Abstract
Although various studies supported the notion that leukemia cells in chronic myeloid leukemia (CML) may be recognized by the immune system, direct evidence showing the immunogenicity in vivo of proteins derived from the leukemia cells is lacking. In this study, we have constructed an expression cDNA library from the leukemia cells of a patient with CML and used the autologous serum to screen for high-titer IgG antibodies directed at the leukemia-derived proteins. We isolated eight distinct clones from the library, suggesting that multiple immune responses were elicited in the autologous host. Sequence analysis showed high degrees of homology to known gene sequences in six of the eight clones. Neither bcr-abl nor proteinase 3 sequences were isolated. Using Northern blot analysis, seven of the eight clones showed ubiquitous expression in normal bone marrow, leukemia cell lines, fresh leukemia cells, and normal tissues. However, clone no. 4 showed restricted mRNA expression, being only detected in some fresh leukemia cells, K562 cells, and normal testicular RNA. Using bacterial lysates in dot blot analysis, a panel of sera from normal individuals and patients with CML and other hematological malignancies were screened for high-titer antibodies against these eight clones. There were, among the CML patients, signficantly higher prevalence of antibodies against seven of the eight clones. They were observed even after omitting from the analysis patients with multiple myeloma whose associated immune paresis may impair immune responses to these proteins. Interestingly, antibodies against these proteins were also detected in a small number of normal individuals. Although the precise clinical significance of our findings remains to be determined, this study provides evidence in support of the potential immunogenicity of leukemia-derived proteins in the autologous host. It also provides basis for further investigations to characterize these proteins, especially clone no. 4, and determine their potential for immune targeting in CML.
VARIOUS CLINICAL AND laboratory observations suggest that the leukemia cells in chronic myeloid leukemia (CML) are potentially immunogenic.1 Although some of the antileukemic effect observed following allogeneic bone marrow transplant (BMT) arises as part of a more global graft-versus-host reaction (GVHD) due to minor histocompatibility differences between the donor and the recipient, it is likely that additional leukemia eradication is mediated through effector cells directed at leukemia antigens. Such T-cell populations have been dissected from the GVH effect in studies involving the autologous hosts. Not unlike tumor infiltrating lymphocytes in solid tumors, the repertoire of T-cell receptor Vβ gene usage by T cells associated with CML was restricted when compared with that in healthy individuals.2Furthermore, leukemia-reactive T cells could be isolated from the autologous host.3 More recently, leukemic cells from patients with CML have been shown to be able to generate autologous T-cell responses after the in vitro induction of costimulatory molecules, CD80, and CD86 on the leukemia cells.4-6
There are potentially many protein molecules in CML from which processed peptides may be presented by major histocompatibility complex (MHC) molecules to elicit host immune mechanisms. They include overexpressed normal antigens and leukemia-restricted antigens. The latter occur due to leukemogenesis (p210bcr-abl) or are associated with disease progression (eg, gene products of mutatedras oncogene or p53 tumor-suppressor gene). Thebcr-abl junctional peptides7-9 and peptides from proteinase 310 have both been shown to elicit in vitro antileukemic T-cell responses. A recent study analyzing autologous peptides bound by HLA class I and II molecules of leukemia cells in CML confirmed that some of these antigens are indeed naturally processed and presented.11 This technique has additionally isolated other peptides that could potentially be targeted by T cells. However, the immunogenicity in vivo of these antigens in the autologous host remains to be determined.
A technique has recently been developed that can be used to identify immunogenic protein antigens in vivo.12 In this approach, expression cDNA libraries are prepared from tumor specimens and immunoscreened using absorbed and diluted autologous serum for the detection of antigens that have elicited a high-titer IgG antibody response. Such a humoral response may represent a B-cell response with cognitive help from helper T cells. Therefore, even though the antigens are intracellular and are initially identified by antibodies, this technique identifies tumor products that can be investigated in the context of cell-mediated immunity. Applying the serological identification of antigens by recombinant expression cloning (SEREX) to various solid tumors, a combination of known and novel T-cell antigens have been isolated.12-14 In this study, we have applied a modification of this approach to CML to determine any B-cell responses against proteins derived from leukemia cells in patients with CML. This study may therefore form a useful basis for future investigation into identification of leukemia antigens that may be suitable for immune targeting.
MATERIALS AND METHODS
RNA extraction and construction of cDNA expression library.
Total RNA was extracted from the presentation peripheral blood mononuclear cells of a patient (C.D.) with CML using the RNAzol method.15 Poly(A)-RNA was isolated from total RNA using the Poly-A Tract mRNA isolation systems (Promega, Southampton, UK) and a cDNA expression library constructed from 5 μg of poly(A)-RNA. First-strand cDNA synthesis was performed using an oligo(dT) primer that contained an internal Xho I restriction site. The cDNA was ligated to EcoRI adaptors and digested with Xho I before being size-selected, cloned directionally into λZAP expression vector (Stratagene, Cambridge, UK), packaged into phage particles, and transfected into XLO1-blue Escherichia coli.
Immunoscreening of cDNA library.
Immunoscreening was performed on nitrocellulose membranes containing the phage plaques using autologous serum after protein induction with Isopropyl β-D-Thiogalactopyranoside (IPTG; 1.5 mmol/L). Before use, the autologous serum was first preabsorbed with lysate from E coli transfected with the blue phage. The preabsorbed serum, at a final concentration of 1:1,000, was then incubated at room temperature with the nitrocellulose membranes for 1 hour, washed in Tris-buffered saline/0.01% Triton-X 100 (TBS-T) solution, and then incubated in horseradish peroxidase-conjugated goat antihuman IgG antibody for an additional 1 hour. Reactive phage plaques on the nitrocellulose membrane were finally visualized, after further washing in TBS-T solution, using the chemoluminiscence technique according to the manufacturer’s recommendation (Boehringer Mannheim, East Sussex, UK). False-positive clones encoding normal human IgG fragments derived from contaminating circulating B lymphocytes were eliminated by a round of membrane screening using only horseradish peroxidase-conjugated goat antihuman IgG antibody.
Sequence analysis of reactive clones.
Positive clones were subcloned, purified, and excised in vivo to pBK-CMV plasmid forms (Stratagene). Plasmid DNA was prepared and the DNA inserts evaluated by EcoRI and Xho I restriction mapping. Nucleotide sequences of clones showing different cDNA inserts were analyzed. The sequencing reactions were performed by Oswell DNA Sequencing Services (Southampton University, Southampton, UK) using AB1 PRISM (Perkin-Elmer, Norwalk, CT) automated sequencers.
Northern blot analysis.
Northern blot analysis was performed to confirm the origin of the cDNA, estimate the size and copy number of the transcripts, and to determine the expression of the transcripts in normal and leukemia RNA. RNA was isolated from normal bone marrow, fresh CML cells, and leukemia cell lines (CEM, HL60, K562, Molt 4, and p39). RNA from normal brain, lungs, small intestine, muscle, spleen, and testis was obtained from a commercial source (Invitrogen, Leek, The Netherlands). For Northern blot analysis, 30 μg of total RNA was resuspended in a loading buffer containing formamide and formaldehyde, heated at 65°C for 2 minutes, and electrophoresed on a 1.2% agarose/formaldehyde gel before being transferred by capillary method onto nitrocellulose membranes. Subsequent hybridization to32P-labeled probes (derived from EcoRI/XhoI restriction digest of recombinant phagemids and labeled by random priming) and washing were performed under high-stringency conditions. Hybridization was performed at 60°C overnight and final washes of the membrane at 60°C with 0.1× SSC in 0.1% (wt/vol) sodium dodecyl sulfate solution. RNA loading in each case was determined using a probe for the GAPDH transcript.
Reverse transcription-polymerase chain reaction (RT-PCR).
To evaluate the mRNA expression, total RNA was first reverse transcribed using oligo-dT primers and used with gene-specific oligonucleotide primers for PCR amplification of cDNA segments of 300 to 400 bp in length. All PCR reactions were performed using the following cycling conditions: 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute for 30 cycles. RT-PCR was performed in a thermal cycler (Perkin-Elmer), and the PCR products were visualized on an ethidium bromide agarose gel for DNA of expected size. The positive control amplified the plasmid DNA containing the DNA insert, and the negative control was composed of the PCR reaction mixture lacking cDNA.
Preparation of crude fusion proteins.
Freshly prepared E coli cultures containing the appropriate phagemid constructs were grown at 37°C to an absobance at 600 nm of 0.8 to 1.0 in 200 mL of NZY bacterial broth containing 50 μg/mL kanamycin. IPTG was added to induce the expression of the fusion proteins. The E coli were harvested by centrifugation at 3,600 rpm for 15 minutes at 4°C. After removal of the supernatant, cells were weighed and 3 mL of lysis buffer (50 mmol/L Tris, pH 8.0, 1 mmol/L EDTA, 100 mmol/L NaCl) was added to each gram (wet weight) of E coli pellet. The cells were then lysed using the lysozyme-deoxycholate method.16 The cell debris was removed by centrifugation at 10,000 rpm for 15 minutes at 4°C.
Detection of antigen-reactive antibodies in allogeneic sera.
This procedure was performed using dot blot analysis. Briefly, bacterial supernatant containing the crude fusion proteins was dotted onto nitrocellulose membranes and dried at 37°C for 15 minutes. After blocking, the membranes were incubated for 1 hour with 1:800 diluted sera, which had been preabsorbed with lysate from E coli transfected with the blue phage, followed by an additional 1 hour of incubation with horseradish peroxidase-conjugated rabbit antihuman IgG antibody (Sigma, Poole, UK). The membranes were extensively washed with TBS-T solution between each incubation step. Binding of the antibodies to the fusion proteins was visualized by incubating with DAB peroxidase substrate (Sigma FastTM3,3′-diaminobenzidine set; Sigma) for 2 minutes. Each set of blots contained a control consisting of the bacterial lysate from E coli transfected with the blue phage. Sera were analyzed in a random order and with the observer blinded to their origin.
Statistics.
χ2 was used to determine the power of significance between the analyzed groups.
RESULTS
SEREX analysis of CML cDNA library.
To isolate clones expressing antigens reactive with the autologous serum, an expression cDNA library containing 1.8 × 106 primary phage clones, constructed from the leukemic cells of a patient with CML, was used for immunoscreening. After excluding false-positive clones encoding the human Ig fragments, eight positive plaques were identified. They were subcloned, purified, and excised into pBK-CMV phagemid. The inserts were analyzed by restriction mapping, which suggested that they represented eight different clones. The size of the DNA inserts ranged from approximately 800 bp to greater than 3 kb.
Molecular analysis of the identified antigens.
Nucleotide sequence analysis of the cDNA inserts was performed to determine the identity and sequence homology of the clones using the BLAST software on US National Molecular Biology Laboratory and GenBank data bases (released January 1998; Table1). Clones no. 2, 3, 5, and 8 showed strong homology to human nuclear chloride ion channel protein (NCC27), heterogenous nuclear RNA binding protein M4, human modulator recognition factor I, and human MAF oncogene, respectively. Interestingly, although high degrees of homology (>95%) were observed, there appeared to be an internal nucleotide deletion of more than 100 bp, respectively, in clones no. 2 and 3 when compared with their homologue genes. No significant sequence homology was observed for clones no. 4 and 7. Clones no. 1 and 6 appeared to be fusion genes previously not reported. Further work is ongoing to confirm that they are not cloning artefacts.
Sequences Isolated From a CML Library by Autologous Antibody Screening
| Clone No. . | Size (bp) . | Sequence Homology . | Comments . |
|---|---|---|---|
| 1 | 3872 | Human nuclear envelope protein (NEP) lamin C; phosphoglycerate knase (PGK) | Fusion between NEP lamin C and PGK genes |
| 2 | 1104 | Human nuclear chloride ion channel protein (NCC27) | Internal nucleotide deletion of 132 bp between nucleotide 36 and 168 of normal NCC27 |
| 3 | 2122 | Heterogeneous nuclear binding protein (M4) | Internal nucleotide deletion of 117 bp between nucleotide 485 and 602 of normal M4 |
| 4 | 2221 | No strong homology | |
| 5 | 792 | Human modulator recognition factor I | |
| 6 | 3323 | Human poly-A binding protein (PABP); GATA-2 transcription factor | Fusion between PABP and GATA-2 genes |
| 7 | 931 | No strong homology | |
| 8 | 1847 | Human MAF oncogene |
| Clone No. . | Size (bp) . | Sequence Homology . | Comments . |
|---|---|---|---|
| 1 | 3872 | Human nuclear envelope protein (NEP) lamin C; phosphoglycerate knase (PGK) | Fusion between NEP lamin C and PGK genes |
| 2 | 1104 | Human nuclear chloride ion channel protein (NCC27) | Internal nucleotide deletion of 132 bp between nucleotide 36 and 168 of normal NCC27 |
| 3 | 2122 | Heterogeneous nuclear binding protein (M4) | Internal nucleotide deletion of 117 bp between nucleotide 485 and 602 of normal M4 |
| 4 | 2221 | No strong homology | |
| 5 | 792 | Human modulator recognition factor I | |
| 6 | 3323 | Human poly-A binding protein (PABP); GATA-2 transcription factor | Fusion between PABP and GATA-2 genes |
| 7 | 931 | No strong homology | |
| 8 | 1847 | Human MAF oncogene |
Expression of the identified genes in the autologous host.
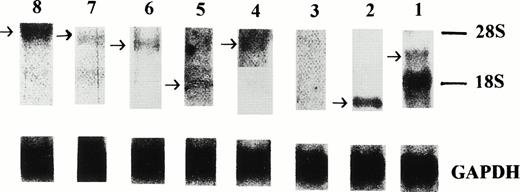
The origin of the isolated sequences was confirmed by Northern blot analysis on total RNA isolated from the primary leukemia cells (Fig 1). Positive signals were obtained using probes derived from seven of the eight clones (no. 1, 2, 4, 5, 6, 7, and 8). However, no signal was obtained in the Northern blot analysis using a probe derived from clone no. 3, for which one potential explanation is a very low level of mRNA expression. The expression of the mRNA from which clone no. 3 was derived was subsequently confirmed by RT-PCR using a pair of sequence-specific oligonucleotide primers (Fig 2).
Northern blot analysis of the expression of the eight isolated sequences in the primary leukemia cells. Signals were obtained using probes derived from clones no. 1, 2, 4, 5, 6, 7, and 8. The level of gene expression of these seven clones was variable compared with signals obtained from a control probe, GAPDH.
Northern blot analysis of the expression of the eight isolated sequences in the primary leukemia cells. Signals were obtained using probes derived from clones no. 1, 2, 4, 5, 6, 7, and 8. The level of gene expression of these seven clones was variable compared with signals obtained from a control probe, GAPDH.
RT-PCR analysis for the expression of clone no. 3 (lane 1, molecular marker; lane 2, positive control consisted of PCR amplification of a recombinant plasmid containing sequence 3; lane 3, RT-PCR amplification of total RNA isolated from the primary leukemia cells; lane 4, negative control). Clone no. 3 PCR primer sequences were as follows: clone no. 3-1, 5′-CAA AGA AGA TCC TGA TGG-3′; clone no. 3-2, 5′-GCA GTG CAT TGG TCT ATC-3′.
RT-PCR analysis for the expression of clone no. 3 (lane 1, molecular marker; lane 2, positive control consisted of PCR amplification of a recombinant plasmid containing sequence 3; lane 3, RT-PCR amplification of total RNA isolated from the primary leukemia cells; lane 4, negative control). Clone no. 3 PCR primer sequences were as follows: clone no. 3-1, 5′-CAA AGA AGA TCC TGA TGG-3′; clone no. 3-2, 5′-GCA GTG CAT TGG TCT ATC-3′.
Expression of the identified genes in leukemia cells.
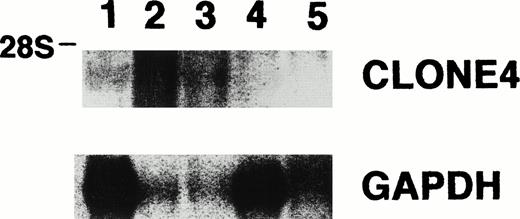
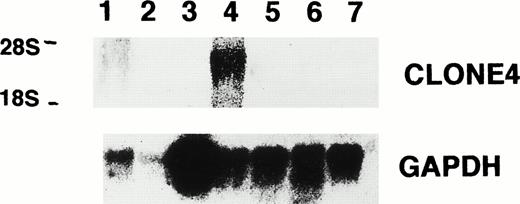
The expression of the isolated gene sequences was evaluated in the other CML patients by Northern blot analysis using total RNA derived from the leukemia cells of four other patients. We found that all of the eight clones were detected in most, but not all, of the four other CML patients (Table 2 and Fig 3). In addition, we also determined the gene sequence expression of six (clones no. 1, 2, 3, 4, 5, and 7) of these eight clones in five leukemia cell lines and found that clones no. 1, 2, 3, 5, and 7 were detected in RNA from all cell lines, except for P39 cells, in which gene sequence 7 was not detected (Table 2). Interestingly, in contrast to clones no. 1, 2, 3, 5, and 7, clone no. 4 was only detected in K562 cells and not CEM, HEL, Molt 4, and P39 cells (Fig 4). The results provide supporting evidence for the preferential expression of gene sequence from clone no. 4 in CML.
mRNA Expression in Fresh CML Cells and Leukemia Cell Lines
| Clone No. . | Fresh CML Cells . | CEM . | HEL . | K562 . | Molt 4 . | P39 . |
|---|---|---|---|---|---|---|
| 1 | 5/5 | Positive | Positive | Positive | Positive | Positive |
| 2 | 5/5 | Positive | Positive | Positive | Positive | Positive |
| 3 | 5/5* | Positive | Positive | Positive | Positive | Positive |
| 5 | 2/5 | Positive | Positive | Positive | Positive | Positive |
| 6 | 5/5 | NT | NT | NT | NT | NT |
| 7 | 4/5 | Positive | Positive | Positive | Positive | Positive |
| 8 | 5/5 | NT | NT | NT | NT | NT |
| Clone No. . | Fresh CML Cells . | CEM . | HEL . | K562 . | Molt 4 . | P39 . |
|---|---|---|---|---|---|---|
| 1 | 5/5 | Positive | Positive | Positive | Positive | Positive |
| 2 | 5/5 | Positive | Positive | Positive | Positive | Positive |
| 3 | 5/5* | Positive | Positive | Positive | Positive | Positive |
| 5 | 2/5 | Positive | Positive | Positive | Positive | Positive |
| 6 | 5/5 | NT | NT | NT | NT | NT |
| 7 | 4/5 | Positive | Positive | Positive | Positive | Positive |
| 8 | 5/5 | NT | NT | NT | NT | NT |
Abbreviation: NT, not tested.
Weakly positive.
Northern blot analysis for the expression of clone no. 4 and GAPDH, showing clone no. 4 mRNA in leukemia cells derived from two of the four other CML patients (lane 1, RNA from leukemia cells derived from patient C.D.; lanes 2, 3, 4, and 5, RNA from leukemia cells derived from four other CML patients).
Northern blot analysis for the expression of clone no. 4 and GAPDH, showing clone no. 4 mRNA in leukemia cells derived from two of the four other CML patients (lane 1, RNA from leukemia cells derived from patient C.D.; lanes 2, 3, 4, and 5, RNA from leukemia cells derived from four other CML patients).
Northern blot analysis for the expression of clone no. 4 and GAPDH in leukemia cell lines, normal bone marrow, and patient C.D., showing clone no. 4 mRNA in K562 cells (lane 1, P39; lane 2, Molt 4; lane 3, K562; lane 4, HEL; lane 5, CEM; lane 6, normal bone marrow; lane 7, patient C.D.).
Northern blot analysis for the expression of clone no. 4 and GAPDH in leukemia cell lines, normal bone marrow, and patient C.D., showing clone no. 4 mRNA in K562 cells (lane 1, P39; lane 2, Molt 4; lane 3, K562; lane 4, HEL; lane 5, CEM; lane 6, normal bone marrow; lane 7, patient C.D.).
Expression of the identified genes in normal tissues.
The mRNA expression of these eight gene sequences was also evaluated by Northern blot analysis using total RNA derived from a small panel of normal tissues, namely, bone marrow, brain, lungs, small intestine, muscle, spleen, and testis. Table 3 shows that clones no. 1, 2, 3, 5, 6, 7, and 8 are widely expressed amongst the normal tissues. However, expression of gene sequence from clone no. 4 appears to be restricted to only normal testis (Fig 5). In particular, we did not detect expression of the gene sequence in normal bone marrow.
mRNA Expression in Normal Tissues
| Clone No. . | Bone Marrow . | Brain . | Lungs . | Small Intestine . | Muscle . | Spleen . | Testis . |
|---|---|---|---|---|---|---|---|
| 1 | Positive | Positive | Positive | Positive | Positive | Positive | Positive |
| 2 | Positive | Negative | Positive | Positive | Weak positive | Positive | Positive |
| 3 | Positive | Positive | Positive | Positive | Positive | Positive | Positive |
| 5 | Positive | Positive | Positive | Positive | Positive | Positive | Positive |
| 6 | Positive | Negative | Positive | Positive | Weak positive | Positive | Strong positive |
| 7 | Positive | Negative | Positive | Positive | Positive | Positive | Positive |
| 8 | Positive | Positive | Positive | Positive | Positive | Positive | Positive |
| Clone No. . | Bone Marrow . | Brain . | Lungs . | Small Intestine . | Muscle . | Spleen . | Testis . |
|---|---|---|---|---|---|---|---|
| 1 | Positive | Positive | Positive | Positive | Positive | Positive | Positive |
| 2 | Positive | Negative | Positive | Positive | Weak positive | Positive | Positive |
| 3 | Positive | Positive | Positive | Positive | Positive | Positive | Positive |
| 5 | Positive | Positive | Positive | Positive | Positive | Positive | Positive |
| 6 | Positive | Negative | Positive | Positive | Weak positive | Positive | Strong positive |
| 7 | Positive | Negative | Positive | Positive | Positive | Positive | Positive |
| 8 | Positive | Positive | Positive | Positive | Positive | Positive | Positive |
Northern blot analysis for the expression of clone no. 4 and GAPDH showing clone no. 4 mRNA in normal testis (lane 1, lungs; lane 2, spleen; lane 3, muscle; lane 4, testis; lane 5, small intestine; lane 6, brain; lane 7, normal bone marrow).
Northern blot analysis for the expression of clone no. 4 and GAPDH showing clone no. 4 mRNA in normal testis (lane 1, lungs; lane 2, spleen; lane 3, muscle; lane 4, testis; lane 5, small intestine; lane 6, brain; lane 7, normal bone marrow).
Prevalence of high-titer antibodies against the identified antigens amongst sera from patients and healthy donors.
Screening was performed using diluted sera from a group of patients with CML and other hematological diseases and normal healthy donors. The clinical characteristics of these patients are shown in Table 4. Because the prevalence of autoantibody is influenced by the sex and age of the patients, we first determined any differences in the distribution of these two parameters in each of the sample groups. There was no difference in the sex distribution of patients with CML compared with the groups of normal healthy donors or patients with other hematological malignancies. Whereas there was no difference in the age distribution of CML patients and normal healthy donors, CML patients were significantly younger than those with other hematological malignancies (P < .0003).
Clinical Characteristics of Study Groups
| Diagnosis . | Sex . | Age (yrs) . | |||
|---|---|---|---|---|---|
| Male . | Female . | Mean . | Median . | Range . | |
| CML | 10 | 5 | 52 | 58 | 16-72 |
| Normal | 13 | 11 | 59.3 | 61 | 21-82 |
| Other hematological malignancies | 15 | 23 | 70.8 | 72 | 33-87 |
| Diagnosis . | Sex . | Age (yrs) . | |||
|---|---|---|---|---|---|
| Male . | Female . | Mean . | Median . | Range . | |
| CML | 10 | 5 | 52 | 58 | 16-72 |
| Normal | 13 | 11 | 59.3 | 61 | 21-82 |
| Other hematological malignancies | 15 | 23 | 70.8 | 72 | 33-87 |
Other hematological malignancies consisted of multiple myeloma (13), myeloproliferative diseases (8), non-Hodgkin’s lymphoma (7), myelodysplasia (4), monoclonal gammopathy of unknown significance (3), chronic lymphocytic leukemia (2), and large granular lymphocytic leukemia (1). No differences between sex distribution in each group and no differences in the age distribution between CML patients and normal healthy donors were found. However, CML patients were significantly younger than patients with other hematological malignancies (P < .0003).
To perform screening for the presence of high-titer antibodies against the identified antigens by dot blot analysis, crude fusion proteins were prepared from lysates of the bacterial transformants. Because the cDNAs were cloned into the β-galactosidase gene in the plasmid, successful preparation of fusion proteins was demonstrated by the ability of the proteins to bind polyclonal anti–β-galactosidase antibodies, ie, the production of fusion proteins between β-galactosidase and leukemia proteins (Fig 6). All sera were then used for screening at a dilution of 1:800 and a negative control consisting of bacterial lysate from a blue phage was included in all cases. Those sera that did not show any reactivity at this serum dilution were confirmed at a serum dilution of 1:200. Table 5 shows the distribution of serum reactivity to these antigens in the various groups of individuals. There was a significantly higher prevalence of antibodies directed against the leukemia-derived antigens in patients with CML compared with normal healthy donors. This was observed for seven of the eight antigens (clones no. 1, 2, 3, 4, 6, 7, and 8). Such differences were observed even when four CML patients who were on interferon-α (IFN-α) therapy were excluded from the analysis (.001 < P< .01). Interestingly, one normal donor exhibited antibodies to all eight and another to seven of the eight antigens.
Dot blot analysis using crude fusion protein extracts. (A) Bacterial extracts from all eight clones contained fusion proteins capable of binding to anti–β-galactosidase antibodies. (B) Dot blot screening of sera for antibodies to leukemia derived proteins. (Top) Negative reactivity to all eight proteins. (Middle) Positive reactivity to all eight proteins. (Bottom) Positive reactivity to proteins derived from clone no. 5 only. B1, bacterial extract from a clone of blue phage; B2, bacterial extract from a nontransformed E coli.
Dot blot analysis using crude fusion protein extracts. (A) Bacterial extracts from all eight clones contained fusion proteins capable of binding to anti–β-galactosidase antibodies. (B) Dot blot screening of sera for antibodies to leukemia derived proteins. (Top) Negative reactivity to all eight proteins. (Middle) Positive reactivity to all eight proteins. (Bottom) Positive reactivity to proteins derived from clone no. 5 only. B1, bacterial extract from a clone of blue phage; B2, bacterial extract from a nontransformed E coli.
Prevalence of IgG Antibodies Directed at 8 Leukemia-Derived Antigens (P Value Compared With CML Patients)
| Diagnosis . | Antigen . | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | |
| CML | 9/15 | 9/15 | 8/15 | 8/15 | 9/15 | 11/15 | 8/15 | 9/15 |
| Normal | 2/24 (P < .01) | 2/24 (P < .01) | 2/24 (P < .01) | 2/24 (P < .01) | 9/24 NS | 2/24 (P < .001) | 2/24 (P < .01) | 1/24 (P < .01) |
| Other hematological malignancies | 7/38 (P < .01) | 7/38 (P < .01) | 7/38 (P < .01) | 7/38 (P < .01) | 8/38 (P < .02) | 7/38 (P < .001) | 7/38 (P < .01) | 7/38 (P < .01) |
| Diagnosis . | Antigen . | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | |
| CML | 9/15 | 9/15 | 8/15 | 8/15 | 9/15 | 11/15 | 8/15 | 9/15 |
| Normal | 2/24 (P < .01) | 2/24 (P < .01) | 2/24 (P < .01) | 2/24 (P < .01) | 9/24 NS | 2/24 (P < .001) | 2/24 (P < .01) | 1/24 (P < .01) |
| Other hematological malignancies | 7/38 (P < .01) | 7/38 (P < .01) | 7/38 (P < .01) | 7/38 (P < .01) | 8/38 (P < .02) | 7/38 (P < .001) | 7/38 (P < .01) | 7/38 (P < .01) |
Abbreviation: NS, not significant.
The high prevalence of antibodies appears to be disease-related. Only 7 of the 38 patients with other hematological malignancies (1 myeloma, 1 myeloproliferative disease, 1 non-Hodgkin’s lymphoma, 2 monoclonal gammopathy of unknown significance, 1 myelodysplastic syndrome, and 1 acute myeloid leukemia) also exhibited antibodies to most of the eight antigens. Despite being of a younger patient group, more CML patients produced antibodies to these antigens when compared with the group of older patients with other hematological malignancies (.001 <P < .02). This was observed even after excluding patients with multiple myeloma (.01 < P < .05) whose associated immune-paresis may have limited any antibody responses.
To determine the durability of the B-cell responses, sera from a number of individuals were tested on two or three separate occasions. Sera were collected between 3 and 6 months apart. In all cases, the results were identical.
In 20 randomly selected samples, we also compared our method of antibody screening with screening using phage plaques as described in previous studies.12 13 We found concordance between results obtained by these two techniques. However, in general, results obtained with phage plaques tended to be less clear, probably reflecting lower concentrations of the antigens present in the plaques compared with bacterial lysate.
DISCUSSION
Recent advances in tumor immunology suggest the potential immunogenicity of various tumors in the autologous setting. Furthermore, with the successful generation of autologous cytotoxic T-lymphocytes (CTLs) against tumor cells, an array of antigenic targets on the tumor cells has been defined, either through the screening of target cells transfected with recombinant tumor DNA libraries17 or biochemical purification of peptides eluted from the MHC molecules.18 Unfortunately such approaches are limited by the ability to generate in vitro tumor-reactive CTLs. Therefore, these techniques cannot be easily adapted to define antigens in most tumors. Based on the knowledge of overexpressed normal proteins or novel fusion proteins in tumor cells, workers in CML have identified immunogenic epitopes within the bcr-abl7-9 and proteinase 310 proteins. Peptides within these proteins are able to stimulate the generation of specific CTLs that also recognize HLA-matched fresh leukemia cells. However, such an approach does not provide any information regarding the immunogenicity of these peptides in vivo. It remains to be determined if the CTL reactivity with fresh leukemia cells represents true immune responses against peptides from these proteins that have been endogenously processed and presented on the leukemia cells or cross-reactivity with other endogenously processed peptides expressed on leukemia cells. In this study, we have used the SEREX approach to investigate any B-cell responses to antigens derived from the leukemia cells in CML. This work therefore provides foundation for future identification of leukemia antigens in CML.
We demonstrated that multiple B-cell immune responses occur in the autologous host to proteins derived from the leukemia cells in CML. A total of eight cDNA clones encoding the proteins were identified. Six of these eight sequences showed strong homology to known genes, whereas the other two were novel sequences. Interestingly, although the six sequences showed strong homology to known genes, there was an internal nucleotide deletion in two of these sequences. Two fusion sequences were also identified that are currently under further investigations to exclude that they are cloning artefacts. It remains to be determined if the antibody responses arose as a result of these genetic anomalies. Using sequence comparison, we have so far not identified any mutation in these gene fragments. Surprisingly, potential CML antigens such as the bcr-abl fusion protein and proteinase 3 were not represented.
We then determined the tissue distribution of these gene sequences in CML cells, leukemia cell lines, and a limited panel of normal tissue. Although seven of the eight clones showed ubiquitous expresssion, clone no. 4 appeared to exhibit restricted tissue expression, being detected only in normal testis and in CML cells. Interestingly, it was also detected in CML cell line, K562. Although the restricted expression of clone no. 4 requires to be confirmed using a bigger panel of normal tissues, clone no. 4 with a novel gene sequence may be a new member of the group of cancer-testicular antigens in which normal testicular antigens are aberantly expressed in tumor cells. Work is ongoing to confirm this and to express the cDNA in mammalian cells for further characterization of the protein.
The observation that antibodies against seven of the eight antigens were found predominantly in disease-bearing patients suggests that the immune responses may be disease-related. This was observed even after excluding CML patients on IFN-α therapy, because IFN-α therapy may potentially induce autoimmunity. However, we also detected antibodies in two healthy donors. Although it is possible that the antibodies detected in healthy donors may not be directed at the same B-cell epitopes on these proteins, one possible explanation for the presence of antibodies in these individuals is that the antigen may also be expressed under nonmalignant conditions such as viral infections or other inflammatory processes. Another explanation may be the presence of malignant CML cells in these apparently normal individuals, because a previous study19 indicated the ability to detect thebcr-abl transcripts in apparently normal individuals.
The antibodies against the leukemia-derived proteins detected by our screening system were of the IgG subclass, implying the possible involvement of CD4 cognitive help in the B-cell responses. The coexistence of cellular and humoral responses have indeed been reported in tumor antigens such as NY-ESO-1 in esophageal carcinoma14 and HER-2/neu in breast cancer.20In CML, this is particularly relevant, because, based on works involving selective T-cell depletion for allogeneic BMT,21CD4 T cells appear to have an important role in mediating antileukemic immune responses. Obviously such antileukemic immune responses would only occur if the antigens are also processed and presented by the leukemia cells in the context of MHC class II molecules.
The clinical significance of B-cell responses to leukemia-derived proteins is unclear. In breast cancer, the presence of antibodies directed to p53 protein has been associated with a poor clinical outcome.22 It is therefore important to determine if the presence of antibodies to these proteins influences the responses to IFN-α therapy and outcome after allogeneic BMT in CML. However, this could only be achieved through analysis of many more CML patients. Further work would also permit the determination of any correlation between antibody production and mRNA expression by the leukemia cells.
Supported by the Leukaemia Research Fund, UK, the Leukaemia Research Appeal for Wales, and the Welsh Bone Marrow Transplant Research Fund.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Seah H. Lim, MD, PhD, Myeloma and Transplantation Research Center, University of Arkansas for Medical Sciences, 4301 W Markham, Slot 776, Little Rock, AR 72205.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal