Abstract
Interleukin-10 (IL-10) selectively inhibited lipopolysaccharide (LPS)-induced chemoattractant cytokine gene expression: levels of IP-10 mRNA were markedly suppressed in IL-10–treated mouse peritoneal macrophages, whereas the expression of the RANTES mRNA was only modestly reduced. IL-10 inhibited IP-10 mRNA accumulation by reducing IP-10 gene transcription as demonstrated by nuclear run-on analysis. Interestingly, the ability of IL-10 to inhibit expression of IP-10 was dependent on the inducing stimulus; IL-10 did not suppress interferon γ (IFNγ)- or IFNβ-stimulated IP-10 transcription or mRNA accumulation. These results suggested that IL-10 might act indirectly to suppress IP-10 expression by inhibiting LPS-induced class I IFN production. This hypothesis was supported by the following observations. First, LPS-induced IP-10 mRNA expression was blocked in cells cotreated with cycloheximide. Second, IL-10 inhibited the production of IFN/β-mediated antiviral activity. Finally, the IL-10–mediated suppression of LPS-stimulated IP-10 production could be rescued by cotreatment with IFNβ.
MONONUCLEAR phagocytes play an important role in the orchestration of acute and chronic inflammation.1-3 The behavioral potential of macrophages during an inflammatory process is determined, in part, by multiple signals encountered in the tissue microenvironment. These may include bacterial cell wall products such as lipopolysaccharide (LPS) and secreted cytokines from antigen-stimulated T cells such as interferon γ (IFNγ). Because the inflammatory functions of macrophages are potentially damaging to normal tissue, precise control of the response is necessary and is frequently provided by anti-inflammatory cytokines such as interleukin-10 (IL-10).
IL-10 has been shown to have profound effects on monocytes and macrophages, causing inhibition of a number of LPS-induced proinflammatory cytokines such as IL-1α, IL-6, IL-8, and tumor necrosis factor α (TNFα).4-8 The molecular mechanisms through which this inhibitory function is achieved have been the subject of multiple studies, the results of which are diverse.9-18 Thus, IL-10 has been reported to inhibit stimulus-induced monocyte/macrophage gene expression by blocking transcription, by altering the stability of mRNAs, and by reducing the translation of mRNAs.
This mechanistic heterogeneity could reflect differences in the signaling events induced by the stimuli that promote their expression. To determine if differential signaling from distinct inducing stimuli might account for differential sensitivity to IL-10, we have chosen to evaluate the mechanisms involved in IL-10–mediated control of the gene encoding the chemoattractant cytokine (chemokine) IP-10. The IP-10 gene is an appealing model for this purpose, because the mechanisms involved in controlling induced expression are well characterized.19-23 More importantly, the gene can be induced in macrophages in response to multiple stimuli, including virus, poly IC, LPS, IFNγ, and IFNα/β.19 22-26 Our findings demonstrate that IL-10 suppresses IP-10 expression in a stimulus-dependent fashion: response to LPS is sensitive, whereas that to IFNγ or IFNβ is unaffected by IL-10. The suppression of IP-10 appears to be indirect and depends on the immediate inhibition of LPS-induced type I IFN.
MATERIALS AND METHODS
Reagents.
Brewer’s Thioglycollate broth (TG) was purchased from Difco Laboratories (Detroit, MI). LPS prepared from the Escherichia coliserotype 0111:B4 was purchased from Sigma Chemical Co (St Louis, MO). Recombinant IFNγ, RPMI 1640, antibiotics, and glutamine were purchased from GIBCO BRL Life Technologies (Gaithersburg, MD). IFNβ and IL-10 were obtained from Genzyme Inc (Cambridge, MA). Fetal bovine serum (FBS) was obtained from Hyclone (Logan, UT) and was heat-inactivated before use. All cell culture reagents were specified to be endotoxin free. Cesium chloride, guanidine thiocyanate, agarose, sodium dodecyl sulfate (SDS), Tris, proteinase K, RNase-free DNase, and random priming kits were purchased from Boehringer Mannheim Corp (Indianapolis, IN). Formamide was obtained from US Biochemical (Cleveland, OH). Dextran sulfate was obtained from Pharmacia (Uppsala, Sweden). Nylon transfer membrane was purchased from Micron Separations Inc (Westborough, MA). Dupont NEN Research Products (Boston, MA) was the source of α[32P]-dCTP and α[32P]-UTP.
Mice.
Specific pathogen-free, inbred C57BL/6 mice, 9 to 12 weeks of age, were purchased from Jackson Laboratories (Bar Harbor, ME) and housed in microisolator cages with autoclaved food and bedding to minimize exposure to viral and microbial pathogens and to ensure that the degree of spontaneous activation of tissue macrophages would be minimal.27
Cell culture.
TG-elicited macrophages were obtained as reported previously.28 29 Peritoneal lavage was performed using 10 mL of cold Hanks’ balanced salt solution (HBSS) containing 10 U/mL heparin. Macrophages were plated in 100- or 150-mm tissue culture dishes, incubated for 2 hours at 37°C in an atmosphere of 5% CO2, and then washed three times with HBSS to remove nonadherent cells. The macrophages were cultured overnight in RPMI 1640 at 37°C in 5% CO2 and then cultured in the presence or absence of stimuli for the indicated times.
Preparation of plasmid DNA.
Preparation of RNA and Northern hybridization analysis.
Total cellular RNA was prepared by the guanidine thiocyanate-cesium chloride method.33 Equal amounts of RNA (10 μg) were used in each lane of the gel. The RNA was denatured, separated by electrophoresis in a 1% agarose-formaldehyde gel, and transferred to a nylon membrane as previously described.28 29 The blots were prehybridized for 12 to 24 hours at 42°C in 50% formamide, 1% SDS, 5× SSC, 1× Denhardt’s (0.02% Ficoll, 0.02% bovine serum albumin [BSA], and 0.02% polyvinylpyrrolidone), 0.25 mg/mL denatured herring testis DNA, and 50 mmol/L sodium phosphate buffer, pH 6.5. Hybridization was performed at 42°C for 18 hours with 1 × 107 cpm of denatured plasmid DNA containing appropriate specific cDNA inserts. The filters were rinsed with a solution of 0.1% SDS-0.2× SSC, washed at 42°C for 1 hour, and washed at 65°C for 15 minutes. The blots were dried and exposed by using XAR-5 x-ray film (Eastmann Kodak Co, Rochester, NY) with Dupont (Wilmington, DE) Cronex Lightening Plus intensifying screens at −70°C. Blots were quantified by phosphoresence analysis using an instrument from Molecular Dynamics (Sunnyvale, CA).
Nuclear transcription assay.
Cultures of 5 × 107 macrophages were treated as indicated in the text and nuclei isolated as described previously.24,34 Transcription initiated in intact cells was allowed to complete in the presence of α-[32P]-UTP and the RNA was isolated and hybridized to slot-blotted plasmids containing specific cDNA (7 μg DNA/slot) essentially as described elsewhere.24 34 Blots were hybridized for 72 hours and exposed to x-ray film for 2 to 4 days. The α-tubulin gene was used as an internal standard. The expression of specific transcripts was quantified by phosphorimage analysis. Numerical values for specific transcripts were normalized to α-tubulin transcript level in the same sample. This ratio in untreated samples was arbitrarily set to unity. Experimental values are presented as fold induction relative to untreated samples.
IFNα/β-mediated antiviral activity.
Macrophages were stimulated in complete medium for 4 hours with LPS in the presence or absence of IL-10. Supernatant medium was collected, centrifuged to pellet residual cells and cell debris, and used to measure antiviral activity as described previously.35 Mouse embryo fibroblasts (MEF) were plated at a density of 5 × 104 per well in 24-well plates. Cultures were treated with medium alone or medium containing various dilutions of macrophage culture supernatants or known quantities of IFNβ. After 24 hours, MEF cultures were washed with fresh medium, infected with endomyocarditis (EMC) virus (multiplicity of infection [MOI] = 2; gift of Dr Robert Silverman, Cleveland Clinic Foundation, Cleveland, OH) and incubation continued for an additional 24 hours. The cell controls included MEF cells cultured with supernatants alone or with EMC infection. Cell viability was measured by staining wells with neutral red in PBS and elution in 50% ethanol in 0.1 mol/L NaH2PO4, and the absorbance at 540 nm was determined. The results are presented as the percentage of protection calculated as follows: [A540(sample) − A540(virus control)]/[A540(cell control) − A540(virus control)] × 100.
RESULTS
IL-10 selectively inhibits expression of LPS-induced chemokine mRNAs.
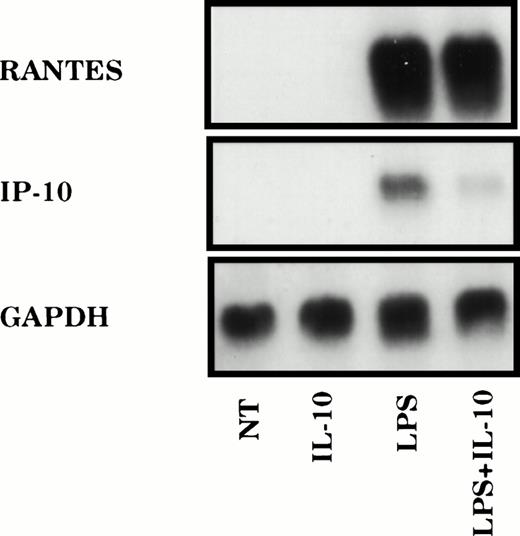
Previous work from multiple laboratories has demonstrated that IL-10 is a potent suppressor of induced gene expression in mononuclear phagocytes.4-8 The mechanisms through which such inhibitory effect is achieved are diverse.9-18 To distinguish the importance of stimulus specificity in determining the mechanisms involved in IL-10–mediated suppression, we have examined the effects of IL-10 on the chemokine gene IP-10, the control of which has been extensively studied.20-23 Peritoneal macrophages were treated with LPS for 2 hours in the presence or absence of IL-10, and expression of mRNAs encoding IP-10 or RANTES was analyzed by Northern hybridization. In agreement with previous results, LPS treatment stimulated strong expression of IP-10 mRNA, which was markedly reduced (>80% as quantified by phosphorimage analysis) in macrophages treated simultaneously with IL-10 (Fig 1). This effect was gene selective, because levels of RANTES mRNA were only modestly reduced (<25%).
IL-10 selectively suppresses LPS-induced chemokine gene expression. TG-elicited macrophages (1 × 107) were treated with LPS (10 ng/mL) in the absence or presence of IL-10 (25 ng/mL) for 2 hours. Total RNA was prepared and the levels of IP-10, RANTES, and GAPDH mRNA were analyzed using northern hybridization. Similar results were obtained in three separate experiments.
IL-10 selectively suppresses LPS-induced chemokine gene expression. TG-elicited macrophages (1 × 107) were treated with LPS (10 ng/mL) in the absence or presence of IL-10 (25 ng/mL) for 2 hours. Total RNA was prepared and the levels of IP-10, RANTES, and GAPDH mRNA were analyzed using northern hybridization. Similar results were obtained in three separate experiments.
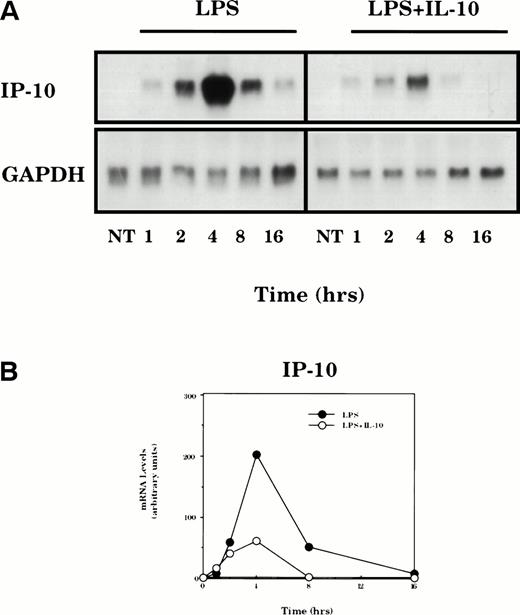
The time dependence for IL-10–mediated suppression of LPS-induced IP-10 mRNA expression was determined in macrophages treated for various times (Fig 2). LPS-stimulated expression of IP-10 mRNA was transient: the peak levels were obtained within 4 hours, declined markedly by 8 hours, and returned to nearly baseline after 16 hours of stimulation. IL-10 suppressed the production of IP-10 mRNA, although the inhibition was not evident until 2 hours of IL-10 exposure.
Time dependence of IL-10–mediated suppression of LPS-induced IP-10 mRNA expression. (A) TG-elicited macrophages were treated with LPS (10 ng/mL) in the absence or presence of IL-10 (25 ng/mL) for the indicated times. Total RNA was prepared and levels of IP-10 and GAPDH mRNA were analyzed by northern hybridization. (B) The levels of specific mRNA on the blot shown in (A) were quantified by phosphorimage analysis. Levels of IP-10 mRNA in each sample are normalized for the levels of GAPDH mRNA. Similar results were obtained in two separate experiments.
Time dependence of IL-10–mediated suppression of LPS-induced IP-10 mRNA expression. (A) TG-elicited macrophages were treated with LPS (10 ng/mL) in the absence or presence of IL-10 (25 ng/mL) for the indicated times. Total RNA was prepared and levels of IP-10 and GAPDH mRNA were analyzed by northern hybridization. (B) The levels of specific mRNA on the blot shown in (A) were quantified by phosphorimage analysis. Levels of IP-10 mRNA in each sample are normalized for the levels of GAPDH mRNA. Similar results were obtained in two separate experiments.
Mechanisms of IL-10–mediated suppression of IP-10 mRNA expression.
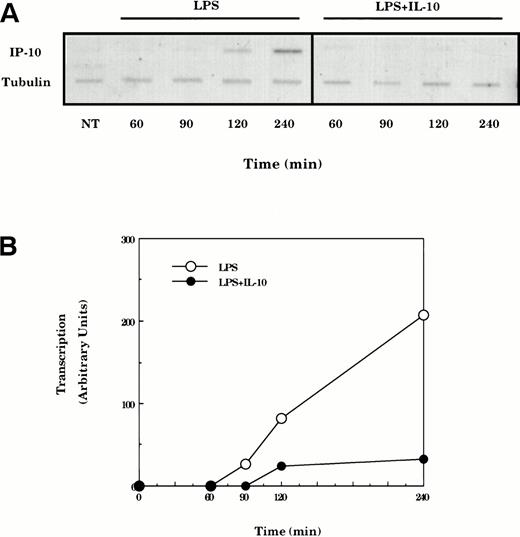
LPS-induced IP-10 mRNA expression is mediated by increases in gene transcription.20-22 To determine if the suppression of LPS-induced IP-10 mRNA by IL-10 involved inhibition of transcription, nuclear run-on experiments were performed. Cultures of peritoneal macrophages were treated with LPS for various times in the absence or presence of IL-10, the nuclei were harvested, and the RNA transcripts initiated in vivo were allowed to elongate in vitro in the presence of32P-UTP. The radiolabeled RNA products were hybridized to slot-blotted cDNAs encoding IP-10 and α-tubulin and the resulting blots were quantified by phosphorimage analysis (Fig 3). The transcriptional activity of the IP-10 gene was elevated within 2 hours and appeared to be optimally active after stimulation with LPS for 4 hours. This LPS-dependent stimulation of transcription of the IP-10 gene was almost fully suppressed in cells that had been cotreated with IL-10.
IP-10 mRNA expression is controlled at the level of transcription. (A) TG-elicited macrophages were treated with LPS (10 ng/mL) for various times in the absence or presence of IL-10 (25 ng/mL) before isolation of nuclei and analysis of transcription by nuclear run-on. Radiolabeled nuclear RNA was hybridized with nylon membranes containing equivalent amounts of denatured plasmid DNA encoding IP-10 and tubulin (top panel). (B) Slot blots shown in the top panel were quantified by phosphorimage analysis and levels of IP-10 transcript normalized for levels of tubulin. Similar results were obtained in three separate experiments.
IP-10 mRNA expression is controlled at the level of transcription. (A) TG-elicited macrophages were treated with LPS (10 ng/mL) for various times in the absence or presence of IL-10 (25 ng/mL) before isolation of nuclei and analysis of transcription by nuclear run-on. Radiolabeled nuclear RNA was hybridized with nylon membranes containing equivalent amounts of denatured plasmid DNA encoding IP-10 and tubulin (top panel). (B) Slot blots shown in the top panel were quantified by phosphorimage analysis and levels of IP-10 transcript normalized for levels of tubulin. Similar results were obtained in three separate experiments.
IL-10–mediated suppression of IP-10 mRNA is stimulus-dependent.
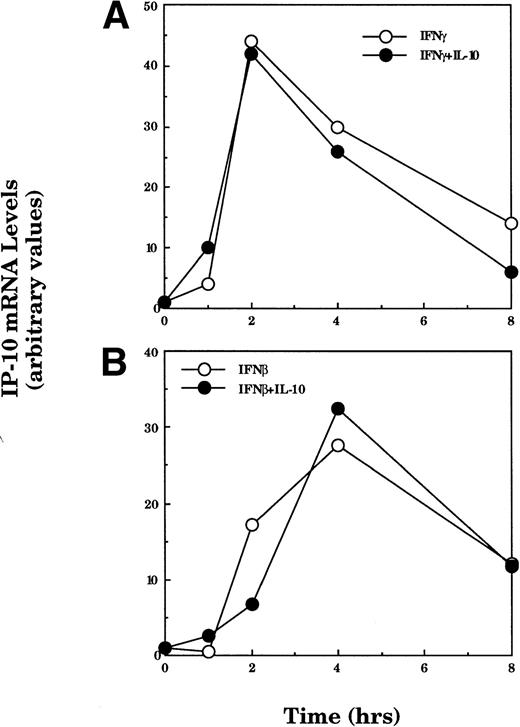
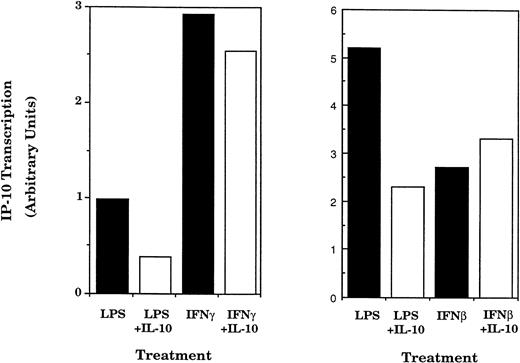
IFNγ and IFNβ are also important stimuli of IP-10 gene expression.22,24 36 To determine whether the effect of IL-10 on IP-10 expression varies with the inducing stimulus, peritoneal macrophages were incubated with IFNγ or IFNβ in the absence or presence of IL-10 for various times and IP-10 mRNA levels were measured by Northern hybridization and phosphorimage analysis (Fig 4). IFNγ-stimulated IP-10 mRNA expression reached maximum levels within 2 hours, whereas IFNβ-induced IP-10 mRNA levels were maximal at 4 hours. Neither IFNγ nor IFNβ stimulated the expression of the RANTES gene (data not shown). In contrast to the marked suppression of LPS-induced IP-10 levels seen in Figs 1 and 2, IL-10 had no significant effect on IP-10 mRNA levels stimulated by either IFNγ or IFNβ. This difference was confirmed at the level of transcription; IP-10 gene transcription induced by either IFNβ or IFNγ was insensitive to IL-10 treatment, whereas transcription in response to LPS was selectively inhibited (Fig 5).
The suppressive action of IL-10 is stimulus-dependent. TG-elicited macrophages were treated with IFNγ (50 U/mL; A) or IFNβ (500 U/mL; B) in the absence or presence of IL-10 (25 ng/mL) for the indicated times. Total RNA was prepared and IP-10 and GAPDH mRNA were analyzed using northern hybridization. Levels of expression in each lane were quantified using phosphorimage analysis. The data presented for IP-10 mRNA are normalized to the level of GAPDH in each sample. Similar results were obtained in two separate experiments.
The suppressive action of IL-10 is stimulus-dependent. TG-elicited macrophages were treated with IFNγ (50 U/mL; A) or IFNβ (500 U/mL; B) in the absence or presence of IL-10 (25 ng/mL) for the indicated times. Total RNA was prepared and IP-10 and GAPDH mRNA were analyzed using northern hybridization. Levels of expression in each lane were quantified using phosphorimage analysis. The data presented for IP-10 mRNA are normalized to the level of GAPDH in each sample. Similar results were obtained in two separate experiments.
IL-10 selectively inhibits transcription of the IP-10 gene. TG-elicted macrophages (1 × 107 cells/sample) were stimulated with LPS (10 ng/mL) or IFNβ (500 U/mL) in the presence or absence of IL-10 (25 ng/mL) for 2 hours. In a separate experiment, macrophages were treated with LPS or IFNγ plus or minus IL-10. Nuclei were prepared and used in nuclear run-on assay as described in the legend to Fig 3. Similar results were obtained in two separate experiments.
IL-10 selectively inhibits transcription of the IP-10 gene. TG-elicted macrophages (1 × 107 cells/sample) were stimulated with LPS (10 ng/mL) or IFNβ (500 U/mL) in the presence or absence of IL-10 (25 ng/mL) for 2 hours. In a separate experiment, macrophages were treated with LPS or IFNγ plus or minus IL-10. Nuclei were prepared and used in nuclear run-on assay as described in the legend to Fig 3. Similar results were obtained in two separate experiments.
The inhibition of LPS-induced IP-10 expression by IL-10 is mediated by the suppression of LPS-induced IFNβ production.
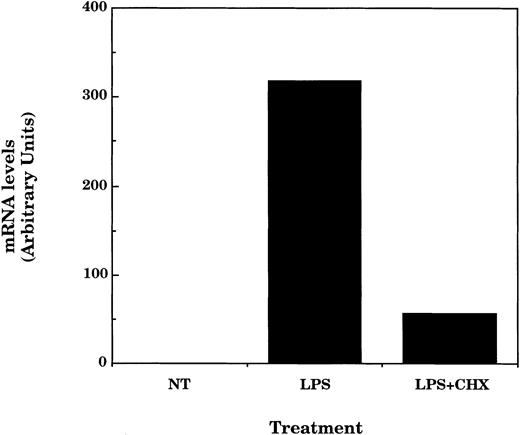
The finding that IP-10 expression induced by IFNγ or IFNβ was insensitive to inhibition by IL-10 suggested the possibility that the effects of IL-10 on LPS-induced IP-10 expression might involve an indirect mechanism. In a previous report we observed that the ability of LPS to stimulate transcription from the promoter of the IP-10 gene was blocked by including antibody against type I IFNs in the culture medium.21 Thus, IFNβ, induced in response to LPS, might be the direct mediator of IP-10 expression and the immediate target of IL-10 suppression; IL-10 could inhibit IFNβ expression and indirectly the LPS-induced expression of IP-10. To test this possibility, we have performed three distinct experiments. First, we have determined if IP-10 expression in response to LPS depends on intermediate protein synthesis. Macrophages were treated with LPS in the presence or absence of cycloheximide for 4 hours and the total RNA was then analyzed for IP-10 mRNA content. Consistent with our hypothesis and previous results, LPS-induced IP-10 expression was blocked in cells treated with cycloheximide (Fig 6).
LPS-induced IP-10 mRNA expression is dependent on protein synthesis. TG-elicited macrophages (1 × 107 cells/sample) were treated with LPS (10 ng/mL) in the absence or presence of cycloheximide (1 μg/mL) for 4 hours before measurement of IP-10 and GAPDH mRNA levels by northern hybridization and phosphorimage analysis as described in the legend to Fig 2. Similar results were obtained in two separate experiments.
LPS-induced IP-10 mRNA expression is dependent on protein synthesis. TG-elicited macrophages (1 × 107 cells/sample) were treated with LPS (10 ng/mL) in the absence or presence of cycloheximide (1 μg/mL) for 4 hours before measurement of IP-10 and GAPDH mRNA levels by northern hybridization and phosphorimage analysis as described in the legend to Fig 2. Similar results were obtained in two separate experiments.
In the second experiment we determined if LPS-induced IFNβ is sensitive to suppression by IL-10. Macrophages were treated with LPS in the presence or absence of IL-10 for 4 hours. Culture supernatants were harvested and the IFNβ content was measured on the basis of IFNβ-dependent antiviral activity (Table1). LPS stimulated significant antiviral activity in two separate experiments, and this was suppressed by 75% when IL-10 was included in the culture medium. The antiviral activity was identifiable as IFNβ on the basis of complete neutralization with antibody specific for type I mouse IFNs (data not shown).
IL-10 Inhibits LPS-Induced Production of Type I IFN
| Treatment . | Protection (%) . | Inhibition (%) . |
|---|---|---|
| Experiment no. 1 | ||
| None | 0 | NA |
| LPS | 51.3 | 0 |
| LPS + IL-10 | 12.8 | 75 |
| Experiment no. 2 | ||
| None | 0 | NA |
| LPS | 20.1 | 0 |
| LPS + IL-10 | 5.8 | 71 |
| Treatment . | Protection (%) . | Inhibition (%) . |
|---|---|---|
| Experiment no. 1 | ||
| None | 0 | NA |
| LPS | 51.3 | 0 |
| LPS + IL-10 | 12.8 | 75 |
| Experiment no. 2 | ||
| None | 0 | NA |
| LPS | 20.1 | 0 |
| LPS + IL-10 | 5.8 | 71 |
TG-elicited peritoneal macrophages (1 × 107) were stimulated with LPS (10 ng/mL) without or with IL-10 (25 ng/mL) for 4 hours. Supernatants were harvested and used to pretreat cultures of MEF cells for 24 hours before infection with EMC virus at an MOI of 2. Control cultures were treated with supernatant but not infected with virus. After 24 hours, MEF cell viability was assayed. The percentage of protection was calculated as described in Materials and Methods. Antiviral activity was completely inhibited by anti-IFNα/β antibody (data not shown).
Abbreviation: NA, not applicable.
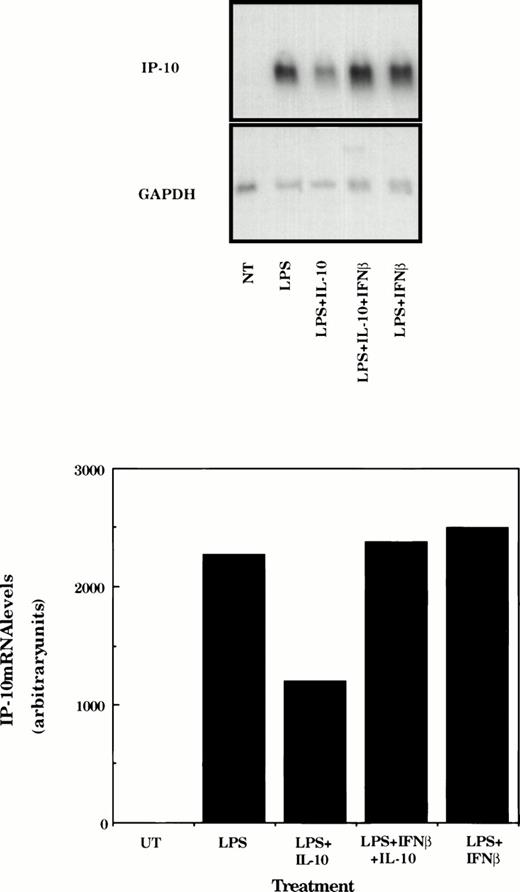
The final experiment was to determine if IL-10–mediated suppression of LPS-induced IP-10 could be rescued by inclusion of IFNβ in the culture medium. Macrophages were stimulated with LPS or LPS and IFNβ in the absence or presence of IL-10 for 4 hours before determination of IP-10 mRNA levels by Northern hybridization. As expected, LPS stimulated strong expression of IP-10, which was suppressed in this experiment by approximately 50% in the presence of IL-10 (Fig 7). Inclusion of IFNβ in the medium did not increase the levels of IP-10 mRNA but did protect against IL-10–mediated suppression.
IFNβ rescues IL-10–suppressed IP-10 mRNA expression. TG-elicited macrophages (1 × 107 cells/sample) were treated with LPS (10 ng/mL), IFNβ (500 U/mL), or IL-10 (25 ng/mL) as indicated for 4 hours before measurement of IP-10 and GAPDH mRNA levels using northern hybridization and phosphorimage analysis as described in the legend to Fig 2. Similar results were obtained in two separate experiments.
IFNβ rescues IL-10–suppressed IP-10 mRNA expression. TG-elicited macrophages (1 × 107 cells/sample) were treated with LPS (10 ng/mL), IFNβ (500 U/mL), or IL-10 (25 ng/mL) as indicated for 4 hours before measurement of IP-10 and GAPDH mRNA levels using northern hybridization and phosphorimage analysis as described in the legend to Fig 2. Similar results were obtained in two separate experiments.
DISCUSSION
Multiple studies have demonstrated that IL-10 suppresses cytokine production in various cell types, although the mechanisms involved appear to be diverse. Inhibition may result from decreased gene transcription, reduction in specific mRNA stability, and/or reduced mRNA translation.7,9,10,12,14-16,18,37 Which of these different mechanisms participate is likely to depend on the signaling pathway(s) that controls expression. To evaluate this problem we elected to examine mechanisms involved in IL-10–mediated suppression of a chemokine gene (IP-10) known to be induced in response to several stimuli that use distinct signaling pathways.23-25 The results demonstrate that IL-10 inhibits IP-10 gene transcription through a mechanism that depends on the stimulus. The stimulus selectivity is a result of two related events: (1) LPS-induced IP-10 expression depends on the intermediate induction of class I IFNs and (2) LPS-induced expression of class I IFNs is suppressed by IL-10. These conclusions are supported by the following experimental observations. (1) IL-10 selectively reduces LPS-induced but not IFNγ- or IFNβ-induced IP-10 mRNA levels. (2) Nuclear run-on analysis indicates that LPS-stimulated IP-10 transcription is selectively suppresssed by IL-10. (3) LPS-induced IP-10 expression is blocked by inhibitors of protein synthesis. (4) Antibodies to class I IFNs can block LPS-stimulated transcription from the IP-10 promoter.21 (5) Production of LPS-induced, IFNβ-mediated antiviral activity is suppressed by IL-10. (6) IFNβ treatment rescues IL-10–mediated suppression of LPS-induced IP-10 mRNA.
LPS is known to be a potent inducer of IL-10 in macrophages.38,39 IL-10 produced under such conditions may act in both autocrine and paracrine fashion to provide negative feedback regulation of the LPS-induced inflammatory response. Including antibody to IL-10 in cultures of LPS-treated macrophages enhances some LPS-induced gene expression and, indeed, the autocrine/paracrine action of IL-10 appears to be a component of LPS-induced tolerance to second endotoxin exposure.40 41
Several reports have shown that gene transcription can be a mechanistic target for the suppressive function of IL-10, and our results demonstrating that IL-10 suppresses LPS-stimulated transcription of the IP-10 gene provide further confirmation.10,14 On the basis of electrophoretic mobility shift assays, Wang et al42showed that IL-10 could inhibit the LPS-induced nuclear localization of NFκB in human monocytes and that NFκB is likely to be an important transcription factor controlling the expression of the IP-10 gene.21,26,43-45 The marked sensitivity to IL-10 of LPS-induced but not IFN-induced IP-10 expression suggests the possibility that IL-10 might differentially target the intracellular signalling pathways used by these agents: IP-10 induced by LPS might be more dependent on activation of NFκB than IP-10 induced by IFN and could, therefore, exhibit greater sensitivity to the action of IL-10. Our own studies indicate that IL-10 does not block the activation of NFκB in macrophages stimulated with LPS and thus does not support NFκB as a target of IL-10 (data not shown). Rather, the data indicate that the action of IL-10 on LPS-induced IP-10 is indirect. In support of this, the induction of IP-10 by LPS is apparently indirect; inhibition of protein synthesis blocked the expression of IP-10 when LPS was the stimulus. Furthermore, in a previous report, we observed that neutralizing antibody against IFNα/β could block the ability of LPS to stimulate reporter gene expression driven by the IP-10 promoter.21 LPS is well known to induce class I IFNs,46,47 and our findings now show that LPS-induced class I IFN production is strongly suppressed by IL-10. Finally, inclusion of IFNβ in the culture medium rescues IL-10–suppressed IP-10 expression. Thus, IL-10 acts to suppress the intermediate expression of IFNβ, the result of which is indirect inhibition of IP-10 gene transcription.24
Many studies of IL-10–mediated suppression of gene expression have implicated accelerated mRNA decay as an important contributing mechanism. mRNAs that exhibit shortened half lives in IL-10–treated cells include TNFα, IL-1α, IL-1β, granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-10, IL-6, and the chemokine genes macrophage inflammatory protein-1α (MIP-1α), MIP-1β, and IL-8.9,11,14,15,18 In previous work, we have also observed significant effects of IL-10 on mRNA stability.48 The intracellular pathway(s) through which IL-10 enhances mRNA degradation is currently unknown. AU-rich sequence elements (AREs) in the 3′-untranslated regions (3′UTRs) of short-lived mRNAs have been shown to confer accelerated decay upon stable mRNAs,49-52 and it is noteworthy that mice bearing a mutant TNFα allele, in which the ARE in the 3′-UTR has been altered, have lost regulation of TNFα expression in vivo in response to IL-10, suggesting that this motif may be involved.53 ARE sequences are present in the 3′UTRs of a variety of cytokine mRNAs that are sensitive to suppression by IL-10 (ie, GM-CSF, TNFα, IL-1α, IL-1β, MIP-1α, and KC) but are absent in RANTES mRNA that is insenstive to suppression by IL-1054-61 (see Fig 1). The IFNβ mRNA sequence contains an ARE motif that has been reported to be important in regulating levels of mRNA under various circumstances.62-64 Thus, it is possible that the IL-10–mediated suppression of LPS-induced IP-10 gene transcription depends on destabilization of IFNβ mRNA through a mechanism that may involve the ARE motif.
The IL-10–induced signaling pathways that mediate its inhibitory effects remain incompletely defined. The IL-10 receptor is most closely related to the receptors for class I and class II IFNs. Consistent with this relationship, IL-10 treatment of T cells and monocytes stimulates tyrosine phosphorylation of the Janus family protein kinases, TYK2 and Jak1, and the phosphorylation and nuclear translocation of STAT1α and STAT3.65-67 A recent report indicates that STAT3 docking sites on the intracellular domain of the IL-10 receptor are required for IL-10–mediated suppression, although activation of STAT3 itself may not be necessary.68 In addition, IL-10 has been reported to suppress LPS-induced protein tyrosine phosphorylation.69 However, the specific roles for these various intracellular signaling events will require additional analysis.
Supported by US Public Health Services Grants No. CA39621 and CA62220.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Thomas A. Hamilton, PhD, Department of Immunology, Lerner Research Institute, Cleveland Clinic Foundation, 9500 Euclid Ave, Cleveland, OH 44195.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal