter by Stat6 in Jurkat cells, we assayed for Bcl-6 binding to both the P2 element and a consensus Bcl-6 site using Jurkat nuclear extracts in EMSA. Using conditions known to support Bcl-6 binding,32 we did not detect Bcl-6 using two specific antisera (not shown). Additionally, IL-4 gene expression was not inhibited in Jurkat cells expressing inducible Bcl-6 protein (Dr Riccardo Dalla-Favera, personal communication, Spring 1998). Thus, we conclude that the inability of Stat6 to activate IL-4 transcription in Jurkat T cells is not due to the presence of the specific Stat6 antagonist Bcl-6.
The IL-4R transduces other signals in addition to the Jak-mediated tyrosine phosphorylation of Stat6. For example, the I4R motif of the IL-4Rα subunit leads to the IRS-1–dependent phosphorylation of the nonhistone chromosomal protein HMGI/Y.51 Phosphorylation of HMGI/Y in B cells inhibits its ability to bind DNA and results in the derepression of germline IgE transcription.43,52 HMGI/Y has recently been shown to downregulate the IL-4 promoter by competing with NFAT for binding to the P1 element.53 Thus, activation of this IL-4R signaling pathway would be expected to de-repress the IL-4 promoter. However, our observation that Stat6 also displaces NFAT from the P1 element (Fig 8) might explain why this did not occur in our experiments.
It is worth emphasizing that IL-4–dependent differentiation of naive Th cells into effector Th2 cells involves prolonged exposure to multiple concomitant signals emanating from the T-cell receptor, costimulatory molecules, and possibly other APC-derived cytokines.5,54-56 The precise role of Stat6 in this process requires further study. We have shown that Stat6 does not directly transactivate the IL-4 promoter, which it actually represses in cells transcribing the IL-4 gene. Together with recent analyses of committed Th cells,28 29 our results suggest that Stat6 may rather facilitate the acquisition of an IL-4–producing phenotype in differentiating Th cells in an indirect fashion. In this regard, investigating the regulation of other lineage-specific Th2 transcription factors by Stat6 may be helpful.
ACKNOWLEDGMENT
The authors thank Dr Riccardo Dalla-Favera for assistance with the Bcl-6 experiments, Dr Anjana Rao for the NFATp expression vector, and Dr Marcia Wills-Karp (Johns Hopkins University, Baltimore, MD) for helpful suggestions.
Supported by Grant No. AI01152 from the National Institutes of Health and Research Grant No. 056-N from the American Lung Association.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Steve N. Georas, MD, Room 4B.41, The Johns Hopkins Asthma & Allergy Center, 5501 Hopkins Bayview Circle, Baltimore, MD 21224; e-mail: sgeoras@welchlink.welch.jhu.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal