Abstract
We have recently described Poliovirus Receptor Related 2 (PRR2), a new cell surface molecule homologous to the poliovirus receptor (PVR/CD155). Both molecules are transmembrane glycoproteins belonging to the Ig superfamily (IgSF). They contain 3 Ig domains of V, C2, and C2 types in their extracellular regions that share 51% aa identity. The PRR2 gene encodes two mRNA isoforms of 3.0 kb (hPRR2 [short form]) and 4.4 kb (hPRR2δ [long form]), both widely expressed in human tissues, including hematopoietic cells. To further characterize PRR2 expression during hematopoiesis and to analyze its function, we have developed a monoclonal antibody (MoAb) directed against its extracellular region (R2.477). PRR2 was expressed in 96% of the CD34+, 88% of the CD33+, and 95% of the CD14+ hematopoietic lineages and faintly in the CD41 compartment. Ectopic expression of both PRR2 cDNAs induced marked cell aggregation. A soluble chimeric receptor construct with the Fc fragment of human IgG1 (PRR2-Fc) as well as a fab fragment of the anti-PRR2 MoAb (R2.477) inhibit aggregation. PRR2-Fc binds specifically to PRR2-expressing cells. These results suggest that PRR2 is a homophilic adhesion receptor. PRR2 was also expressed at the surface of endothelial cells at the intercellular junctions of adjacent cells but not at the free cellular edges. Homophilic interactions are associated with dimerization of isoforms of PRR2 and lead to the tyrosine phosphorylation of PRR2δ. Altogether, these results suggest that homophilic properties of PRR2 could participate to the regulation of hematopoietic/endothelial cell functions.
ADHESION MOLECULES play a fundamental role in the regulation of different biological processes such as embryonic development, neuronal ontogeny and physiology, immune response, and hematopoietic differentiation. Most adhesion molecules mediate cell/cell or cell/substratum interactions through heterophilic mechanisms. In some cases, adhesion molecules interact with themselves through homophilic mechanisms. Adhesion molecules are classified in different families and, among them, the Ig superfamily (IgSF) includes molecules involved in homophilic adhesion processes.1
We have recently cloned two new human genes encoding transmembrane glycoproteins that contain 3 Ig domains of V, C2, and C2 types in their extracellular regions. The encoded proteins are homologous to the poliovirus receptor (PVR; CD155),2 and we named them PRR1 and PRR2 (poliovirus receptor related).3,4 The PVR and PRR2 genes are located in the same human chromosomal region (19q13); they encode proteins sharing 51% identity in their extracellular regions. The PRR1 gene is located on chromosome 11q23 and encodes protein sharing 30% and 32% identity with the PVR and PRR2, respectively.3 Among the three members of this family, PVR and PRR2 proteins are more closely related.
At least four PVR transcripts, resulting from alternative splicing, have been cloned: two transmembrane forms (PVRα and PVRδ) and two soluble forms (PVRβ and PVRγ) with common Ig domain sequences.5 Two different PVR monkey cDNAs (AGMα1 and AGMα2) have been isolated and encode receptors to poliovirus.6 Two murine PVR homolog (MPH) cDNAs have been cloned, but the corresponding proteins do not bind poliovirus.7,8 By cloning the human PRR2 gene, we showed that the true human homolog of MPH was hPRR2 and not hPVR. MPH was renamed mPRR2.4 Two hPRR2 cDNAs (hPRR2α [short form] and hPRR2δ [long form]) were identified. These two cDNAs encode alternative forms of the same gene sharing identical extracellular regions but different transmembrane and intracytoplasmic regions. The mPRR2 gene encodes also two glycoproteins, mPRR2α and mPRR2δ, sharing 69% and 64% identities with hPRR2α and hPRR2δ, respectively, along the whole amino acid sequence, but only 51% identity with PVR in the extracellular region. Finally, no cDNAs encoding soluble forms of hPRR2 or mPRR2 have been identified.
In the present report, we demonstrate that hPRR2 is expressed at the surface of hematopoietic cells of the myelo-monocytic and megakaryocytic lineages. hPRR2 is also detected at the surface of endothelial cells; its localization is restricted to intercellular borders. Both hPRR2 isoforms could mediate homophilic intercellular adhesion, confirming another recently published report.9Homophilic adhesion mediated by PRR2 correlates with dimerization of the molecule at the cell surface and phosphorylation of the long form on tyrosine residues. Altogether, these results suggest that hPRR2 is a new homophilic adhesion molecule that could be involved not only in cell-cell interactions, but also in the regulation of intracellular functions through signal transduction.
MATERIALS AND METHODS
Cells.
The murine hematopoietic cell line Da-1 and the human erythroleukemic cell line TF110 were cultivated in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). Murine interleukin-3 (IL-3; supernatant from transfected X63Ag8-653 myeloma) was added to the Da-1 culture and 5 ng/mL of recombinant human granulocyte-macrophage colony-stimulating factor (huGM-CSF; Sandoz, Rueil Malmaison, France) was added to the TF1 culture. The murine cell line expressing the CD28 antigen (DWT6.11) was cultivated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS.11 The recombinant CHO DG44 cell line (kindly provided by Dr L. Chasin, Columbia University, New York, NY) was cultivated in CHOSFMII serum-free medium (LifeTechnologies, Grand Island, NY).
Adult bone marrow was aspirated from the posterior iliac crest of healthy volunteers after informed consent had been obtained. ECV304 endothelial cell line (umbilical cord origin; ATCC, Rockville, MD) was cultivated in DMEM supplemented with 10% FCS. Eahy924 cell line (kindly provided by Dr C.J. Edgell, University of North Carolina, Chapel Hill, NC) was cultivated in DMEM supplemented with 10% FCS and hypoxanthine aminopterin thymidine (HAT) medium.12 Endothelial cells were harvested from human umbilical cord vein (HUVEC) as previously described.13 The cells were maintained in RPMI containing 20% FCS.
Antibodies.
Flow cytometric analyses were performed using fluorescein isothiocyanate (FITC)-labeled monoclonal antibodies (MoAbs). CD34 (HPCA-2) was purchased from Becton Dickinson (Mountain View, CA). CD33 (D3HL60.251), CD14 (RMO52), CD41 (P2), glycophorin A (11E4B-7-6), and CD19 (J4.119) were purchased from Immunotech (Marseille, France).
The MoAbs directed against PRR2 (R2.477 and R2.525) and PVR (PV.404) were obtained in the laboratory and submitted at the VIth International Workshop on Human Leukocyte Differentiation Antigens.14 Fab fragments were obtained after papain digestion according to the manufacturer’s protocol (Pierce, Rockford, IL). The MoAb against PRR2 (BC-12) was kindly provided by Dr Claude Clement (Diaclone, Berançon, France).
Immunesera directed against the intracellular domain of PRR2α (anti-PRR2α) and δ (anti-PRR2δ) were obtained after rabbit immunization with C-terminal peptides SQLDGSLISRRAVYV and SYQGKGFVMSRAMYV, respectively.
The PECAM-1/CD31 MoAb (clone JC/70A) was purchased from Dako (Glostrup, Denmark).
The 4G10 MoAb (UBI; Euromedex, Souffelweyersheim, France) was used in Western blotting to analyze the tyrosine phosphorylation status of PRR2.
Immunohistochemistry.
Immunodetection of PRR2 and PECAM-1/CD31 were performed on frozen sections (5 μm) of human placenta using the R2.477 and the PECAM-1/CD31 MoAbs. Specimen were processed with the Universal Dako Kit ChemMate according to the supplier’s recommendations and counterstained for 5 minutes in Harris hematoxylin and mounted in Dako glycergel mounting medium.
Cell transfection.
The PRR2α and δ cDNAs were cloned in the LXSN vector and then transfected in Da-1 cells. The extracellular region of PRR2 (aa 1 to 347) was cloned in frame with the Fc fragment of the human IgG1 sequence using the Cos Fc Link vector (SmithKline Beecham Pharmaceuticals, King of Prussia, PA). CHO DG44 cells were then transfected with PRR2-Fc plasmid to produce an Fc-tagged PRR2 soluble receptor.
Transfection of Da-1 and CHO DG44 cells was performed with Lipofectin according to the manufacturer’s protocol (Life Technologies, Inc).
Production and purification of soluble chimeric PRR2-Fc protein.
CHOSFMII medium was harvested every 4 days and stored at −20°C. Approximately 1 L of supernatant was filtered and loaded on a 5-mL Affigel protein A column according to the manufacturer’s protocol (Bio-Rad, Hercules, CA). After washing, the PRR2-Fc protein was eluted with a 0.1 mol/L citrate buffer, pH 3.5, concentrated, and dialyzed against phosphate-buffered saline (PBS). Purification steps were monitored by enzyme-linked immunosorbent assay (ELISA) using a sandwich revelation system composed with coated antibody against human Fc (Sigma, St Louis, MO) and biotinylated R2.477 antibody.
Purity and quality of the protein were controlled by gel electrophoresis followed by silver gel staining (Bio-Rad). As shown in Fig 6B, Western blot analysis of purified PRR2-Fc molecule showed a molecular weight of 80 kD.
The B7.1-Fc protein, used as an irrelevant control, was kindly provided by Dr R. Sweet (SmithKline Beecham Pharmaceuticals) and was produced similar to PRR2-Fc.
Immunoprecipitation and Western blot analyses.
Cells (5 × 106) were lysed in ice-cold lysis buffer containing 1% Triton X100, 10% glycerol, 0.1% sodium dodecyl sulfate (SDS), 50 mmol/L HEPES buffer, pH 7.5, 150 mmol/L NaCl, 1.5 mmol/L MgCl2, 1 mmol/L EGTA, 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 1 mmol/L phenylmethylsulfonyl fluoride. Orthovanadate (1 mmol/L) was added in the case of phosphotyrosine analysis. Cell lysates were clarified by centrifugation at 13,000 rpm for 15 minutes at 4°C. They were incubated for 1 hour at 4°C with 5 μg of R2.477 antibody and 50 μL of protein A Sepharose (Pharmacia, Uppsala, Sweden). Immune complexes were washed three times with cold lysis buffer, heated in SDS-sample buffer, separated by gel electrophoresis, semi-dry transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Boston, MA), and immunoblotted with a 1:10,000 dilution anti-PRR2 immunesera.
PRR2-Fc protein was shown by immunoblotting using a peroxidase-conjugated antibody directed against human Fc fragment (Jackson ImmunoResearch, West Grove, PA).
Chemical cross-linking.
Before cross-linking, 5 × 106 Da-1/PRR2 aggregated cells were dissociated by agitation and then incubated with 1 mmol/L of Bis (sulfosuccinimidyl) suberate (BS3) cross-linker (Pierce). This concentration gives maximum cross-linking efficiency. After 15 minutes at 14°C, the reaction was quenched by the addition of 10 mmol/L ammonium acetate. Cells were washed twice with PBS and counted to confirm that no aggregation of suspended cells had occurred. Cells lysis, immunoprecipitation, and Western blot analyses were performed as described in the previous paragraph.
Aggregation assays.
Experiments were performed as previously described.15Briefly, Da-1/PRR2 cells were suspended in complete medium at 1 × 106 cells/mL and transferred to polystyrene tubes. Incubation was performed at 37°C and aliquots were taken every 15 minutes after gentle mixing. The number of aggregates (>3 cells) were counted in a hemocytometer. Mixed-cell aggregation experiments were performed with Da-1 cells stained with 0.5 μmol/L of CM-DiI dye as recommended by the manufacturer (Molecular Probes, Eugene, OR) and Da-1/PRR2 cells at a 1:1 ratio.
PRR2-Fc binding assays.
Three different populations of Da-1/PRR2 cells were sorted according to the membrane levels of PRR2 and further put into culture. After 10 days, cells were controlled for the expression of PRR2 and the three populations were named Da-1/PRR2 lo (low), Da-1/PRR2 me (medium), and Da-1/PRR2 hi (high).
Each population of Da-1/PRR2 cells was analyzed in a test of PRR2 soluble receptor binding. Briefly, Da-1/PRR2 cells (1 × 105) were washed three times with cold PBS/bovine serum albumin (BSA) and incubated for 1 hour at 4°C with 40 μg/mL of PRR2-Fc protein. After two washes with the PBS/BSA buffer, cells were incubated for 30 minutes at 4°C with a 1:60 dilution of phycoerythrin-labeled goat antihuman IgG (Immunotech). Cells were washed three times in the PBS/BSA buffer, and pellets were resuspended in 200 μL of PBS. PRR2-Fc binding was analyzed using a FacScan cytometer (Becton Dickinson).
Adhesion assays.
Experiments were performed as previously described.16Briefly, 3.5-mm tissue culture Petri dishes were first coated with methanol-solubilized nitrocellulose and then with polyornithin (1.5 μg/mL; Sigma). Three-microliter spots of different soluble proteins (PRR2-Fc or B7-1-Fc) at concentrations of 200 μg/mL were applied in triplicate to the nitrocellulose/polyornithin-coated surfaces and incubated for 12 hours at 37°C in a humidified atmosphere. Spots were washed three times with Ca2+- and Mg2+-free PBS. Cells were then incubated for 15 minutes at 37°C, washed three times with PBS, and then fixed with PBS containing 2% formaldehyde (Fluka, St Quentin Fallavier, France). Cell adhesion was analyzed by microscopy and photomicrographs were taken at the edges of the spots of soluble proteins.
Confocal immunofluorescence analysis of PRR2 expression.
HUVEC were cultured on coverslips, fixed with 3% paraformaldehyde for 20 minutes, and indirectly stained with the R2.477 MoAb and goat antimouse Ig conjugated to FITC (Immunotech). Serial optical sections were collected using a TCS 4D Leica laser scanning confocal microscope (Heidelburg, Germany). Microscope settings were adjusted so that black level values were obtained with a mouse IgG1 isotypic control.
RESULTS
PRR2 expression in hematopoiesis.
Northern blot analysis has shown that PRR2 is expressed in various tissues, including bone marrow cells.4 This approach does not allow us to define precisely which hematopoietic cells express PRR2. We therefore developed MoAbs against PRR2 molecules. FACS analysis of bone marrow cells demonstrated that PRR2 was expressed in 96% of the progenitor compartment defined by the expression of the CD34 antigen (Fig 1A). Similar results were obtained with CD34+ cells purified from mobilized peripheral blood (data not shown). PRR2 expression was also detected in the myelo-monocytic compartment, ie, in 88% of CD33+ and in 95% of CD14+ cells. PRR2 was also expressed at a low level in the megakaryocytic lineage (50%). Expression of PRR2 on CD19-expressing cells was found to be negative or low. No expression was detected in the glycophorin A-expressing cells. Among peripheral blood cells, PRR2 expression was predominantly detected on monocytes (89%) and slightly on polymorphonuclear cells (data not shown).
Expression of PRR2 in human bone marrow cells and endothelial cells. (A) Bone marrow cells were doubled stained with the indicated MoAbs and the MoAb R2.477 as described in Materials and Methods. Fluorescence density plots were gated on specific regions defined on scatter plot. The bottom-left quadrant was defined according to an isotypic-matched control antibodies. The percentage represents the number of PRR2-expressing cells in each lineage. (B) FACS analysis of PRR2 and PECAM-1 expression in HUVEC. HUVEC were stained either with the MoAb R2.477 or the MoAb anti–PECAM-1. Each fluorescence distribution was superposed and compared with the background fluorescence distribution of an isotypic-matched control antibody.
Expression of PRR2 in human bone marrow cells and endothelial cells. (A) Bone marrow cells were doubled stained with the indicated MoAbs and the MoAb R2.477 as described in Materials and Methods. Fluorescence density plots were gated on specific regions defined on scatter plot. The bottom-left quadrant was defined according to an isotypic-matched control antibodies. The percentage represents the number of PRR2-expressing cells in each lineage. (B) FACS analysis of PRR2 and PECAM-1 expression in HUVEC. HUVEC were stained either with the MoAb R2.477 or the MoAb anti–PECAM-1. Each fluorescence distribution was superposed and compared with the background fluorescence distribution of an isotypic-matched control antibody.
PRR2 was expressed in greater than 98% of HUVEC. FACS analysis showed that PRR2 expression is homogenous and weaker than PECAM-1 expression (Fig 1B). PRR2 was equally expressed in HUVEC, EaHy926, and ECV304 cells (data not shown). The R2.477 MoAb was used in immunohistochemistry (IHC) on frozen sections of normal human placenta to confirm that PRR2 was actually expressed in endothelial cells. PECAM-1 expression was analyzed, as a control, by IHC in parallel on serial sections. PRR2 immunostaining was detected in endothelial cells from vessels located inside the placental villi (Fig 2B) and was weaker than PECAM-1 immunostaining in the same vessels (Fig 2C). These differences in expression levels are in agreement with the profiles of expression detected by FACS analysis on cultured endothelial cells (Fig 1B). A faint PRR2 immunostaining was also detected by IHC in trophoblastic cells lining the placental villi (Fig 2B), indicating that PRR2 expression is probably not restricted to endothelial cells.
Immunohistochemical detection of PRR2 in endothelial cells. Frozen sections of human placenta were stained with either an isotypic-matched control antibody (A), the R2.477 MoAb (B), or the PECAM-1/CD31 MoAb (C) as described in Materials and Methods. Slides were observed by light microscopy at 40× magnification.
Immunohistochemical detection of PRR2 in endothelial cells. Frozen sections of human placenta were stained with either an isotypic-matched control antibody (A), the R2.477 MoAb (B), or the PECAM-1/CD31 MoAb (C) as described in Materials and Methods. Slides were observed by light microscopy at 40× magnification.
Biochemical characterization of the two PRR2 isoforms (PRR2δ and α).
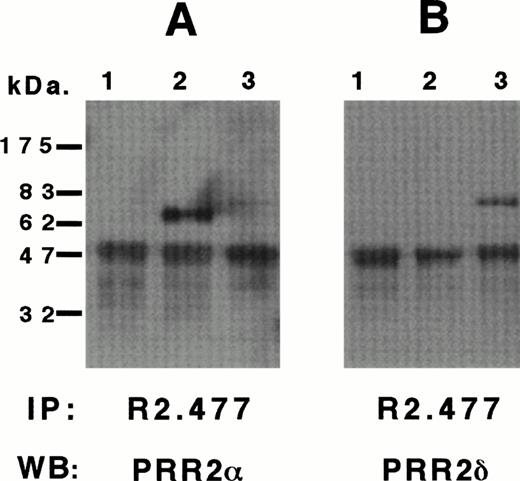
We previously identified two transcripts of 3.0 and 4.4 kb corresponding to PRR2α and δ cDNAs. Cells of a murine hematopoietic cell line Da-1 were transfected with the PRR2α and δ cDNAs to analyze the proteins encoded by the two transcripts. Western blot analyses of Da-1/PRR2α and Da-1/PRR2δ cells are represented in Fig 3A and B, respectively. After immunoprecipitation of PRR2 with the R2.477 MoAb, a unique band of 64 kD was detected with the immuneserum directed against the C terminal region of PRR2α in Da-1/PRR2α cells (Fig 3A, lane 2), but not in control cells or Da-1/PRR2δ cells. The immuneserum directed against the C terminal region of PRR2δ detected a 72-kD band in Da-1/PRR2δ cells only (Fig 3B, lane 3). Identical results were obtained with the R2.525 and BC-12 MoAbs (data not shown). Tunicamycin treatment of both transfectants reduced the molecular weight of both proteins, suggesting that PRR2 is N-glycosylated (data not shown).
Biochemical characterization of huPRR2 and PRR2δ. Da-1 (1), Da-1/PRR2 (2), and Da-1/PRR2δ (3) cell lines were lysed and immunoprecipitated with MoAb R2.477 as described in Materials and Methods. Blots were then incubated with a 1/10,000 rabbit immune serum directed against PRR2 (A), PRR2δ (B), or both (C). and δ isoforms of PRR2 were indicated and the calculated molecular weights were 64 and 72 kD, respectively. The band at 50 kD corresponds to the heavy chain of MoAb R2.477. Molecular weight markers were from Biolabs.
Biochemical characterization of huPRR2 and PRR2δ. Da-1 (1), Da-1/PRR2 (2), and Da-1/PRR2δ (3) cell lines were lysed and immunoprecipitated with MoAb R2.477 as described in Materials and Methods. Blots were then incubated with a 1/10,000 rabbit immune serum directed against PRR2 (A), PRR2δ (B), or both (C). and δ isoforms of PRR2 were indicated and the calculated molecular weights were 64 and 72 kD, respectively. The band at 50 kD corresponds to the heavy chain of MoAb R2.477. Molecular weight markers were from Biolabs.
PRR2 mediates intercellular homophilic adhesion.
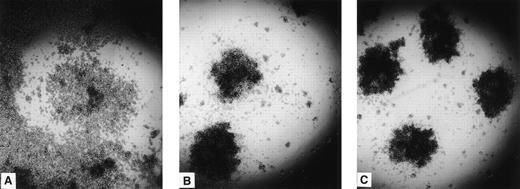
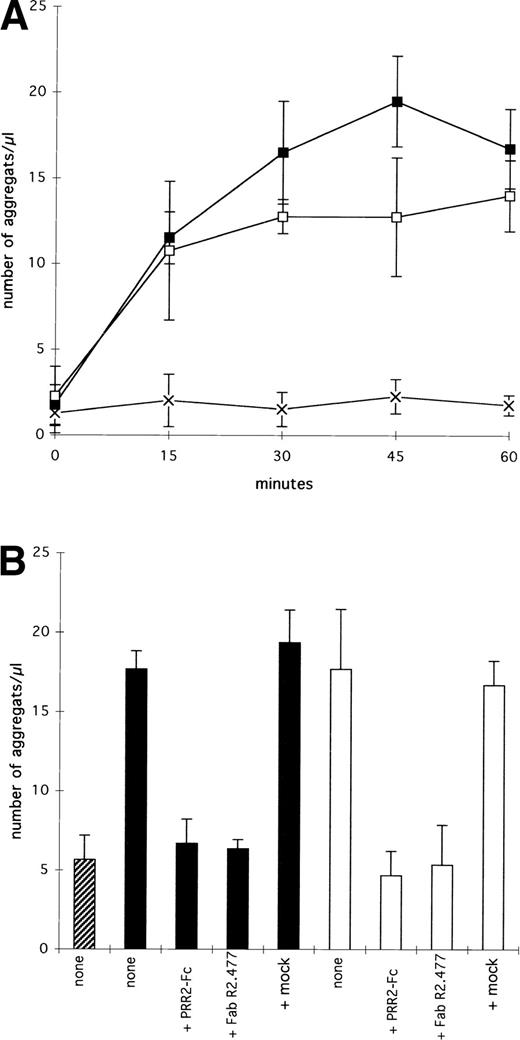
Both Da-1/PRR2α and δ cells formed aggregates with a similar phenotype, whereas control Da-1 cells did not (Fig 4). To link cell aggregation with PRR2 expression, aggregation experiments were performed with Da-1/PRR2α and Da-1/PRR2δ and compared with Da-1 cells (Fig 5). For both transfectants, an increase in the number of aggregates was observed as a function of time, with a maximum reached between 30 and 45 minutes. Da-1 control cells do not aggregate in the same conditions (Fig 5A). To precisely measure the contribution of both forms of PRR2, aggregation experiments were performed in the presence of either the soluble PRR2-Fc protein or the fab fragment of the MoAb R2.477. In both cases, aggregation of transfectants was strongly decreased, demonstrating that PRR2 mediates cell aggregation (Fig 5B). Mixed-cell aggregation experiments between transfectants and CM-diI–stained Da-1 cells demonstrated that aggregates were exclusively formed with unstained cells (data not shown). These results strongly suggest that PRR2 mediates aggregation via a homophilic mechanism.
Aggregation of PRR2 transfectants. Da-1 cells transfected either with the LXSN vector (A), PRR2 (B), or PRR2δ (C) constructions were then plated on to 96 mutliwell plates. After 2 hours, aggregation was observed by light microscopy. Control cells did not aggregate in these conditions.
Aggregation of PRR2 transfectants. Da-1 cells transfected either with the LXSN vector (A), PRR2 (B), or PRR2δ (C) constructions were then plated on to 96 mutliwell plates. After 2 hours, aggregation was observed by light microscopy. Control cells did not aggregate in these conditions.
Aggregation assays of PRR2 transfectants. (A) Short-term kinetic. Values are the mean ± SE for three independant experiments and correspond to the number of aggregates (n > 3 cells) per microliter. (▪) Da1/PRR2 cells; (□) Da1/PRR2δ cells; (×) Da1 cells. (B) PRR2-Fc (20 μg/mL) or fab fragment of R2.477 MoAb (20 μg/mL) inhibits aggregation. The number of aggregates was counted after 60 minutes of incubation. (▨) Da1; (▪) Da1/PRR2; (□) Da1/PRR2δ. The mock control corresponds to a murine irrelevant IgG1 (20 μg/mL).
Aggregation assays of PRR2 transfectants. (A) Short-term kinetic. Values are the mean ± SE for three independant experiments and correspond to the number of aggregates (n > 3 cells) per microliter. (▪) Da1/PRR2 cells; (□) Da1/PRR2δ cells; (×) Da1 cells. (B) PRR2-Fc (20 μg/mL) or fab fragment of R2.477 MoAb (20 μg/mL) inhibits aggregation. The number of aggregates was counted after 60 minutes of incubation. (▨) Da1; (▪) Da1/PRR2; (□) Da1/PRR2δ. The mock control corresponds to a murine irrelevant IgG1 (20 μg/mL).
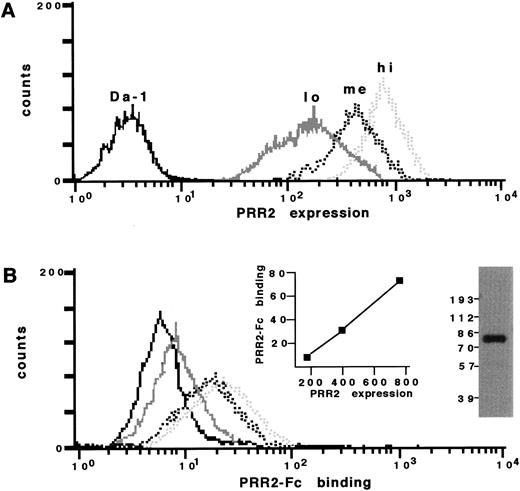
To demonstrate further the homophilic properties of PRR2, two additional experiments were performed. First, PRR2-Fc binding was analyzed by FACS analysis on three populations derived on the basis of their level of PRR2 expression (Da-1/PRR2α lo, me, and hi and Da-1/PRR2δ lo, me, and hi). Expression analyses of Da-1/PRR2α lo, me, and hi populations are represented in Fig 6A. Their respective mean fluorescence intensities (MFI) were 180, 395, and 750. As shown in Fig6B, the binding of PRR2-Fc was proportional to the expression of PRR2α at the cell surface, whereas the binding to the control LXSN cells was lower than in transfected cells. Analysis of PRR2-Fc binding as a function of PRR2 expression showed a linear relationship between the respective MFI signals (Fig 6B). The differences of fluorescence signal intensity between PRR2 expression and PRR2 binding could result from a difference in affinities between the MoAb R2.477 and the PRR2-Fc protein. Similar results were obtained with PRR2δ transfectants. Second, adhesion experiments of transfectants to coated PRR2-Fc plates were performed. Da-1/PRR2α hi and Da-1/PRR2δ hi cells adhered specifically to PRR2-Fc but not to B7.1-Fc proteins used as a negative control (Fig 7C through F). This binding was independent of the presence of divalent cations (data not shown). The CD28-transfected cells (DWT6.11) were used as a positive control for B7.1-Fc–coated plates (Fig 7G and H). Preincubation of Da-1/PRR2α hi cells with the fab fragment of the MoAb R2.477 totally inhibited adhesion to PRR2-Fc–coated plates (data not shown).
PRR2-Fc soluble receptor binds to PRR2 transfectants. (A) FACS analysis of Da-1 (continuous black) and Da-1/PRR2 lo (continuous gray), me (dashed black), and hi (dashed gray) stained with R2.477 MoAb. (B) FACS analysis of these transfectants stained with 2 μg of PRR2-Fc. Inserts show Western blot analysis of the PRR2-Fc soluble receptor using a peroxidase-conjugated goat antihuman Fc antibody and a plot corresponding to the MFI of PRR2-Fc binding as a function of the MFI of PRR2 expression.
PRR2-Fc soluble receptor binds to PRR2 transfectants. (A) FACS analysis of Da-1 (continuous black) and Da-1/PRR2 lo (continuous gray), me (dashed black), and hi (dashed gray) stained with R2.477 MoAb. (B) FACS analysis of these transfectants stained with 2 μg of PRR2-Fc. Inserts show Western blot analysis of the PRR2-Fc soluble receptor using a peroxidase-conjugated goat antihuman Fc antibody and a plot corresponding to the MFI of PRR2-Fc binding as a function of the MFI of PRR2 expression.
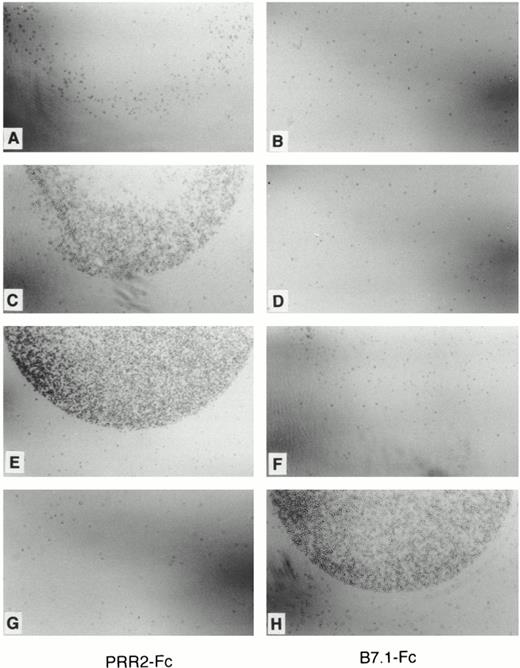
Adhesion assays of PRR2 transfectants. Adhesion assays of Da-1/PRR2 hi and Da1/PRR2δ hi on either PRR2-Fc (A, C, E, and G) or B7.1-Fc (B, D, F, and H) –coated petri dishes as described in Materials and Methods. Adhesion was observed by light microscopy. No or slight binding was observed with control Da-1 cells (A and B), whereas Da-1/PRR2 hi adhered specifically on PRR2-Fc–coated petri dish (C and D), similar to Da1/PRR2δ hi (E and F). The CD28-expressing cells adhere specifically on B7.1-Fc protein (G and H).
Adhesion assays of PRR2 transfectants. Adhesion assays of Da-1/PRR2 hi and Da1/PRR2δ hi on either PRR2-Fc (A, C, E, and G) or B7.1-Fc (B, D, F, and H) –coated petri dishes as described in Materials and Methods. Adhesion was observed by light microscopy. No or slight binding was observed with control Da-1 cells (A and B), whereas Da-1/PRR2 hi adhered specifically on PRR2-Fc–coated petri dish (C and D), similar to Da1/PRR2δ hi (E and F). The CD28-expressing cells adhere specifically on B7.1-Fc protein (G and H).
PRR2 is expressed on endothelial cells.
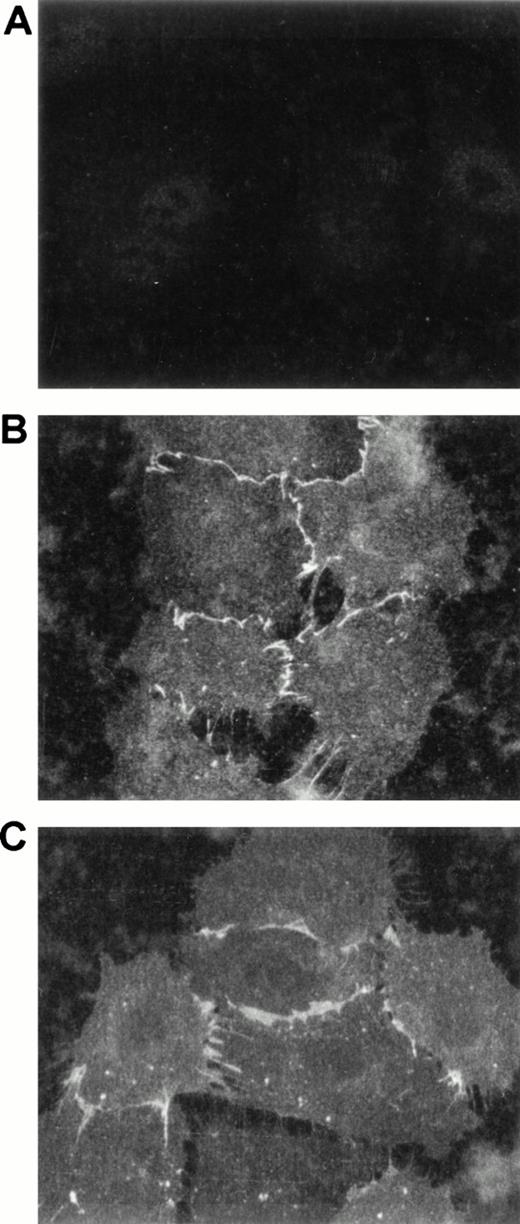
Because PRR2 is a homophilic adhesion molecule expressed on endothelial cells, the distribution of the molecule was investigated by confocal immunofluorescence microscopy. The localization of PRR2 on cultured HUVEC was compared with that of PECAM-1/CD31. Analysis of the distribution in nonconfluent HUVEC showed that PRR2 expression (Fig 8B), like that of PECAM-1/CD31 (Fig8C), was concentrated at the intercellular borders of cultured cells but was absent at the free cellular edges. These observations suggest that PRR2 could be involved in the physiological functions of vascular endothelium as described for other homophilic adhesion molecules (for review, see Dejana17).
PRR2 expression on HUVEC by confocal microscopy. After trypsinization, HUVEC were plated on coverslips for 6 hours. Nonconfluent HUVEC were stained with an isotypic-matched control antibody (A), the MoAb R2.477 (B), and the MoAb against PECAM-1/CD31 (C) and then with goat antimouse FITC antibody. Confocal acquisitions were then performed at a 63× magnification.
PRR2 expression on HUVEC by confocal microscopy. After trypsinization, HUVEC were plated on coverslips for 6 hours. Nonconfluent HUVEC were stained with an isotypic-matched control antibody (A), the MoAb R2.477 (B), and the MoAb against PECAM-1/CD31 (C) and then with goat antimouse FITC antibody. Confocal acquisitions were then performed at a 63× magnification.
Dimerization and phosphorylation of PRR2.
As shown in Fig 3A and B, the 64- and 72-kD bands correspond to monomeric forms of PRR2α and PRR2δ, respectively. To detect multimeric forms of PRR2, cell surface proteins were chemically cross-linked with BS3 before immunoprecipitation. As shown in Fig 9A, cell surface cross-linking of PRR2 transfectants showed additional bands at approximately 118 and 136 kD that probably correspond to the dimeric forms of PRR2δ and PRR2α, respectively. Because cross-linking was performed after dissociation of aggregated cells, this suggests that the observed PRR2 homodimerization results from cis-interactions within the membranes of individual cells rather than from trans-interactions.
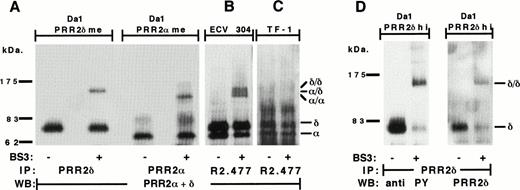
PRR2 dimerization and phosphorylation. (A) Da-1/PRR2δ me and Da-1/PRR2 me transfectants were immunoprecipitated with PRR2δ and PRR2 antisera, respectively. The ECV304 (B) and the TF-1 (C) cell lines were immunoprecipitated with the MoAb R2.477. Immunoprecipitation was performed in the absence (−) or presence (+) of BS3 cross-linker. Blots were then hybridized with a mix of PRR2 and PRR2δ immunesera. (D) Da-1/PRR2δ hi cells were immunoprecipitated with the PRR2δ immuneserum and blot was first analyzed with the 4G10 MoAb, stripped, and incubated with PRR2δ immuneserum. Molecular weight markers were from Biolabs. / and δ/δ represent PRR2 and PRR2δ homodimeric forms, respectively. /δ represents the PRR2 and PRR2δ heterodimeric form.
PRR2 dimerization and phosphorylation. (A) Da-1/PRR2δ me and Da-1/PRR2 me transfectants were immunoprecipitated with PRR2δ and PRR2 antisera, respectively. The ECV304 (B) and the TF-1 (C) cell lines were immunoprecipitated with the MoAb R2.477. Immunoprecipitation was performed in the absence (−) or presence (+) of BS3 cross-linker. Blots were then hybridized with a mix of PRR2 and PRR2δ immunesera. (D) Da-1/PRR2δ hi cells were immunoprecipitated with the PRR2δ immuneserum and blot was first analyzed with the 4G10 MoAb, stripped, and incubated with PRR2δ immuneserum. Molecular weight markers were from Biolabs. / and δ/δ represent PRR2 and PRR2δ homodimeric forms, respectively. /δ represents the PRR2 and PRR2δ heterodimeric form.
Dimerization was further investigated with ECV304 endothelial cells that constitutively express PRR2 (Fig 1B). Two bands could be identified with molecular weights similar to those found in Da-1/PRR2α and Da-1/PRR2δ cells. They correspond probably to the two PRR2 isoforms (Fig 9B). After cell surface cross-linking, three additional bands at 118, 128, and 136 kD were shown and are likely homodimers and heterodimers. The 118- and 128-kD bands were specifically shown with the anti-PRR2α serum, and the 128-and 136-kD bands were shown with the anti-PRR2δ serum (data not shown). Thus, the intermediate 128-kD band corresponds to a heterodimeric PRR2α/PRR2δ form as it is recognized by both sera, whereas the 118- and 136-kD bands represent the homodimeric forms of PRR2α and PRR2δ, respectively. Although these results were in agreement with those presented in Fig 9A, it could be noted that, even after a prolonged trypsinization (10 minutes), ECV304 cells were not completely dissociated. Thus, in ECV304 cell line, we cannot exclude that PRR2 dimerization occurs exclusively through cis-interactions. Dimeric forms of PRR2 in ECV304 were only shown after a long-time exposure of Western blots, suggesting that they were less abundant than in PRR2 transfectants. A relationship between PRR2 dimerization and homophilic aggregation was further established by cross-linking experiments performed on the hematopoietic TF-1 cell line that also expressed PRR2 but did not form aggregates during cell culture. As for the ECV304 cell line, the two PRR2 isoforms were detected at similar molecular weights, but no dimeric forms of PRR2 could be detected after cross-linking, even after a long exposure time (Fig 9C).
Intracytoplasmic phosphorylation of adhesion receptors has been frequently described in heterophilic and homophilic aggregation processes. Because the cytoplasmic regions of the two hPRR2 forms contain tyrosine residues (2 for hPRR2α and 8 for hPRR2δ), we tested a possible tyrosine phosphorylation in Da-1/PRR2αhi and PRR2δhi. Western blot analysis performed with an antiphosphotyrosine MoAb showed that PRR2δ was strongly phosphorylated on tyrosine residues in both monomeric and dimeric forms (Fig 9D). No phosphorylation was detected for PRR2α (data not shown).
DISCUSSION
We previously described PRR2, a new cell surface molecule homologous to the poliovirus receptor (PVR/CD155). This molecule belongs to the IgSF and is composed of three Ig domains of V, C2, and C2 types. Two cDNA isoforms were cloned and both transcripts were ubiquitously found among the various human tissues tested,4 including bone marrow cells and HUVEC.
The present study demonstrates that, in hematopoiesis, PRR2 expression is restricted to the myelo-monocytic and megakaryocytic lineages and detected at the surface of the hematopoietic progenitor cells expressing the CD34 antigen (Fig 1A). However, Aoki et al9reported that the mouse homologue of PRR2 (mPRR2) was expressed in greater than 90% of B cells purified from the spleen, and no expression was detected on monocytes, T cells, and NK cells. The discrepancy between our results and those reported by Aoki et al9 could be explained by the fact that analyses were performed on cells isolated from different tissues. Alternatively, as shown for other cell surface receptors, the human and murine PRR2 molecules could have different expression patterns. Recently, PRR2 expression was described on human monocytes.18
Western blot analysis of immunoprecipitated PRR2 isoforms showed two bands of 64 and 72 kD corresponding to PRR2α and PRR2δ, respectively (Fig 3). The same molecular weights were observed with TF-1 and ECV304 cells (Fig 9).
Transfection of either the short or the long form of PRR2 in the murine hematopoietic cell line Da-1 induced aggregation of the cells in culture (Fig 4). We demonstrate that both isoforms of PRR2 could mediate adhesion by an homophilic mechanism (Figs 5, 6, and 7).
PRR2 isoforms form dimers at the surface of Da1/PRR2 cells, probably bycis-interactions (Fig 9A). Interestingly, no dimeric forms of PRR2 were detected in the TF-1 cell line (Fig 9C). This could explain the failure of hematopoietic cells that constitutively express PRR2 to form aggregates. Because the expression level of PRR2 in TF-1 is lower than that in the transfectants and because the aggregation of Da1/PRR2 cells was directly correlated with the PRR2 expression level, we can speculate that aggregation could be proportional to the cell surface expression level of dimeric PRR2. The functional significance of this dimerization is unknown. One can speculate thatcis-dimerization is correlated with cell surface expression levels and that it contributes to strengthen adhesion, as suggested for other adhesion molecules.19-21
Preliminary long-term culture experiments using a human bone marrow stromal layer showed no differences in colony formation in the presence or absence of the blocking MoAbs R2.477 (results not shown). The contribution of PRR2 could be masked by the presence of many other adhesion molecules expressed at the cell surface of the CD34+ progenitors. Other homophilic receptors, such as the CD66, which belongs to the carcinoembryonic family, and the ALCAM/HCA antigen, expressed on hematopoietic cells, could mediate homophilic interactions with no clear associated function.22,23 Only the PECAM-1/CD31 molecule mediates homotypic homophilic interactions between endothelial cells and heterotypic homophilic interactions between leukocytes and endothelial cells leading to leukocyte transendothelial migration.24 We showed that PRR2 is also expressed at the surface of endothelial cells isolated from umbilical cord (HUVEC; Fig 1B), on endothelial cell lines (ECV304 and EAHy924), and on placental endothelial cells (Fig 2). Confocal microscopy showed that PRR2 was concentrated at the intercellular borders of adjacent endothelial cells (Fig 8B). No PRR2 concentration was observed at the free edges of the cell layer. Because both isoforms of PRR2 could homodimerize or heterodimerize at the cell surface of endothelial cells (Fig 9B), we speculate that dimers are only formed at the cellular junction, thus mediating homophilic interactions. Altogether, these observations could suggest that PRR2 is involved in leukocyte transendothelial migration.
Most of the adhesion receptors may also reflect interactions that are important for signal transduction as described for the CD66 and PECAM-1/CD31 antigens. These molecules can be tyrosine phosphorylated in their cytoplasmic region.25 26 We observed that the long form of PRR2 was phosphorylated on tyrosine residues when cells are aggregated in culture (Fig 9D). At the present time, we do not know whether phosphorylation is directly linked to the homophilic engagement of the molecules either in an outside-in mechanism or, indirectly, via other conjugated molecules in an inside-out mechanism.
As for CD66 and CD31, hPRR2δ contains a YxxL sequence in its cytoplasmic region. This sequence is conserved between hPRR2δ and mPRR2δ and could serve as recognition sequence for SH2 domains of cytoplasmic kinases and phosphatases.
Whereas PRR2 belongs to a new family of at least three receptors related to the PVR/CD155 molecule (PVR, PRR1, and PRR2) and expressed in hematopoiesis,27 recent reports demonstrated that these molecules were also receptors for herpes simplex viruses.28 29
The identification of the role of this new antigen family is currently under investigation and should contribute to the understanding of cellular interactions in normal and pathological hematopoiesis.
ACKNOWLEDGMENT
The authors are grateful to Elisabeth Devilard, Dr Nathalie Bardin, and Dr Véronique Francès for expert technical assistance and helpful discussions. We acknowledge Drs Luc Xerri, Françoise Birg, and Claude Mawas for critically reviewing the manuscript.
Supported by INSERM, the Association pour la Recherche Contre le Cancer (ARC), and the Ligue Nationale Française Contre le Cancer (LNFCC).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Marc Lopez, PhD, INSERM U.119, 27, Bd Leı̈ Roure, 13009 Marseille, France; e-mail:lopez@marseille.inserm.fr.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal