Abstract
This work reports the molecular genetic study of a patient who suffered from Glanzmann thrombasthenia (GT). Structural analysis of the glycoprotein (GP) IIb and GPIIIa genes showed the presence of a homozygous G1846→T transversion in exon 11 of GPIIIa that changes Glu616→Stop. Cytometric and immunochemical analysis indicated that platelet GPIIb-IIIa was absent in the proband but present at normal levels in the heterozygous relatives. The following observations indicate that this mutation is responsible for the thrombasthenic phenotype of the proband. (1) We failed to detect mutations other than [T1846]GPIIIa in the coding region of both GPIIb and GPIIIa genes. (2) The G1846→T mutation was observed in either parent and a brother of the proband, but none of 100 unrelated individuals carried this defect. (3) Pulse-chase and immunoprecipitation analysis of GPIIb-IIIa complexes in cells transiently cotransfected with cDNAs encoding normal GPIIb and [T1846]GPIIIa showed neither maturation of GPIIb nor complex formation and surface exposure of GPIIb-▵GPIIIa. These observations indicate that the sequence from Glu616 to Thr762 in GPIIIa is essential for heterodimerization with GPIIb. Polymerase chain reaction-based analysis demonstrated the presence of normal levels of full-length GPIIIa-mRNA in the proband and in heterozygous relatives. In addition, a shortened transcript, with a 324-nucleotide deletion, resulting from in-frame skipping of exons 10 and 11, was detectable upon reamplification of the DNA. Thus, unlike other nonsense mutations, [T1846]GPIIIa does not lead to abnormal processing or reduction in the number of transcripts with the termination codon.
GLANZMANN described in 1918 a bleeding disorder manifested immediately after birth, characterized by a prolonged bleeding time and abnormal clot retraction, a reason why it was named thrombasthenia.1 Later on, it was recognized that platelets from these patients would not spread or aggregate and their fibrinogen content was low or absent,2-5 indicating that the fibrinogen receptor was absent or functionally defective.6,7 The Glanzmann thrombasthenia (GT) has been classified into types I and II.8 In type I GT, platelets lack fibrinogen and clot retraction and they show absence of glycoprotein (GP) IIb-IIIa complexes at their surface. In type II GT, the platelets contain detectable amounts of fibrinogen, the clot retraction capability may vary from low to moderate, and the expression of GPIIb-IIIa complexes is 10% to 20% of the control values. In cases of GT termed variants, the platelets possess normal or near normal (60% to 100%) expression of dysfunctional receptors.7,9These differences may justify the clinical heterogeneity of this bleeding disorder.10 Knowledge of the structural organization of the GPIIb and GPIIIa genes11-15 has made it possible to establish the association of thrombasthenic phenotypes with distinct genetic lesions in one or both genes (for a recent review, see French and Coller16).
The present investigation is aimed at elucidating the molecular genetic lesion in a 20-year-old Caucasian woman who suffered from GT. Structural analysis of the GPIIb and GPIIIa genes showed the existence of a novel homozygous G1846→T transversion in exon 11 of GPIIIa that changes Glu616→Stop. Reverse transcription-polymerase chain reaction (RT-PCR) analysis demonstrated that full-size [T1846]GPIIIa-mRNA was the only detectable transcript. Transfection and immunoprecipitation analysis demonstrated that the translational product of [T1846]GPIIIa does not complex with GPIIb; therefore, it is the molecular lesion underlying the lack of platelet expression of GPIIb-IIIa receptor and the thrombasthenic phenotype of the patient.
MATERIALS AND METHODS
Clinical data.
The proband is a 20-year-old Caucasian woman clinically diagnosed of GT who referred a history of mucocutaneous bleeding episodes and unprovoked bruising that started soon after birth and copious menstrual hemorrhages. Her parents neither suffer from any hematological disorder nor recognize consanguinity. The hematological studies of the propositus showed a bleeding time of 18 minutes. The platelet count was 305,000/μL, the platelet adhesion capacity was reduced (7%), and no aggregation was detected either spontaneously or in response to agonists. Plasma fibrinogen content was 250 mg/dL and there was no clot retraction.
Preparation of platelets and analysis of GPIIb-IIIa and fibrinogen content.
Blood samples from the patient and relatives were obtained after informed consent was obtained by Dr G. Iruı́n (Department of Hematology, Hospital Cruces, Bilbao, Spain). Platelets were separated by differential centrifugation at 120g for 20 minutes at room temperature, sedimented at 1,000g for 10 minutes, and washed several times with an isoosmotic buffer (10 mmol/L Tris, 150 mmol/L NaCl, 1 mmol/L EDTA, pH 7.4). The platelets were sonicated at 4°C and centrifuged at 100,000g for 1 hour to separate the soluble and particulate fractions. Competitive solid-phase enzyme immunoassays17 were performed to determine the platelet content of GPIIb, GPIIIa, and fibrinogen using monoclonal antibodies (MoAbs) for GPIIb (M3), GPIIIa (P37 and P95.2), and fibrinogen.18,19 Fibrinogen was determined in the 100,000g supernatant of sonicated platelets, and GPIIb-GPIIIa in detergent (3% sodium dodecyl sulfate [SDS]) solubilized particulate fraction. The mean ± SD of control platelet fibrinogen content was 6% ± 0.6% (wt/wt) of the total 100,000g supernatant protein. The mean ± SD of the GPIIb-IIIa content in control platelets was as follows: for GPIIb, 3.7 ± 0.7 (n = 17); for GPIIIa, 2.4 ± 0.7 (n = 18), expressed as the percentage of the total protein of the solubilized particulate fraction. The total protein content was determined using the method of Markwell et al.20
The detergent-solubilized particulate fraction of sonicated platelets was also used for Western blot analysis of the GPIIb-IIIa content. Protein (2.5 to 10 μg) was electrophoresed on 7.5% SDS-polyacrylamide gels under reducing and nonreducing conditions and electrotransferred on nitrocellulose membranes. The blotted membranes were blocked by incubation in phosphate-buffered saline (PBS)-10% defatted milk powder and then incubated with anti-GPIIIa (P37 or P95.2) and anti-GPIIb (M3) specific antibodies. After incubation in a 1:3,000 dilution of antimouse IgG-horseradish peroxidase (Bio-Rad Laboratories, Hercules, CA), immuno-reactive bands were shown with H2O2/4-chloro-1-naphtol and quantitated by computerized image analyzer.
Flow cytometry.
Platelets were harvested using 0.5 mmol/L EDTA in PBS, washed twice with the same buffer, resuspended at a density of 106cells/100 μL, and incubated with a MoAb directed against either GPIIb (M3), GPIIIa (P37 or P95.2), or GPIb at 4°C for 20 minutes. Cells were then washed and resuspended in 50 μL of PBS containing a 1:20 dilution of fluorescein isothiocyanate (FITC)-conjugated F(ab′)2 fragment of rabbit antimouse Ig (Dako A/S, Glostrup, Denmark), followed by incubation at 4°C for 20 minutes. Finally, after three washes with PBS, the cell suspension was adjusted to a density of 2.5 × 106 cells/mL and the surface fluorescence was analyzed in a Coulter flow cytometer, model EPICS XL (Coulter, Hialeah, FL).
Single-stranded conformational polymorphism (SSCP) analysis, cloning, and sequencing of PCR-amplified genomic DNA fragments of the GPIIb and GPIIIa genes.
Genomic DNA was isolated from peripheral blood specimens. PCR amplification of DNA fragments encompassing one or more exons of GPIIIa or GPIIb was performed with oligonucleotides complementary to the intronic flanking regions using Taq polymerase according to the protocol recommended by Perkin-Elmer Cetus (Norwalk, CT). MgCl2 concentration and annealing temperatures were optimized for each pair of primers. Screening for mutations in GPIIb and GPIIIa was performed using cold SSCP analysis.21,22 A fragment of 198 bp comprising exon-11 of GPIIIa was amplified with the following oligonucleotides: sense, 5′-GGGATACGCTTAGGCTTGCT-3′; antisense, 5′-AACCTGGGTGTGTGCAACTCT-3′. To increase the sensitivity of the method, the amplification products were digested with HinfI to yield two fragments of 140 and 58 bp, and the digests were electrophoresed at 12°C in a nondenaturing 16% acrylamide gel containing 8.7% glycerol. DNAs showing altered electrophoretic mobility patterns of single-stranded bands were cloned directly into a T vector,23 and at least 10 positive clones from each amplification were pooled and their sequence was determined24 with the T7 sequencing kit of Pharmacia Biotech (Uppsala, Sweden). Sequence analyses were performed as described by Marck.25
The carrier status for the G1846→T mutation in the kindred was determined by allele-specific PCR (ASPCR)26analysis. Genomic DNA was used as template for the amplification of a 148-bp fragment comprising exon 11, using as sense primer either 5′-TCCTTCAGAGAATGTGTGG-3′ (normal) or 5′-TCCTTCAGAGAATGTGTGT-3′ (mutant), whose 3′ ends are complementary to the normal or the mutated base, respectively, and the antisense primer 5′-AACCTGGGTGTGTGCAACTCT-3′. Each amplification cycle consisted of 30 seconds of denaturation at 94°C, 1 minute of annealing at 64°C, and 2 minutes of extension at 72°C. Portions of the PCR products were analyzed by agarose gel electrophoresis.
Construction of mammalian expression vectors containing normal or mutant [T1846]GPIIIa cDNA.
[T1846]GPIIIa cDNA was prepared by the splicing by overlap extension procedure (SOE).27 Human GPIIIa cDNA contained in the plasmid pBluescript KS was used as template to generate two PCR overlapping fragments, herewith referred to as 5′ and 3′ segments: the 5′ segment was amplified using the oligonucleotide sense IIIa (1670-1689), 5′-ACTGCAACTGTACCACGCGT-3′, and the mutated antisense primer (1861-1839), 5′-AACTTCTTACACTACACACAT-3′; the 3′ segment was obtained using as sense mutant primer (1837-1857), 5′-GAATGTGTGTAGTGTAAGAAG-3′, and the primer Xho I antisense IIIa (2319-2296), 5′-TGATAATGACTCGAGGATGACTGC-3′. Bases substituted to generate mutation in overlapping primers are underlined. The 5′ and 3′ PCR products were used as template in a new round of PCR amplification with the oligonucleotides sense (1670-1689) andXho I antisense (2319-2296) described above; the amplified DNA was digested with Xho I and partially with Not I, because another internal Not I site is found in the cDNA sequence. The 581-bp digestion product was then exchanged for the wild sequence in the pBluescript KS+-GPIIIa. Both wild-type and mutated GPIIIa-containing plasmids were subjected to total digestion with Xho I and partial digestion with BamHI, and the released complete cDNA sequences were subcloned into the expression vector pcDNA3 digested with the same enzymes. Nucleotide sequence analysis was performed to confirm the proper insertion of the amplified mutant product and the absence of errors potentially caused by the Taq polymerase.
Wild-type GPIIb-cDNA in pBluescript, obtained from Dr. D. Phillips (COR Therapeutics, Inc, San Francisco, CA), was subcloned into theHindIII site of pcDNA3.
Cell culture and transfection.
CHO cells were grown in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal calf serum in 5% CO2-95% air at 37°C. Cells were incubated with 3.5 μg of the plasmid pcDNA3-GPIIb and/or 3.5 μg of either pcDNA3-GPIIIa or pcDNA3-[T1846]GPIIIa in the presence of 100 μg/mL diethyl aminoethyl (DEAE)-dextran and 100 μmol/L chloroquine diphosphate.28 After 4 hours, cells were exposed to 10% dimethyl sulfoxide (DMSO) in PBS for 6 minutes and rinsed with PBS; completed medium was then added and the incubation was continued for 48 or 72 hours.
Biotin labeling and immunoprecipitation analysis of GPIIb-IIIa complexes from CHO cells cotransfected with cDNAs encoding GPIIb and either normal or mutant [T1846]GPIIIa.
Biotin labeling and immunoprecipitation of cell surface GPIIb-IIIa complexes was performed as follows. CHO cells transfected with cDNA encoding normal GPIIb and either normal or mutated [T1846]GPIIIa were washed twice with PBS and incubated in 2 mL of PBS containing 5 mmol/L biotin-NHS (D-biotin-N-hydroxy succinimidester; Boehringer Mannheim, Mannheim, Germany). At the end of the incubation, the cells were washed with PBS and treated 30 minutes at 4°C with lysis buffer (50 mmol/L Tris, pH 7.4, 150 mmol/L NaCl, 2 mmol/L phenylmethyl sulfonyl fluoride [PMSF], 1% Triton X-100, 0.05% Tween 20, and 0.03% sodium azide) and GPIIb, GPIIIa, or GPIIb-IIIa complexes were immunoprecipitated as described before.29 The immunoprecipitates were separated by electrophoresis at 40 V in 0.1% SDS-7.5% polyacrylamide slab gels. Proteins were transferred to a nitrocellulose membrane in the presence of Towbin buffer (25 mmol/L Tris, 192 mmol/L glycine) containing 15% methanol and 1.3 mmol/L SDS. The membranes were blocked by incubation in PBS-10% defatted milk powder, washed with PBS-0.1% Tween 20, and incubated in a 1:3,000 dilution of avidin-horseradish peroxidase (Bio-Rad Laboratories, Hercules, CA) for 1 hour at room temperature. The color of biotin-containing materials was developed by incubation in PBS containing 0.015% H202 and 0.5 mg/mL of 4CN (4-chloro-1-naphtol).
For biotin labeling and detection of total (surface and intracellular) GPIIb-IIIa complexes, detergent lysates of transfected CHO cells were incubated with 5 mmol/L biotin-NHS for 2 hours at room temperature, followed by immunoprecipitation with specific MoAbs against GPIIIa (P37), GPIIb (M3), or GPIIb-IIIa (CII), as described above.
Metabolic labeling of transfected CHO cells.
CHO cells were transiently transfected as described above. Forty-eight hours after transfection, the cells were incubated in DMEM without methionine for 30 minutes and then pulsed for 30 minutes with [35S]methionine (200 μCi/mL). The plates were washed 3 times with PBS containing 1 mg/mL cold methionine and incubated with DMEM containing unlabeled methionine for 0, 0.5, 2, and 8 hours. Cells were washed 5 times with PBS containing cold methionine and extracted after 30 minutes in 1 mL of lysis buffer. Immunoprecipitation analysis was performed as described for biotin-labeled cells. The immunoprecipitates were electrophoresed at 50 V in 0.1% SDS-7.5% polyacrylamide slab gels. The gels were vacuum dried and exposed to hypersensitive x-ray film.
RT-PCR analysis of GPIIb and GPIIIa mRNAs.
Lymphocytes from the proband, her father, and normal individuals were immortalized with Epstein-Barr virus as previously described.30 Total RNA was extracted using the guanidinium thiocyanate method,31 reverse-transcribed with the primer IIIa-AS (2329-2309; 5′-TGGCACAGGCTGATAATGATC-3′), and used as template for the PCR amplification of two overlapping GPIIIa cDNA fragments. The 5′ segment was amplified using the oligonucleotides IIIa-S (63-84), 5′-ATGTGTGCCTGGTGCTCTGAT-3′, and IIIa-AS (1439-1419), 5′-GGGCGATAGTCCTCCTCTGA-3′; the 3′ cDNA fragment was amplified with oligo IIIa-S (1366-1385), 5′-GAGTGTGGGGTATGCCGTTG-3′, and IIIa-AS (2329-2309). To search for alternative spliced forms of GPIIIa-mRNA, portions of the PCR products were reamplified with the same primers. The amplified DNA was analyzed by electrophoreses in agarose gels and cloned in a T vector23 for further sequence analysis.
PCR-based quantitation of platelet GPIIIa-mRNA with the TaqMan system.
The instrumentation and the fluorogenic probes of the Perkin-Elmer Cetus (Norwalk, CT) LS-50B TaqMan System were used for PCR-based quantitation of GPIIIa mRNA in platelets from the proband, her heterozygous relatives, and normal individuals, as previously described.32 Specific oligonucleotide probes, R-CTCTGGCGCGTTCTTCCTCAAATTTAGC-Q and R-ATGCCCT-Q-CCCCCATGCCA TCCTGCGT, were designed to anneal to targets located within PCR-amplified fragments of 132 bp (2157-2288) of GPIIIa or 295 bp (2141-2435) of β-actin cDNA, respectively. The location of the reporter and quencher dyes are indicated by R and Q, respectively. Because of the scant amount of material made available to us, only two different amounts of RNA were used for amplification of GPIIIa and β-actin DNA fragments using the rTth polymerase XL from Perkin-Elmer Cetus. Briefly, in a first step, mRNA was reverse-transcribed with the antisense primers in the presence of 1.1 mmol/L Mn(OAc)2 for 30 minutes at 60°C. The PCR amplification was then performed by chelating the Mn2+ and adding 0.8 mmol/L Mg(OAc)2, the sense primer, and the specific TaqMan probe. Thirty amplification cycles were performed, consisting of 15 seconds at 95°C and 15 seconds at 65°C. After PCR cycling, 25-μL portions were taken from each sample and the fluorescence was measured using a 488-nm excitation wavelength and 518- and 580-nm emission wavelengths for the reporter and quencher dyes, respectively. Values were corrected for internal quenching and expressed as GPIIIa/β-actin fluorescence ratios.
Materials.
Restriction enzymes were obtained from Boehringer (Mannheim, Germany) and DNA sequencing reagents were from Pharmacia Biotech (Uppsala, Sweden). The pcDNA3 expression vector was from Invitrogen (San Diego, CA). Most other reagents were purchased from Sigma Chemical Co (St Louis, MO) or from Merck (Darmstadt, Germany). [35S]-methionine (specific activity [SA], 1,000 Ci/mmol) and [35S]-dATP (SA, 1,000 Ci/mmol) were obtained from Amersham Ibérica (Madrid, Spain). MoAbs specific for GPIIIa, GPIIb, GPIIb-IIIa heterodimer, and fibrinogen were provided by Dr J. González (Instituto Roscasolano (CSIC), Madrid, Spain). Anti-GPIb/IX MoAb was purchased in Sigma Hispania (Madrid, Spain).
RESULTS
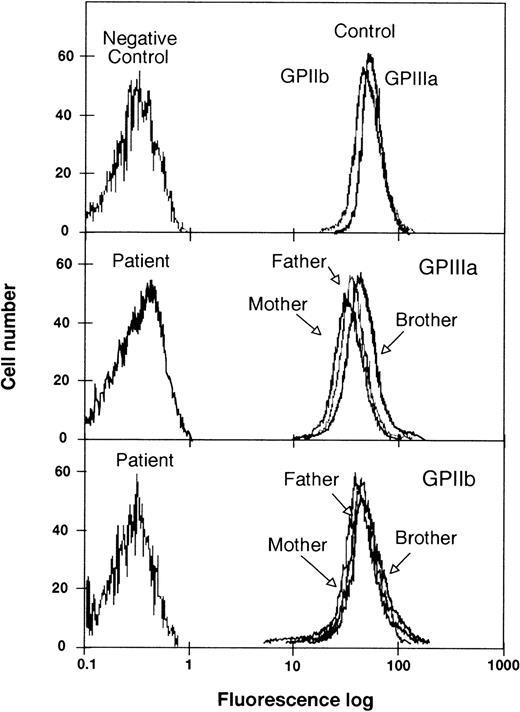
The proband refers a history of frequent bleeding episodes, particularly mucocutaneous, that started immediately after birth. However, no family history of hemorrhage disorders or consanguinity was recognized by her parents. Analytical features were compatible with a diagnostic of GT. The absence of platelet GPIIb-IIIa complexes by flow cytometric analysis using specific MoAbs (Fig 1) confirmed that the proband was a type I case of GT. Her parents and her brother showed platelet expression of GPIIb-IIIa within the range of normal individuals. Moreover, enzyme immunoassay and immunoblotting analysis using a different anti-GPIIIa (P95.2) MoAbs confirmed the absence of platelet GPIIb-IIIa in the proband and levels within the normal range in her brother and her parents (Table 1). In agreement with the latter observation, intraplatelet fibrinogen content was 15% of the platelet control in the proband and within normal levels in the other family members. The apparently normal, or near normal, levels of platelet GPIIb-IIIa in the brother and parents of the proband contrast with previous reports showing a marked reduction in the surface exposure of this heterodimer in heterozygous individuals for other mutations.
Flow cytometric analysis of GPIIb-IIIa content in platelets from the proband, her parents, and her brother. The fluorescence analysis was performed in a Coulter cytometer, model Epics XL. Results are expressed as semilog plots of cell number versus fluorescence intensity. The upper panel shows the fluorescence signals of GPIIIa and GPIIb from control platelets. The negative control represents the fluorescent signal of platelets without antibodies. The middle panel shows the labeling of GPIIIa with the MoAb P95.2 in platelets from the proband, her parents, and her brother. The lower panel shows the results of labeling GPIIb with the MoAb M3 in platelets from the proband, her parents, and her brother. For the sake of clarity, the original plots have been redrawn.
Flow cytometric analysis of GPIIb-IIIa content in platelets from the proband, her parents, and her brother. The fluorescence analysis was performed in a Coulter cytometer, model Epics XL. Results are expressed as semilog plots of cell number versus fluorescence intensity. The upper panel shows the fluorescence signals of GPIIIa and GPIIb from control platelets. The negative control represents the fluorescent signal of platelets without antibodies. The middle panel shows the labeling of GPIIIa with the MoAb P95.2 in platelets from the proband, her parents, and her brother. The lower panel shows the results of labeling GPIIb with the MoAb M3 in platelets from the proband, her parents, and her brother. For the sake of clarity, the original plots have been redrawn.
Quantitation of Platelet GPIIb and GPIIIa
| . | Flow Cytometry (mean fluorescence channel) . | Western Blotting (% of control) . | Enzyme Immunoassay (% of control) . | ||||
|---|---|---|---|---|---|---|---|
| GPIIb . | GPIIIa . | GPIb . | GPIIb . | GPIIIa . | GPIIb . | GPIIIa . | |
| Control | 38 ± 7 (10) | 46 ± 9 (10) | 24 ± 5 (10) | 100 | 100 | 100 | 100 |
| Proband | ND | ND | 18 | ND | ND | ND | ND |
| Brother | 37 | 45 | 22 | 96 ± 7 | 84 ± 7 | 81 | 91 |
| Father | 30 | 36 | 21 | 86 ± 10 | 78 ± 10 | 85 | 80 |
| Mother | 30 | 37 | 26 | 72 ± 10 | 85 ± 14 | 85 | 72 |
| . | Flow Cytometry (mean fluorescence channel) . | Western Blotting (% of control) . | Enzyme Immunoassay (% of control) . | ||||
|---|---|---|---|---|---|---|---|
| GPIIb . | GPIIIa . | GPIb . | GPIIb . | GPIIIa . | GPIIb . | GPIIIa . | |
| Control | 38 ± 7 (10) | 46 ± 9 (10) | 24 ± 5 (10) | 100 | 100 | 100 | 100 |
| Proband | ND | ND | 18 | ND | ND | ND | ND |
| Brother | 37 | 45 | 22 | 96 ± 7 | 84 ± 7 | 81 | 91 |
| Father | 30 | 36 | 21 | 86 ± 10 | 78 ± 10 | 85 | 80 |
| Mother | 30 | 37 | 26 | 72 ± 10 | 85 ± 14 | 85 | 72 |
Flow cytometry, Western blotting analysis, and enzyme immunoassays were performed as described in Materials and Methods. In the flow cytometry analysis, GPIIb, GPIIIa, and GPIb were determined by using the M3, P95.2, and anti-GPIb/IX MoAbs, respectively. The values are means ± SD of the mean fluorescence channel of the number of observations indicated in parentheses. In Western blotting and enzyme immunoassay analysis, the GPIIb and GPIIIa content in the detergent solubilized particulate fraction of sonicated platelets was detected using the M3 and P95.2 MoAbs, respectively. Western blotting data were quantitated by computerized image analysis. The mean values ± SD in control platelets were as follows: for GPIIb, 58 ± 7 (n = 6); for GPIIIa, 40 ± 7 (n = 6), expressed in arbitrary units per microgram of protein. The values for the proband and her relatives are the mean ± SE of six different analyses of the same platelet preparation run in duplicates and are expressed as the percentage of the control values. The mean ± SD values for the enzyme immunoassay of GPIIb/IIIa in control platelet were as follows: for GPIIb, 3.7 ± 0.7 (wt/wt; n = 18); for GPIIIa, 2.4 ± 0.7 (wt/wt; n = 17), expressed as the percentage of the total protein content.
Abbreviation: ND, not detectable.
Structural analysis of the GPIIb and GPIIIa genes.
GPIIb and GPIIIa genes were screened for mutations by the cold SSCP21 22 of PCR-amplified exons from genomic DNA. Only the amplification product of exon 11 of GPIIIa showed a distinct pattern of bands in the proband. Figure 2 depicts the electrophoretic analysis of HinfI digests of exon 11. All of the analyzed members of the kindred showed a distinct band, indicated by an arrow, that is not observed in the control. The relatives, but not the proband, show an additional band that is also present in the control. This observation indicated that the three members of the kindred appeared to be heterozygous for the mutation carried by the proband. Sequence analysis showed the presence of a homozygous G1846→T transversion that changes Glu616→Stop (Fig 3). The predicted translational product of this messenger is a truncated protein (ΔGPIIIa) in which a segment encompassing 76 amino acid residues of the extracellular domain, the transmembrane domain, and the intracellular carboxyterminal end of the protein would be absent. The carrier status of the kindred members was further verified by allele-specific PCR amplification (Fig4). As expected, amplification from the proband’s DNA was only observed with the mutant primer.
SSCP analysis of exon 11 of GPIIIa amplified from genomic DNA. Genomic DNA fragment of 198 bp comprising exon 11 of GPIIIa and intronic flanking regions was amplified as described in Materials and Methods. The PCR products were digested with HinfI to yield fragments of 140 and 58 bp and electrophoresed in nondenaturing 16% acrylamide slab gels containing 8.7% glycerol. The arrow points to a distinct band shown by the patient, her parents, and her brother that is absent in the control (wt) DNA.
SSCP analysis of exon 11 of GPIIIa amplified from genomic DNA. Genomic DNA fragment of 198 bp comprising exon 11 of GPIIIa and intronic flanking regions was amplified as described in Materials and Methods. The PCR products were digested with HinfI to yield fragments of 140 and 58 bp and electrophoresed in nondenaturing 16% acrylamide slab gels containing 8.7% glycerol. The arrow points to a distinct band shown by the patient, her parents, and her brother that is absent in the control (wt) DNA.
Identification of a G1846→T mutation in exon 11 of GPIIIa. Exon 11 of GPIIIa was amplified from genomic DNA as described in Materials and Methods. The amplification products were cloned in a T-vector and the primary nucleotide sequence of pooled DNA was determined in both directions. The figure shows a fragment of the sequencing ladder of the sense strand. The arrows point to a homozygous G1846→T transversion that changes Glu616→Stop.
Identification of a G1846→T mutation in exon 11 of GPIIIa. Exon 11 of GPIIIa was amplified from genomic DNA as described in Materials and Methods. The amplification products were cloned in a T-vector and the primary nucleotide sequence of pooled DNA was determined in both directions. The figure shows a fragment of the sequencing ladder of the sense strand. The arrows point to a homozygous G1846→T transversion that changes Glu616→Stop.
Specific amplification of normal and [T1846]-GPIIIa alleles. A DNA fragment of 148 bp encompassing exon 11 and intronic flanking regions of GPIIIa was amplified from genomic DNA of a control, the proband, her parents, and her brother. Each DNA was amplified using a sense primer whose 3′ end was complementary to either the normal sequence (Wt) or to the mutant base (Mut). The amplification products were electrophoresed in a 3% agarose gel and the DNA bands were stained with ethidium bromide.
Specific amplification of normal and [T1846]-GPIIIa alleles. A DNA fragment of 148 bp encompassing exon 11 and intronic flanking regions of GPIIIa was amplified from genomic DNA of a control, the proband, her parents, and her brother. Each DNA was amplified using a sense primer whose 3′ end was complementary to either the normal sequence (Wt) or to the mutant base (Mut). The amplification products were electrophoresed in a 3% agarose gel and the DNA bands were stained with ethidium bromide.
PCR-based analysis of mRNA-GPIIIa.
Because of difficulties in obtaining platelets from this kindred when this study was started and because GPIIIa is expressed in lymphocytes,33 to have an endless source of genetic material, we immortalized cells from the proband and her father with the Epstein-Barr virus. In agreement with the cytometric analysis performed in platelets (Fig 1), the lymphoblasts from the proband showed absence of GPIIIa when compared with her father who is heterozygous for the same mutation (results not shown).
PCR amplification of reverse-transcribed RNA from the proband and one heterozygous relative was performed with oligonucleotides encompassing the entire coding sequence of GPIIIa. Sequence analysis showed that [T1846]GPIIIa was the only form of mRNA found in the proband, whereas apparently similar proportions of mutant [T1846] and normal [G1846]GPIIIa messengers were found in heterozygous individuals. To search for less represented alternative spliced transcripts, we reamplified portions of the first PCR with the same primers. In these conditions, we detected an additional shortened product in the proband and her father that was not observed in the control (results not shown). Sequencing of this shortened transcript showed an in-frame internal deletion of 324 nucleotides as a result of skipping exons 10 and 11; in addition, the first codon of exon 12 changed from AAG(Lys) to GAG(Glu). The translation of this transcript should yield a protein with a deletion of residues 538 to 645.
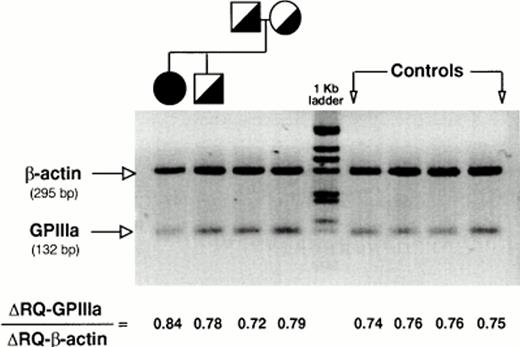
To determine the level of platelet GPIIIa-mRNA, we performed a quantitative TaqMan PCR-based determination of messenger.32The assays were performed under predetermined conditions of cycle number and amount of RNA in which the TaqMan fluorescence emission was a function of the DNA target concentration. Figure 5 depicts the results obtained at one of the RNA concentrations used. RQ represents the ratio of fluorescence emission of the reporter dye to that of a quencher that is a passive internal standard. The fluorescence dye is located in a probe that hybridizes within the target sequence and its signal intensity is proportional to the number of amplification cycles and amount of PCR product. The ΔRQ is calculated by substracting the RQ value obtained by a PCR amplification without template. The ΔRQ-GPIIIa to ΔRQ-β-actin ratio in the proband was virtually identical to the values of the other kindred members and to the normal controls (Fig 5, bottom). This observation indicates that mutant [T1846]GPIIIa allele was expressed at similar rates as the normal one.
PCR-based determination of platelet mRNA-GPIIIa. PCR-based quantitation of β-actin and GPIIIa mRNAs in platelets from the proband, heterozygous relatives, and normal individuals was performed using the instrumentation and the fluorogenic probes of the Perkin-Elmer Cetus LS-50B TaqMan System as described in Materials and Methods. R and Q denote the fluorescence of the reporter and the quencher dyes, respectively. Values were corrected for internal quenching and expressed as GPIIIa/β-actin fluorescence ratios.
PCR-based determination of platelet mRNA-GPIIIa. PCR-based quantitation of β-actin and GPIIIa mRNAs in platelets from the proband, heterozygous relatives, and normal individuals was performed using the instrumentation and the fluorogenic probes of the Perkin-Elmer Cetus LS-50B TaqMan System as described in Materials and Methods. R and Q denote the fluorescence of the reporter and the quencher dyes, respectively. Values were corrected for internal quenching and expressed as GPIIIa/β-actin fluorescence ratios.
Heterologous expression of normal or [T1846]GPIIIa.
CHO cells were transiently cotransfected with the plasmid pcDNA3-GPIIb and either pcDNA3-GPIIIa or pcDNA3-[T1846]GPIIIa. To investigate the surface exposure of GPIIb-IIIa, we labeled intact cells with biotin, and the GPIIb-IIIa complexes were immunoprecipitated with anti-GPIIb (M3) or anti-GPIIIa (P37) MoAbs. Bands migrating as native GPIIbH (the disulfide-linked heavy chain of GPIIb) and GPIIIa were found in the immunoprecipitates from cells transfected with normal GPIIb and GPIIIa cDNAs (Fig 6A); however, no biotin-labeled products were found in cells transfected with the void pcDNA3 plasmid or cotransfected with normal GPIIb and mutant [T1846]GPIIIa.
Immunoprecipitation of biotin-labeled surface or total (surface and intracellular) proteins from CHO cells cotransfected with cDNAs encoding GPIIb and either normal or [T1846]GPIIIa. CHO cells were transiently cotransfected with cDNAs encoding GPIIb and either normal or [T1846]GPIIIa in the expression plasmid pcDNA3. (A) Protein labeling was performed by exposure of intact cells to biotin-NHS, and the GPIIb-IIIa complexes were immunoprecipitated with either anti-GPIIIa (P37) or anti-GPIIb (M3) MoAbs as described in Materials and Methods. (B) The experimental design was similar to that described in (A), except that total cell lysates rather intact cells were exposed to biotin-NHS and the immunoprecipitation was also performed with a GPIIb-IIIa complex-specific antibody (CII). Mock cells were transfected with void pcDNA3 plasmid.
Immunoprecipitation of biotin-labeled surface or total (surface and intracellular) proteins from CHO cells cotransfected with cDNAs encoding GPIIb and either normal or [T1846]GPIIIa. CHO cells were transiently cotransfected with cDNAs encoding GPIIb and either normal or [T1846]GPIIIa in the expression plasmid pcDNA3. (A) Protein labeling was performed by exposure of intact cells to biotin-NHS, and the GPIIb-IIIa complexes were immunoprecipitated with either anti-GPIIIa (P37) or anti-GPIIb (M3) MoAbs as described in Materials and Methods. (B) The experimental design was similar to that described in (A), except that total cell lysates rather intact cells were exposed to biotin-NHS and the immunoprecipitation was also performed with a GPIIb-IIIa complex-specific antibody (CII). Mock cells were transfected with void pcDNA3 plasmid.
To examine the intracellular presence of GPIIb and/or GPIIIa as either monomers or heterodimers, we labeled total cell lysates with biotin, and GPIIb, GPIIIa, or GPIIb-IIIa complexes were immunoprecipitated as described for the biotin surface-labeled cells using anti-GPIIb (M3), anti-GPIIIa (P37), or GPIIb-IIIa complex-specific (CII) MoAbs. Immunoprecipitates with apparent molecular weights corresponding to proGPIIb, GPIIIa, or ΔGPIIIa were observed in extracts of cells transfected with either normal GPIIb, normal GPIIIa, or mutant ΔGPIIIa, using antibodies directed against each subunit, but not with a complex-specific antibody (Fig 6B). As expected, whether subunit- or complex-specific antibodies were used, GPIIbH and GPIIIa were detected in extracts of cells coexpressing normal GPIIb and GPIIIa (Fig 6B); proGPIIb was detectable only using the anti-GPIIb MoAb, suggesting that it was present mainly as a monomer. Immunoprecipitation using a MoAb against either GPIIb or GPIIIa yielded proGPIIb or ΔGPIIIa, respectively, in cells coexpressing normal GPIIb and mutant ΔGPIIIa. Unlike in extracts from cells coexpressing normal subunits, we failed to immunoprecipitate heterodimers using a complex-specific MoAb (Fig 6B), suggesting that the mutant ΔGPIIIa does not complex with GPIIb. The absence of GPIIb cleavage into heavy and light chains suggested either that ΔGPIIIa did not complex with GPIIb or that heterodimers had not been transported into the Golgi complex, where this process takes place.34 35 To test whether the lack of transport of GPIIb-ΔGPIIIa out of the endoplasmic reticulum was due to formation of unstable heterodimers, we performed a pulse-chase analysis of cells transiently cotransfected with both subunits. Cells were pulse-labeled for 30 minutes with [35S]methionine and then chased with medium containing unlabeled methionine for 0.5, 2, or 8 hours before GPIIb-IIIa complexes were immunoprecipitated with an anti-GPIIb MoAb. Figure 7 shows the reciprocal changes in the labeling of proGPIIb and GPIIbH as a function of the chasing time in cells coexpressing normal GPIIb and GPIIIa proteins, indicating stability and normal transport of GPIIb-IIIa complexes into the Golgi apparatus. In contrast, proGPIIb was immunoprecipitated with anti-GPIIb in cells cotransfected with normal GPIIb and mutant ΔGPIIIa, but GPIIbH or GPIIIa did not. However, labeled material migrating like ΔGPIIIa was detected upon precipitation with an anti-GPIIIa antibody. These observations further verify that the mutant ΔGPIIIa does not complex to GPIIb.
Pulse-chase analysis of the stability of normal or ▵GPIIIa-IIb complexes. CHO cells were transiently cotransfected with cDNAs encoding GPIIb and either normal or [T1846]GPIIIa in the expression plasmid pcDNA3. Cells were pulse-labeled with [35S]-methionine for 30 minutes and then chased with medium containing unlabeled methionine. At the indicated times, the cells were lysed and labeled proteins were immunoprecipitated with the indicated antibodies. The immunoprecipitates were analyzed by electrophoresis as described in Materials and Methods.
Pulse-chase analysis of the stability of normal or ▵GPIIIa-IIb complexes. CHO cells were transiently cotransfected with cDNAs encoding GPIIb and either normal or [T1846]GPIIIa in the expression plasmid pcDNA3. Cells were pulse-labeled with [35S]-methionine for 30 minutes and then chased with medium containing unlabeled methionine. At the indicated times, the cells were lysed and labeled proteins were immunoprecipitated with the indicated antibodies. The immunoprecipitates were analyzed by electrophoresis as described in Materials and Methods.
DISCUSSION
Pathophysiological significance of the G1846→T mutation of GPIIIa.
Clinical and analytical studies supported the conclusion that the proband suffered from a type I GT. We screened the GPIIb and GPIIIa genes for mutations and the only structural change found was a novel mutation, a G1846→T transversion, lying within exon 11 of GPIIIa, that changes Glu616 to Stop. Heterozygosity of both parents and the brother of the proband for this mutation was verified by different analytical procedures. Because both ancestors denied consanguinity, their heterozygosity for the [T1846]GPIIIa mutation could indicate a high incidence of this mutation. However, inasmuch as the ancestors of either parent were from two little villages in the northeastern part of Spain, separated only by 10 Km, it is probable they could be consanguineous without knowing. Moreover, our inability to find [T1846]GPIIIa in more than 100 DNAs from unrelated individuals indicated that this substitution was not a polymorphism and, together with the transfection data, strongly suggested that it was associated with the thrombasthenic phenotype of the patient.
RT-PCR analysis of mRNA GPIIIa.
Because lymphocytes express GPIIIa associated with α subunits other than GPIIb,33 we decided to use lymphoblasts as an endless source of genetic material as well as a convenient mean of investigating the functional repercussion of structural changes in GPIIIa.36 37 Lymphoblasts from the proband did not show surface GPIIIa. In contrast, her father, heterozygous for the G1846→T mutation, showed exposure of GPIIIa similar to either a normal control or a GT patient carrying a mutation in GPIIb (results not shown).
Nonsense mutations are known to force alternate forms of splicing with the frequent result of skipping of one or more exons.38PCR-based analysis of mRNAs indicate that the G1846→T mutation does not alter the processing of GPIIIa-mRNA, because both normal and mutant alleles seem to be expressed at similar rates. The scarce representation of a shortened messenger, with an in-frame deletion of 324 nucleotides, found upon reamplification, suggests that its translational product may not reach a sufficient level as to play a significant functional role. Intraexonic sequences are known to influence splice site selection. The importance of the first two and the last three bases of the exon in the splicing process has been recognized,39 but the role of more internal sequences, as it is the case in the present investigation, has not been yet elucidated. Moreover, whether a nonsense mutation would alter the amount of transcript carrying the stop codon is unclear. Nonsense mutations in GPIIb have been reported to cause either no change40,41 or marked reduction42 in the amount of messenger. These differences appear to be consistent with the proposal that the effect of nonsense mutations is position related, in that the closer to the 5′ end of the coding sequence, the lower the amount of transcript.43
Heterologous expression of normal or [T1846]GPIIIa cDNA.
In principle, the lack of surface exposure of GPIIb-ΔGPIIIa could be the result of either a lack of heterodimerization or a perturbation in the processing and/or intracellular trafficking of heterodimers. Data in this work support the conclusion that the Glu616→Stop mutation in GPIIIa prevents the formation of heterodimers with GPIIb. The lack of immunoprecipitable GPIIbH indicates a deficient association of proGPIIb with ΔGPIIIa and agrees with the postulate that endoproteolytic cleavage of intracellular GPIIb requires its association with GPIIIa and transport from the endoplasmic reticulum into the Golgi complex.44Pulse-chase analysis to determine the stability of GPIIb-ΔGPIIIa heterodimers further verified that the mutated ΔGPIIIa does not complex to GPIIb. The predicted translational product of [T1846]GPIIIa-cDNA is a protein truncated from Glu616 to Thr762, 85% of the normal size, in which the cysteine-rich repeat domain45 is conserved but lacks the transmembrane and cytoplasmic regions. Truncated forms of GPIIb and GPIIIa seem to form soluble ΔGPIIb-ΔGPIIIa heterodimers capable of binding ligand.46,47 The transmembrane region of GPIIIa was reported to be required for cellular retention of the monomeric subunit.44 However, in agreement with a previous observation on the αβ1 integrin,48 a truncated GPIIIa from Ile693 to Thr762, lacking the transmembrane and cytoplasmic domains, was capable of assembly and surface exposure of GPIIb-ΔGPIIIa complexes.46 Based on this observation, it was concluded that regions located in the extracellular domain of GPIIIa were sufficient to achieve a correct processing and surface expression of GPIIb-ΔGPIIIa complexes, indicating that transmembrane interactions were not essential. These observations seem to be in conflict with the data reported in this work demonstrating that neither platelet from the proband nor CHO cells coexpressing normal GPIIb and mutant ΔGPIIIa showed surface exposure of GPIIb-ΔGPIIIa. Because the proband showed normal levels of platelet [T1846]GPIIIa-mRNA, it is unlikely that the lack of platelet expression of GPIIb-ΔGPIIIa was the result of a limited availability of ΔGPIIIa subunits. The discrepancy between our case and previous work showing surface exposure of the ΔGPIIIa46 could find a justification in the extension of the truncation in each condition, Δ616-762 in our case versus Δ693-762 in the recombinant protein. The absence of the Glu616 to Ile693 region implies the loss of several cysteines and disruption of the long-range S406-S655 disulfide bridge45 that could result in an abnormal protein folding. However, recent work has demonstrated that disruption of the long-range GPIIIa S406-S655 disulfide bridge does not prevent the surface exposure of functional GPIIb-IIIa heterodimers.49 The Iraqi-Jewish GT is associated with an 11-bp deletion in the exon 12 of GPIIIa that leads to premature protein termination before the transmembrane domain (GPIIIaΔ650-762).50 In agreement with our findings, these patients show detectable proGPIIb51 and absence of platelet GPIIb-IIIa and vitronectin receptors.51 52 The possibility should also be considered that the Glu616 to Ile693 region could encompass a domain(s) not previously recognized that is essential for the subunit dimerization. In any case, the agreement of the data on transfection experiments with the observations made in the patient’s platelets suggests that, in megakaryocytes, residues from Glu616 to Ile693 may be involved in assembly and surface exposure of GPIIb-GPIIIa complexes.
Platelet expression of GPIIb-IIIa in heterozygous individuals.
The present study indicates that platelets from heterozygotes for the [T1846]GPIIIa mutation showed levels of GPIIb-IIIa within the range of the normal population. The analysis of platelet expression of GPIb receptor (Table 1) seemed to rule out that platelet expression of GPIIb-IIIa was inherently high in this kindred. Our finding contrasts with studies on GPIIb mutations associated with thrombasthenic phenotypes in which the heterozygous states consistently showed a marked reduction (≥50%) in the platelet GPIIb-IIIa content. As far as we know, the G1846→T is the first nonsense GPIIIa mutation in which the platelet content of GPIIb-IIIa has been quantitatively determined in the heterozygous states. However, heterozygotes for other GPIIIa mutations have also been reported to show only moderate decreases in the platelet GPIIb-IIIa content.53,54 The existence of only one functional allele could imply a reduction in the availability of mRNA so that it could become limiting for the synthesis of GPIIb and/or GPIIIa. However, the apparent discrepancy between the platelet expression of GPIIb-IIIa in heterozygous states for the [T1846]GPIIIa and other mutations in GPIIb suggests that, at least for this particular mutation, transcripts from one allele may provide sufficient GPIIIa to maintain a normal rate of heterodimerization and surface exposure of GPIIb-IIIa complexes. Alternative explanations could be the influence of the genetic context of this kindred in the behavior of this mutation or, perhaps, that the truncated protein inhibited the degradative pathway of the normal GPIIIa. The possibility should also be considered that, unlike other mutations that can form stable complexes with GPIIb, in our case the lack of complex formation would leave more GPIIb available to complex normal GPIIIa. Overexpression of the normal allele is improbable, because the overall platelet GPIIIa-mRNA levels were similar in the proband and the heterozygous relatives. Thus, our observations seem to suggest that availability of mRNA-GPIIIa may not be the main rate-limiting step for the synthesis and/or processing of this subunit. This interpretation seems to be in conflict with the reported synthesis of a fivefold excess of pre-GPIIb relative to GPIIIa (cited in Calvete55), indicating that availability of GPIIIa rather than GPIIb mRNA could be a limiting step for the surface exposure of GPIIb-IIIa. Precise quantitative analysis of mRNAs GPIIb and GPIIIa under different functional conditions will be required to elucidate this point.
To conclude, we report the finding of an homozygous [T1846]GPIIIa mutation associated with type I GT. This mutation changes Glu616 to Stop, producing a GPIIIa protein truncated from Glu616 to Ile762. No other mutations were found in either GPIIb or GPIIIa genes and [T1846]GPIIIa accounted for virtually all the platelet GPIIIa-mRNA found in the proband. Coexpression of normal GPIIb and ΔGPIIIa in CHO cells showed a lack of GPIIb maturation and surface exposure of GPIIb-ΔGPIIIa complexes. Moreover, immunoprecipitation and pulse-chase analysis demonstrated that the translational product of the [T1846]GPIIIa mutation does not complex to GPIIb. Thus, the homozygous [T1846]GPIIIa mutation is responsible for the thrombasthenic phenotype of the proband.
ACKNOWLEDGMENT
Normal GPIIb/IIIa cDNAs were a gift of Dr D.R. Philips.
Supported in part by grants from Dirección General de Investigación Cientı́fica y Técnica (PB94-1544), Fondo de Investigaciones Sanitarias (96/2014), CAM: CO7191, and European Community concerted action contract BMH1-CT93-1685. M.F. was the recipient of a predoctoral fellowship from the Comunidad Autónoma de Madrid. C.G.M. was recipient of a grant from Fundacion Rodriguez Pascual.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Consuelo González-Manchón, MD, PhD, Centro de Investigaciones Biológicas, Velázquez 144, 28006-Madrid, Spain; e-mail: CIBP255@FRESNO.CSIC.ES.



![Fig. 4. Specific amplification of normal and [T1846]-GPIIIa alleles. A DNA fragment of 148 bp encompassing exon 11 and intronic flanking regions of GPIIIa was amplified from genomic DNA of a control, the proband, her parents, and her brother. Each DNA was amplified using a sense primer whose 3′ end was complementary to either the normal sequence (Wt) or to the mutant base (Mut). The amplification products were electrophoresed in a 3% agarose gel and the DNA bands were stained with ethidium bromide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/12/10.1182_blood.v92.12.4712/5/m_blod42419004w.jpeg?Expires=1765914666&Signature=QYB03SuQ5INzMQ9FqPQKwKSr6AfW2HQ7KMpH44WASJup4ifQXtWut25weQtP0UPQy7SgQBksGr7zy6tNXJLjRy2UnYQYzfLsJh-NEBSxlWolErGzU9Jl3rrrqt1zrgDBg1huEqAGPwFsFxINXdjQAu9yhP4uhu~XTS7swjofIYLaOI6HWZ4sk9gpkkeum9Hgfahpe-YeFEcyXi4s788dcrHfFt0p4lPjw83qSD0jJj1Q9f4~6uu~mj2B4ocXuZ797nJJqZnUPhSlziJZ706q1QvFjsc-JvARxUVmgMsTFqhfKS1PisPm71k5uwWoaP0~GdW5pYXZPeSMzw1llPKHxg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Immunoprecipitation of biotin-labeled surface or total (surface and intracellular) proteins from CHO cells cotransfected with cDNAs encoding GPIIb and either normal or [T1846]GPIIIa. CHO cells were transiently cotransfected with cDNAs encoding GPIIb and either normal or [T1846]GPIIIa in the expression plasmid pcDNA3. (A) Protein labeling was performed by exposure of intact cells to biotin-NHS, and the GPIIb-IIIa complexes were immunoprecipitated with either anti-GPIIIa (P37) or anti-GPIIb (M3) MoAbs as described in Materials and Methods. (B) The experimental design was similar to that described in (A), except that total cell lysates rather intact cells were exposed to biotin-NHS and the immunoprecipitation was also performed with a GPIIb-IIIa complex-specific antibody (CII). Mock cells were transfected with void pcDNA3 plasmid.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/12/10.1182_blood.v92.12.4712/5/m_blod42419006w.jpeg?Expires=1765914666&Signature=gLadKk2PZ-YtehKqmgDGfRsYt7d9iGcMutRRJjeDT1h0EPdERfG7GOBbGVe0KwwcGOxEIlYbr53ou6M9Jidy4cpKlOLMdNMTcM-R2lb6QnWz61axrGPNLcjLwyes~OPeyFjfuKzl5Hv5RrwPF7HwZFx7wcC-5QJsFNmJwY9oaWaRD1QoFv2jXfBWTynUXHfe7zxojnqJQbp2iVco43NN~~m3XuJyX5zXtyH-VccXHOxjsPylysJ3-JHBq0~Y-fwp89lSgIb1zARMcM9YSHL7pVt6vX-kmeLACYUO3XRJI63gMYJkjXO61S0kS1iYqPm1unqlUEDT~yxQB541-E20vw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Pulse-chase analysis of the stability of normal or ▵GPIIIa-IIb complexes. CHO cells were transiently cotransfected with cDNAs encoding GPIIb and either normal or [T1846]GPIIIa in the expression plasmid pcDNA3. Cells were pulse-labeled with [35S]-methionine for 30 minutes and then chased with medium containing unlabeled methionine. At the indicated times, the cells were lysed and labeled proteins were immunoprecipitated with the indicated antibodies. The immunoprecipitates were analyzed by electrophoresis as described in Materials and Methods.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/12/10.1182_blood.v92.12.4712/5/m_blod42419007w.jpeg?Expires=1765914666&Signature=nQxE4kaawpxPD4P6GAW6f98OSJIaGEBplpvXhwHyv~CXeFGmXdhOXWrsUhpMnZsB7trgUQIkb0zEJiaJmBU~m1aq0KnIcHcqAjFD-drUEPGYppYVj9Sb0FaRsJfIa7Gxr9Iz8VENzplrnpQ5puSxM4xxqlKLcy80qgp6hH4rnHKpwraJEDIS5tA0h2TbprDmuqx83jRdiUrDgG09yj4jsTZ7BiMCz4epDGfYiwy0z64d7c03-u9SAzNo8PS8z4VoPUgc4WxjnH8XrdVr6lugvHSBjhTlCsCQSUx60dMELHbKYePb5QXFgcBhVHyk8XYL3fYg5qQflEYwlHDeic6I5w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal