Abstract
We have previously shown that infection of CD4+ T lymphocytes with the T-lymphotropic human herpesvirus 7 (HHV-7) downregulates surface CD4, which represents the high-affinity receptor for HHV-7. In this study, we report that HHV-7 infection also causes a progressive loss of the surface CXC-chemokine receptor 4 (CXCR4) in CD4+ T cells, accompanied by a reduced intracellular Ca2+ flux and chemotaxis in response to stromal cell-derived factor-1 (SDF-1), the specific CXCR4 ligand. Moreover, CXCR4 is downregulated from the surface of HHV-7–infected T cells independently of CD4. Because intracellular CXCR4 antigen and mRNA levels are unaffected in productively HHV-7–infected cells, the downregulation of CXCR4 apparently does not involve a transcritional block. Since CXCR4 functions in association with CD4 to permit entry of several human immunodeficiency virus (HIV) isolates, the potential of HHV-7 to persistently downregulate the surface expression of CXCR4 may provide novel strategies for limiting HIV infection.
THE 7-TRANSMEMBRANE G-protein–coupled CXC-chemokine receptor 4 (CXCR4) is known to be expressed in all hematopoietic cells, starting from immature CD34+hematopoietic progenitors and including CD4+ and CD8+ T-lymphocyte subsets.1-8 It has been shown that CXCR4 plays an essential role as a coreceptor for the T-cell line-adapted isolates of human immunodeficiency virus (HIV), which are highly cytopathic and typically emerge in the late stages of HIV disease.9,10 Because in vitro pretreatment with the high-affinity ligand for CXCR4, stromal cell-derived factor-1α (SDF-1α), inhibits the entry of these viruses into CD4+ T cells,11-13 the issue of CXCR4 modulation and subcellular trafficking has recently raised considerable attention.
CXCR4 undergoes efficient but transient endocytosis after interaction with SDF-1α or treatment with phorbol esters.14-16 In CD4+ T lymphocytes, it appears to respond to similar modulatory signals as CD4,3,16 and it has been proposed that comodulation of CXCR4 and CD4 may play a role in regulating the activity of CD4+ T cells. However, the different constitutive endocytosis rates of CD4 and CXCR4, expressed on the SupT1 CD4+ T-lymphoblastoid cell line, and the selective SDF-1α–induced downmodulation of CXCR4 but not of CD4,14rather suggest that these two proteins do not normally form stable associations. A complex of CD4 and CXCR4 is likely to be induced by the presence of HIV envelope gp120 protein.17 18
MATERIALS AND METHODS
Cells.
The CD4+ human T-cell line SupT1 (AIDS Research and Reference Reagent Program, Manassas, VA) and a CD4−derivative of SupT1 called BC7 (generously provided by Dr J.A. Hoxie, University of Pennsylvania, Philadelphia, PA)23 were routinely cultured in RPMI 1640 (GIBCO BRL, Gaithersburg, MD) containing 10% fetal calf serum (GIBCO BRL). Primary CD8-depleted and/or -enriched CD4+ T cells were derived from the peripheral blood of healthy blood donors, by negative immunomagnetic selection, and phytohemagglutinin (PHA)/interleukin-2 (IL-2)–activated as previously described.24
Viral infections.
The HHV-7 isolates and the preparation procedures of viral stocks have been previously described.20 25 Briefly, HHV-7 infections of both SupT1 and primary cells were performed by incubation with 0.45-μm–filtered infectious supernatants obtained from HHV-7–infected SupT1 cells. Supernatants from uninfected SupT1 cells were used for mock infections. For inhibition assays, target cells were preincubated for 30 minutes at room temperature with SDF-1α (2.5 μg/mL), 12G5 anti-CXCR4 monoclonal antibody (MoAb; 25 μg/mL), or control mouse IgG2a (25 μg/mL) (all from Pharmingen, San Diego, CA). After the addition of HHV-7 inocula (MOI = 0.01), cells were maintained in the presence of SDF-1α or of the indicated antibodies for an additional 40 hours, at which time cells were harvested and used for RNA extraction (see below).
The occurrence of a productive HHV-7 infection was monitored by (1) indirect immunofluorescence staining on acetone-fixed cells by using a specific HHV-7 MoAb (5E1 MoAb; generously provided by Prof E. Campadelli-Fiume, University of Bologna, Bologna, Italy),26,27 as previously described24; and (2) HHV-7 reverse transcriptase-polymerase chain reaction (RT-PCR), by using H7 and H8 primers28 that amplify the HHV-7 U10 ORF.
Infection of SupT1 cells with HIV-1(IIIB) was performed as described20 by using viral inocula derived from HIV-1(IIIB)–infected H9 T-cells. HIV-1 infection was monitored by serial determinations of HIV-1 p24 antigen released into the culture supernatant (p24 ELISA Kit; DuPont-Merck Pharmaceutical Co, Wilmington, DE) and visual inspection for syncytia formation.
Flow cytometry analysis.
Aliquots of 1.5 × 106 cells were subjected to single- or multiple-label staining to examine the presence of surface and/or intracellular Ags, as described previously.22 CXCR4 expression was analyzed either by indirect staining using 12G5 anti-CXCR4 MoAb (Pharmingen) followed by fluorescein isothiocyanate (FITC)-conjugated goat antimouse IgG or by using the phycoerythrin (PE)-conjugated anti-CXCR4 MoAb (Pharmingen). The expression of CD4 was analyzed by using either FITC-conjugated (Becton Dickinson, San Jose, CA) or PE-conjugated anti-CD4 MoAb (Pharmingen). CD3 expression was analyzed by using the Cy5-PE-conjugated MoAb (Becton Dickinson). Briefly, surface staining was performed in 200 μL of phosphate-buffered saline containing 1% fetal calf serum at 4°C for 30 minutes.
For the specific detection of intracellular CXCR4, SupT1 cells were pretreated with 20 μL of unconjugated anti-CXCR4 MoAb for 1 hour at 4°C to saturate surface CXCR4 before the fixing procedure. On the other hand, for the simultaneous detection of surface CXCR4 and intracellular CXCR4, SupT1 cells were first incubated with the anti-CXCR4 MoAb followed by incubation with FITC-goat antimouse IgG. Cells were then fixed in 2% paraformaldehyde and permeabilized with 0.2% Triton X100, as described previously.22 Intracellular staining was then performed by using the PE-conjugated anti-CXCR4 MoAb. Nonspecific fluorescence was assessed by using irrelevant isotype-matched MoAbs (Becton Dickinson and/or Pharmingen). After staining procedures, samples were analyzed using a FACSCalibur flow cytometer (Becton Dickinson).
Calcium measurement.
Agonist-dependent increase in cytoplasmic Ca2+ was determined as described.29 30 Briefly, aliquots of 106 cells were loaded with Fluo-3 reconstituted with Pluronic F-127 (Molecular Probes, Eugene, OR) for 20 minutes at 37°C. After incubation, the samples were washed once with RPMI and resuspended in 1 mL of the same medium. Data were acquired with a FACSCalibur, with excitation at 488 nm. Cells were gated based on forward and side scatter properties. After 20 seconds, SDF-1α (Pharmingen) or ionomycin (Sigma Chemicals, St Louis, MO) was added by using a magnetic device, and calcium mobilization was determined by a two-parameter density-plot measuring linear emission at 550 nm in the FL-1 window for the gated cell population over time.
Chemotaxis assay.
Cell migration assays were performed as described.11Briefly, uninfected and HHV-7–infected cells were resuspended in RPMI 1640 medium plus bovine serum albumin (1 mg/mL). The cell density was adjusted to 5 × 106 cells/mL and 100 μL of the cell suspension was added to the top chamber of a 24-Transwell apparatus (5-μm pore size; Costar, Cambridge, MA). RPMI 1640/bovine serum albumin containing various concentrations of the recombinant SDF-1α was added to the bottom chamber of the Transwell. After 3 hours of incubation at 37°C, the cell numbers in the bottom chamber were counted in a FACSCalibur, and percentages of the transmigration were determined for each concentration of SDF-1α, after subtraction of the background (absence of SDF-1α) transmigration.
RNA analysis.
RNA purification from uninfected and HHV-7–infected cells was performed using the SV total RNA isolation system (Promega, Madison, WI) following the manufacturer’s protocol. Synthesis of first strand cDNA and amplification were performed using the Access RT-PCR system (Promega) following the manufacturer’s protocol. As a control for DNA contamination, equal amounts of RNA were used for PCR without template retrotranscription. The resulting PCR products were separated on 2% Seakem GTG agarose gel (FMC BioProducts, Rockland, ME). Primers and reaction conditions for the amplification of HHV-7 U10 ORF28 (PCR product, 186 bp), CXCR430 (PCR product, 430 bp), and β-actin (PCR product, 661 bp; Stratagene, San Diego, CA) were as described previously.
RESULTS
Downregulation of surface CXCR4 expression in CD4+ T cells after HHV-7 infection.
The levels of surface CXCR4 in the SupT1 lymphoblastoid CD4+ T-cell line (Fig 1A and B) and in primary T lymphocytes (Fig 1C) were evaluated by flow cytometry at various time points after HHV-7–infection, using the 12G5 anti-CXCR4 MoAb, either alone (Fig 1A) or in combination with anti-CD4 MoAb (Fig 1B and C). 12G5 binding was progressively reduced in HHV-7–infected SupT1 cells compared with uninfected controls (Fig 1A). Moreover, double-staining performed with anti-CXCR4 plus anti-CD4 MoAb showed the presence of four distinct cell populations (CXCR4+/CD4+, CXCR4+/CD4−, CXCR4−/CD4+, and CXCR4−/CD4−; Fig 1B), clearly indicating that the two antigens were downmodulated by HHV-7 independently. On the other hand, CD11 and CD45LCA surface markers were unaffected by HHV-7 infection (data not shown). A marked decrease of both CXCR4 and CD4 surface antigens, but not of CD3, was also observed in CD8-depleted primary T lymphocytes after infection with HHV-7 (Fig1C).
Effect of HHV-7 and HIV-1 infection on surface CXCR4 expression in CD4+ T cells evaluated by flow cytometry. In (A) and (B), progressive downregulation of surface CXCR4 expression in HHV-7–infected SupT1 cells. In (B), surface CXCR4 was analyzed in combination with surface CD4. In (C), surface CXCR4 was analyzed either alone (top panels) or in combination with surface CD3 (middle panels) and surface CD4 (bottom panels) in CD8+-depleted primary cells, either uninfected or infected with HHV-7 (8 days PI). In (D), surface CXCR4 was analyzed in combination with surface CD4 in SupT1 cells, either uninfected or infected with HIV-1 (12 days PI). Data shown in (A) through (D) are representative of at least three separate experiments. Horizontal axis (logarithmic scale), relative surface CXCR4 expression detected by PE fluorescence intensity (in [A] through [D]). Vertical axis, relative cell number, isotype-matched control antibody staining, CD4 expression detected by FITC fluorescence intensity or CD3 expression detected by Cy5-PE fluorescence intensity, as indicated. Percentages of cells in the respective quadrants are indicated. Representative negative controls, constituted by cells stained with irrelevant isotype-matched MoAb, are shown in the top panels of (A) and (B).
Effect of HHV-7 and HIV-1 infection on surface CXCR4 expression in CD4+ T cells evaluated by flow cytometry. In (A) and (B), progressive downregulation of surface CXCR4 expression in HHV-7–infected SupT1 cells. In (B), surface CXCR4 was analyzed in combination with surface CD4. In (C), surface CXCR4 was analyzed either alone (top panels) or in combination with surface CD3 (middle panels) and surface CD4 (bottom panels) in CD8+-depleted primary cells, either uninfected or infected with HHV-7 (8 days PI). In (D), surface CXCR4 was analyzed in combination with surface CD4 in SupT1 cells, either uninfected or infected with HIV-1 (12 days PI). Data shown in (A) through (D) are representative of at least three separate experiments. Horizontal axis (logarithmic scale), relative surface CXCR4 expression detected by PE fluorescence intensity (in [A] through [D]). Vertical axis, relative cell number, isotype-matched control antibody staining, CD4 expression detected by FITC fluorescence intensity or CD3 expression detected by Cy5-PE fluorescence intensity, as indicated. Percentages of cells in the respective quadrants are indicated. Representative negative controls, constituted by cells stained with irrelevant isotype-matched MoAb, are shown in the top panels of (A) and (B).
In contrast, infection of SupT1 cells with HIV-1(strain IIIB) resulted in drastic downregulation of CD4, comparable to that observed in HHV-7–infected cells, but with no significant reduction in 12G5 reactivity (Fig 1D).
Because the experiments described above could not distinguish between loss or surface CXCR4 and blocking of the 12G5 epitope, we next evaluated the intracellular Ca2+ flux and chemotaxis in response to SDF-1α. To avoid the possibility that dead cells and cell debris could interfere with these biological assays in both uninfected and HHV-7–infected cultures, analysis was performed on viable cells gated on the basis of forward and side scatter properties. At day 4 postinfection (PI), HHV-7–infected SupT1 cells showed a reduction in the Ca2+ flux mediated by SDF-1α with respect to uninfected cells (Fig 2A). On the other hand, both uninfected and HHV-7–infected SupT1 cells responded in a similar fashion to a nonspecific agonist, such as the Ca2+ionophore ionomycin (Fig 2A), thus confirming that the reduced Ca2+ flux observed in HHV-7–infected cells in response to SDF-1 was specific.
Mobilization of calcium (A) and chemotaxis (B) of uninfected and HHV-7–infected SupT1 cells in response to SDF-1. HHV-7–infected cultures were analyzed at days 4 (A) or 6 (B) PI. (A) shows transient intracellular calcium flux in response to SDF-1 (40 ng/mL; top panels) or to ionomycin (100 ng/mL; bottom panels). Data are representative of three independent experiments. In (B), chemotaxis was analyzed by using Transwells after induction with the indicated concentration of SDF-1. Data are expressed as the percentage of input cells transmigrated. The mean ± SD of three independent experiments is shown.
Mobilization of calcium (A) and chemotaxis (B) of uninfected and HHV-7–infected SupT1 cells in response to SDF-1. HHV-7–infected cultures were analyzed at days 4 (A) or 6 (B) PI. (A) shows transient intracellular calcium flux in response to SDF-1 (40 ng/mL; top panels) or to ionomycin (100 ng/mL; bottom panels). Data are representative of three independent experiments. In (B), chemotaxis was analyzed by using Transwells after induction with the indicated concentration of SDF-1. Data are expressed as the percentage of input cells transmigrated. The mean ± SD of three independent experiments is shown.
In a chemotactic assay, HHV-7–infected SupT1 cultures showed a cell migration rate significantly (P < .01) lower than those uninfected (Fig 2B).
Unaffected levels of intracellular CXCR4 protein and mRNA in HHV-7–infected cultures.
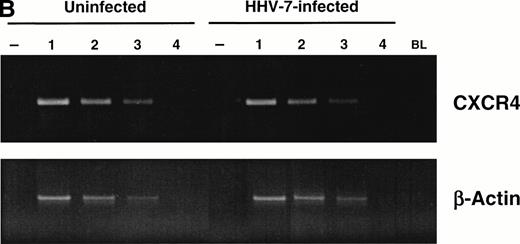
Having previously demonstrated that HHV-7 affects the expression of its primary receptor, CD4, via multiple mechanisms,22 including reduction of the de novo CD4 synthesis, we next evaluated the intracellular levels of CXCR4 in HHV-7–infected and uninfected cells by indirect immunofluorescence shown by flow cytometry (Fig 3A). At day 8 PI, when greater than 50% of surface CXCR4 was already lost in HHV-7–infected cells, all cells still stained positively for intracellular CXCR4 and the fluorescence intensity of intracellular CXCR4 was only modestly decreased in comparison to uninfected controls. Moreover, analysis of CXCR4 mRNA by semiquantitative RT-PCR unequivocally demonstrated that HHV-7 infection did not affect the levels of CXCR4 mRNA (Fig 3B).
Analyses of intracellular CXCR4 protein and CXCR4 mRNA content of uninfected and HHV-7–infected SupT1 cells. (A) Flow cytometric analysis of intracellular CXCR4, either alone (top panels) or in combination with surface CXCR4 (bottom panels), in uninfected and HHV-7–infected (8 days PI) SupT1 cells. Horizontal axis, relative intracellular CXCR4 expression detected by PE fluorescence intensity; vertical axis, isotype-matched antibody staining or relative surface CXCR4 expression detected by FITC fluorescence intensity. Percentages of cells in the respective quadrants are indicated. The profile of the negative controls, represented by SupT1 cells stained with irrelevant isotype matched MoAb, were as shown in Fig 1B (top panel). The data are representative of four experiments from separate infections. (B) Semiquantitative RT-PCR specific for CXCR4 mRNA was applied to analyze CXCR4 mRNA levels in HHV-7–infected SupT1 cultures (8 days PI) as compared with the uninfected cells. β-Actin amplification was used to confirm comparability of the samples. Equivalent amounts of RNA extracted from uninfected and HHV-7–infected cells were used for 1:3 limiting step dilution (lanes 1 through 4) before RT-PCR with the CXCR4 and β-actin primers. Ethidium bromide-stained agarose gel of RT-PCR products is shown. Lanes −, amplification of the indicated RNA template performed before RT; lane BL, blank.
Analyses of intracellular CXCR4 protein and CXCR4 mRNA content of uninfected and HHV-7–infected SupT1 cells. (A) Flow cytometric analysis of intracellular CXCR4, either alone (top panels) or in combination with surface CXCR4 (bottom panels), in uninfected and HHV-7–infected (8 days PI) SupT1 cells. Horizontal axis, relative intracellular CXCR4 expression detected by PE fluorescence intensity; vertical axis, isotype-matched antibody staining or relative surface CXCR4 expression detected by FITC fluorescence intensity. Percentages of cells in the respective quadrants are indicated. The profile of the negative controls, represented by SupT1 cells stained with irrelevant isotype matched MoAb, were as shown in Fig 1B (top panel). The data are representative of four experiments from separate infections. (B) Semiquantitative RT-PCR specific for CXCR4 mRNA was applied to analyze CXCR4 mRNA levels in HHV-7–infected SupT1 cultures (8 days PI) as compared with the uninfected cells. β-Actin amplification was used to confirm comparability of the samples. Equivalent amounts of RNA extracted from uninfected and HHV-7–infected cells were used for 1:3 limiting step dilution (lanes 1 through 4) before RT-PCR with the CXCR4 and β-actin primers. Ethidium bromide-stained agarose gel of RT-PCR products is shown. Lanes −, amplification of the indicated RNA template performed before RT; lane BL, blank.
Inibition of HHV-7 infection by SDF-1α.
Cellular receptors for enveloped viruses are characteristically downregulated from the cell surface after productive infection, rendering infected cells resistant to superinfection by viruses that use the same receptor.31 Indeed, the observation that CD4 was selectively downregulated on HIV-1–infected cells as well as on HHV-7–infected cells provided the initial evidence that CD4 was a receptor for these viruses.20,32 Given the evidence that CXCR4 was downregulated during HHV-7 infection, we used SDF-1α to determine if CXCR4 was also serving as a component of the receptor for HHV-7. As shown in Fig 4A, preincubation with SDF-1α (2.5 μg/mL) led to a drastic reduction of surface CXCR4 associated with a complete block of expression of specific HHV-7 mRNA transcripts, evaluated 40 hours PI. On the other hand, preincubation with 12G5 MoAb (25 μg/mL) or control IgG2a (25 μg/mL) failed to inhibit infection, indicating that the epitope recognized by 12G5 MoAb is not essential for HHV-7 entry. Similar results have been previously reported for several HIV-1 and HIV-2 isolates.33
Effect of SDF-1 on HHV-7 infection. (A) SDF-1–dependent CXCR4 downmodulation (left panel) and inhibition of HHV-7 infection (right panel). SupT1 cells were preincubated with SDF-1 (2.5 μg/mL) for 30 minutes at room temperature and then either remained uninfected or inoculated with HHV-7. Before HHV-7–infection, surface CXCR4 was analyzed by flow cytometry (left panel). Horizontal axis, surface CXCR4 expression detected by PE fluorescence intensity; vertical axis, relative cell number. Negative control (Cont) is represented by cells stained with irrelevant isotype-matched control MoAb. The right panel shows HHV-7 expression determined by specific RT-PCR in SupT1 cells preincubated with nothing (lane 1), 2.5 μg/mL of SDF-1 (lane 2), 25 μg/mL of control mouse IgG2a (lane 3), and 25 μg/mL of 12G5 anti-CXCR4 MoAb (lane 4). Equivalent amounts of RNA, extracted at 40 hours PI, were used as template for RT-PCR using either HHV-7–specific primers (U10 ORF) or β-actin primers. Control reaction, performed by amplifying the same RNA samples before RT (−RT) is also shown; lane BL, blank. The data are representative of three separate experiments. (B) CD4+ SupT1 or CD4− BC7 cells were inoculated with HHV-7 and monitored for HHV-7 expression and replication by specific viral RT-PCR at 40 hours PI (left panels) and by indirect immunofluorescence at 8 days PI (microphotographs). For RT-PCR, equivalent amounts of RNA samples, before (−) and after (+) RT, were used as template for the amplification reactions. PCR products for HHV-7/U10 ORF and β-actin are shown; lane BL, blank. Immunofluorescence microscopy was performed as described by using the 5E1 HHV-7–specific MoAb; negative reactions are counterstained with Evans blue. The data are representative of three experiments from separate infections.
Effect of SDF-1 on HHV-7 infection. (A) SDF-1–dependent CXCR4 downmodulation (left panel) and inhibition of HHV-7 infection (right panel). SupT1 cells were preincubated with SDF-1 (2.5 μg/mL) for 30 minutes at room temperature and then either remained uninfected or inoculated with HHV-7. Before HHV-7–infection, surface CXCR4 was analyzed by flow cytometry (left panel). Horizontal axis, surface CXCR4 expression detected by PE fluorescence intensity; vertical axis, relative cell number. Negative control (Cont) is represented by cells stained with irrelevant isotype-matched control MoAb. The right panel shows HHV-7 expression determined by specific RT-PCR in SupT1 cells preincubated with nothing (lane 1), 2.5 μg/mL of SDF-1 (lane 2), 25 μg/mL of control mouse IgG2a (lane 3), and 25 μg/mL of 12G5 anti-CXCR4 MoAb (lane 4). Equivalent amounts of RNA, extracted at 40 hours PI, were used as template for RT-PCR using either HHV-7–specific primers (U10 ORF) or β-actin primers. Control reaction, performed by amplifying the same RNA samples before RT (−RT) is also shown; lane BL, blank. The data are representative of three separate experiments. (B) CD4+ SupT1 or CD4− BC7 cells were inoculated with HHV-7 and monitored for HHV-7 expression and replication by specific viral RT-PCR at 40 hours PI (left panels) and by indirect immunofluorescence at 8 days PI (microphotographs). For RT-PCR, equivalent amounts of RNA samples, before (−) and after (+) RT, were used as template for the amplification reactions. PCR products for HHV-7/U10 ORF and β-actin are shown; lane BL, blank. Immunofluorescence microscopy was performed as described by using the 5E1 HHV-7–specific MoAb; negative reactions are counterstained with Evans blue. The data are representative of three experiments from separate infections.
Although the downregulation of surface CXCR4 by HHV-7 and the inhibitory effects of SDF-1α on HHV-7 infection are consistent with a role of CXCR4 as a coreceptor for HHV-7, the final proof of this hypothesis would be to show that noninfectable cells that are CD4 and CXCR4 negative are rendered infectable by HHV-7 only when CD4 and CXCR4 are both expressed.
The possibility that CXCR4 might represent an alternative receptor for HHV-7 in the absence of CD4 was ruled out in infection experiments performed on the BC7 cells, a derivative line from SupT1 cells that expresses CXCR4 but not CD4.23 As shown in Fig 4B, BC7 cells were not permissive for HHV-7 infection.
DISCUSSION
Although CXCR4 functions in association with CD4 to permit the entry of different HIV isolates,9,10 the precise mechanism of action of this protein as coreceptor has not yet been fully elucidated. Irrespective of the mechanism of action of CXCR4 in mediating HIV entry, it has been clearly established that CXCR4 is downregulated only by a few HIV-2 isolates, which use CXCR4 as the primary receptor in the absence of CD4.23 On the other hand, CXCR4 is either unaffected or minimally downregulated by HIV-1 strains (Fig 2) or by those HIV-2 strains that still require CD4 for infection and cell fusion.23
We have shown here that CXCR4 interacts with HHV-7 in a unique manner compared with HIV. In fact, both CD4 and CXCR4 are downregulated from the surface of HHV-7–infected CD4+ T cells, although with different kinetics and probably through different mechanisms. After HHV-7 infection, CD4 loss is more rapid and involves both transcriptional and posttranscriptional mechanisms,22whereas downregulation of surface CXCR4 does not depend on a block of transcription.
Although initial studies have suggested that chemokine receptor internalization is not required to block HIV entry, more recent data indicate that SDF-1α is a more effective inhibitor of T-cell line-adapted HIV isolates on cells expressing endocytosis-competent CXCR4.14 15 This suggests that SDF-1α–mediated CXCR4 endocytosis may make a significant contribution to chemokine protection from HIV entry.
Although the mechanisms of HHV-7–mediated downregulation of CXCR4 remain to be elucidated, the possibility that HHV-7 induced CXCR4 downregulation via an autocrine production of SDF-1α was ruled out, because SDF-1α mRNA was undetectable by RT-PCR in both HHV-7–infected and uninfected SupT1 cells (data not shown). It is also particularly remarkable that, whereas SDF-1α induces a rapid but transient (<1 hour) downregulation of surface CXCR4, HHV-7 infection produces a persistent loss of surface CXCR4.
Thus, beside the relevance of these finding for HHV-7 pathogenesis, the identification of the HHV-7 gene product(s) modulating CXCR4 may allow the design of novel strategies to inactivate CXCR4 by blocking its surface expression.34
ACKNOWLEDGMENT
The authors are very grateful to J.A. Hoxie and to G. Campadelli-Fiume for providing the BC7 cell line and the 5E1 MoAb, respectively.
Supported by an University of Maryland School of Medicine Intramural Research Fund Award, by Telethon Foundation Grant, and by the “AIDS project” of the Italian Ministry of Health.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Paola Secchiero, PhD, Institute of Human Virology, University of Maryland, 725 W Lombard St, Baltimore, MD 21201-1192; e-mail: secchier@umbi.umd.edu.

![Fig. 1. Effect of HHV-7 and HIV-1 infection on surface CXCR4 expression in CD4+ T cells evaluated by flow cytometry. In (A) and (B), progressive downregulation of surface CXCR4 expression in HHV-7–infected SupT1 cells. In (B), surface CXCR4 was analyzed in combination with surface CD4. In (C), surface CXCR4 was analyzed either alone (top panels) or in combination with surface CD3 (middle panels) and surface CD4 (bottom panels) in CD8+-depleted primary cells, either uninfected or infected with HHV-7 (8 days PI). In (D), surface CXCR4 was analyzed in combination with surface CD4 in SupT1 cells, either uninfected or infected with HIV-1 (12 days PI). Data shown in (A) through (D) are representative of at least three separate experiments. Horizontal axis (logarithmic scale), relative surface CXCR4 expression detected by PE fluorescence intensity (in [A] through [D]). Vertical axis, relative cell number, isotype-matched control antibody staining, CD4 expression detected by FITC fluorescence intensity or CD3 expression detected by Cy5-PE fluorescence intensity, as indicated. Percentages of cells in the respective quadrants are indicated. Representative negative controls, constituted by cells stained with irrelevant isotype-matched MoAb, are shown in the top panels of (A) and (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/12/10.1182_blood.v92.12.4521/5/m_blod42438001ax.jpeg?Expires=1767746038&Signature=5CGuhDh-Z6ircV0tW9JrBX0D2z3jI43B0V-LAqTfVaQEiJfwlJfA-GRbLcuqtLGritNYc~j9kEGyMv1eK5urhi-u9q0AatjpT3A9F9pGTfiAYC5urDqfCDYSGxFHPCmteRCloQ88e9JEYcL--uCmrrabQj4Rjwh4vbGXbsJqgCCzJrHJfZnDnoCmqYNqfWRwI~b85waziA8R4a2-alq~-x05owVqXtp85BXsRwd4bFjC4PDd7aDvqgT8wrTbdbO6PQOyW6gxE-KcEqlw5p~~2VEbrvVNn0xkMIjNAkvKgowOLFXoMttwrg4caTb0EpOlOx4PE6QRoXId0E2tO4k8nQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 1. Effect of HHV-7 and HIV-1 infection on surface CXCR4 expression in CD4+ T cells evaluated by flow cytometry. In (A) and (B), progressive downregulation of surface CXCR4 expression in HHV-7–infected SupT1 cells. In (B), surface CXCR4 was analyzed in combination with surface CD4. In (C), surface CXCR4 was analyzed either alone (top panels) or in combination with surface CD3 (middle panels) and surface CD4 (bottom panels) in CD8+-depleted primary cells, either uninfected or infected with HHV-7 (8 days PI). In (D), surface CXCR4 was analyzed in combination with surface CD4 in SupT1 cells, either uninfected or infected with HIV-1 (12 days PI). Data shown in (A) through (D) are representative of at least three separate experiments. Horizontal axis (logarithmic scale), relative surface CXCR4 expression detected by PE fluorescence intensity (in [A] through [D]). Vertical axis, relative cell number, isotype-matched control antibody staining, CD4 expression detected by FITC fluorescence intensity or CD3 expression detected by Cy5-PE fluorescence intensity, as indicated. Percentages of cells in the respective quadrants are indicated. Representative negative controls, constituted by cells stained with irrelevant isotype-matched MoAb, are shown in the top panels of (A) and (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/12/10.1182_blood.v92.12.4521/5/m_blod42438001bx.jpeg?Expires=1767746038&Signature=wRRpFHLKCbQ1OM~IsFRJkVIuYhj07hhtC9uF0FsxpzMT4lU58IMXPjD0m81yWMu4haNRm4h9A5pfAh38Fp3ASdFme3VeSOTZHmNP7~IqVFf6v6wWdrYXJn7O13q4fuajKBSDVlUdT8f-secds4b-2GYQplJib7n8cOuyXM0REMTZiWYrfzurAypwj-HBPO1A8PKjRIjM0p2oRzORgRgOh3BfqfS0WbNhJ424xgwOCyLpjGsdI8AM9fdZTo2uVA1szC8g6hbqTcXU1gwbTOkcfJAHnlU41sNEw2ML77PcP-3j0zu0SlJE0G2v7cPF19Fe4z311SyxKM8ZavdxjWOAMQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 1. Effect of HHV-7 and HIV-1 infection on surface CXCR4 expression in CD4+ T cells evaluated by flow cytometry. In (A) and (B), progressive downregulation of surface CXCR4 expression in HHV-7–infected SupT1 cells. In (B), surface CXCR4 was analyzed in combination with surface CD4. In (C), surface CXCR4 was analyzed either alone (top panels) or in combination with surface CD3 (middle panels) and surface CD4 (bottom panels) in CD8+-depleted primary cells, either uninfected or infected with HHV-7 (8 days PI). In (D), surface CXCR4 was analyzed in combination with surface CD4 in SupT1 cells, either uninfected or infected with HIV-1 (12 days PI). Data shown in (A) through (D) are representative of at least three separate experiments. Horizontal axis (logarithmic scale), relative surface CXCR4 expression detected by PE fluorescence intensity (in [A] through [D]). Vertical axis, relative cell number, isotype-matched control antibody staining, CD4 expression detected by FITC fluorescence intensity or CD3 expression detected by Cy5-PE fluorescence intensity, as indicated. Percentages of cells in the respective quadrants are indicated. Representative negative controls, constituted by cells stained with irrelevant isotype-matched MoAb, are shown in the top panels of (A) and (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/12/10.1182_blood.v92.12.4521/5/m_blod42438001cx.jpeg?Expires=1767746038&Signature=T0kuOzj6Nb9Ao57Fkd6lY67TBv8-YMpcR4Z1rK2kGsiHwTLyIiZYY61aCuyqoitrzvoG4gnjtwBx5uqXHJ2Ouv3EwfWTGH2wUwcKxzht4hBFCzZkwTtT93v8vEk9CH2NsaJK5swv9cfIagTzX37hG5F8ehov2FrQzAo7n2VAGqjXyPlJtykNiPU-tgYtf5-hJ9rD8QoaXG5QCQp8sramTw-zwKSYa~4acJea9oz~3xxG4~QILO7jGiirwsZyH5jKGVE1wk13wHxBtxTt6m0ZIazTAT9ICGKcmwdUw7HuZu45TPqRvLkzCA~q0uFveVwGr-LxwbaX4YvhUQOyQJ7Giw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 1. Effect of HHV-7 and HIV-1 infection on surface CXCR4 expression in CD4+ T cells evaluated by flow cytometry. In (A) and (B), progressive downregulation of surface CXCR4 expression in HHV-7–infected SupT1 cells. In (B), surface CXCR4 was analyzed in combination with surface CD4. In (C), surface CXCR4 was analyzed either alone (top panels) or in combination with surface CD3 (middle panels) and surface CD4 (bottom panels) in CD8+-depleted primary cells, either uninfected or infected with HHV-7 (8 days PI). In (D), surface CXCR4 was analyzed in combination with surface CD4 in SupT1 cells, either uninfected or infected with HIV-1 (12 days PI). Data shown in (A) through (D) are representative of at least three separate experiments. Horizontal axis (logarithmic scale), relative surface CXCR4 expression detected by PE fluorescence intensity (in [A] through [D]). Vertical axis, relative cell number, isotype-matched control antibody staining, CD4 expression detected by FITC fluorescence intensity or CD3 expression detected by Cy5-PE fluorescence intensity, as indicated. Percentages of cells in the respective quadrants are indicated. Representative negative controls, constituted by cells stained with irrelevant isotype-matched MoAb, are shown in the top panels of (A) and (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/12/10.1182_blood.v92.12.4521/5/m_blod42438001dx.jpeg?Expires=1767746038&Signature=glEjR-okATUQxy9mAAboRauj-3gMnexgpQg0puK8RJHGEvNxXHnHrjevwDq2GfktEko7RvuiR8LoWoXSbkId2wkma0--OPDJJazES99l-530kplkiPMLaQV76cV3~fEgw~179UMdRQbZV-TIBJ1QIxxAReQkvPVZDGOEQTG-a5g9M61l1YNPAg8Ks8-~GvNpioOhoLwy2hCXBh-yPVcQOD413I9VnqLc1HB-qiTl~qzmAS9JfRdEyihdeRQk-EqvvWFEs1mGVqyZjKXyQAV1~4Wd-0EiFitMqHauAjOdTNXiFwIOo6XmRLu~U1uaqQcBjiH7RPxQEpTHRW-Mn0ZoqQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal