Abstract
Severe immune thrombocytopenia is an idiosyncratic complication of quinine therapy. Although in most cases the responsible antibody is directed against platelet membrane glycoprotein (GP) Ib-IX, specificity for GPIIb-IIIa or both epitopes has also been reported. The objective of this study was to characterize the binding site of GPIb-IX–specific quinine-dependent antibodies. Antibody binding to Chinese hamster ovary cells or mouse L cells stably transfected with various combinations of the three genes (Ibα, Ibβ, or IX) that encode this complex was detected using flow cytometry, monoclonal antibody–specific immobilization of platelet antigens assay, and differential adsorption studies. IgG in sera from 15 patients with quinine-induced thrombocytopenia binding to the cells, in the presence of quinine, showed three distinct patterns. Group 1 sera contained at least two antibody populations, one which binds to GPIbα and another which recognizes GPIX. Group 2 sera contained an antibody which binds drug dependently to GPIX, and Group 3 sera contained an antibody which recognizes a quinine-dependent epitope on GPIbα. Thus, the quinine-dependent antibodies fall into two distinct populations that bind to GPIbα and GPIX independently. Using proteases which cleave GPIbα at specific sites, we have shown that the GPIbα-specific antibody binds to an 11–amino acid (283 to 293) region. Peptide inhibition studies provide confirmatory evidence that this region contains the epitope for the GPIbα-specific quinine-dependent antibody.
DRUG-INDUCED thrombocytopenia is a common hematologic problem in clinical practice.1 Drugs frequently implicated include quinine and its optical isomer, quinidine. In quinine-/quinidine-induced thrombocytopenia, drug-dependent binding of antibodies to platelets leads to increased platelet clearance by the reticuloendothelial system and results in severe thrombocytopenia of acute onset. In the majority of cases the antibody is directed against platelet membrane glycoprotein (GP) Ib-IX complex,2,3although antibodies with specificity against GPIIb-IIIa have also been reported.4 5
GPIb is a major sialoglycoprotein on the platelet cell surface with approximately 25,000 copies per platelet.6 GPIb is composed of two subunits, the α subunit of 143 kD which is disulphide bonded to a smaller β subunit of 24 kD. GPIbα can be cleaved by a Ca2+-dependent protease resulting in a single-chain, water-soluble glycoprotein of 120 kD (glycocalicin) and a small membrane-bound fragment. GPIb is noncovalently linked to GPIX, which has a molecular mass of 20 kD. The three glycoproteins have leucine-rich motifs and they exist as a heterodimeric complex in the platelet membrane.7,8 GPIb-IX is associated noncovalently with GPV, a glycoprotein of 82 kD, which also has leucine-rich motifs.9 The components of GPIb-IX are all encoded by separate genes.10
Kunicki et al11 first showed that GPIb-IX was involved in quinine-induced thrombocytopenia when they reported that the quinine-/quinidine-dependent antibodies failed to react with platelets from an individual with Bernard-Soulier Syndrome. We provided the first direct evidence that the GPIb-IX complex was the platelet autoantigen for the antibody in quinine-induced thrombocytopenia when we showed the drug-dependent immunoprecipitation of GPIb and GPIX using a patient's serum.12 We subsequently showed2 that the portion of GPIb-IX that remains associated with the platelet membrane after removal of glycocalcin by proteases and the NH2terminal of the GPIbα are the components of the complex that is involved with antibody binding. The precise antibody binding site(s) on GPIb-IX has not been reported.
In this study we have shown that antibodies from patients with quinine-induced thrombocytopenia react with only a limited number of epitopes within the GPIb-IX complex. We identified only two binding sites for the quinine-dependent antibodies. One site is on GPIX and the other on GPIbα, which we have narrowed to a stretch of 11 amino acids, encompassing residues 283 to 293.
This is the first study to define the quinine-dependent antibody binding sites on GPIb-IX in such detail. This is the first report that has been able to localize the binding site of one of these antibodies to a region of 11 amino acids on GPIbα.
MATERIALS AND METHODS
Materials.
Bovine serum albumin (BSA), quinine hydrochloride, phenylmethylsulfonyl fluoride (PMSF), EDTA, disodium salt, bacitracin, benzamidine, dithiothreitol (DTT), dimethylsulfoxide, iodoacetamide, propidium iodide, and 2,2′-azinobis 3-ethylbenzthiazolinesulfonic acid (ABTS) were purchased from Sigma (St Louis, MO); 3,3′,5,5′-tetramethylbenzidine dihydrochloride from Kirkegaard & Perry (Gaithersburg, MD); hydrogen peroxide (30% wt/vol), Triton X-100, and Tween-20 from BDH (Poole, Dorset, UK); sulfosuccinimidobiotin from Pierce (Rockford, IL); polyscreen poly vinylidene difluoride (PVDF) transfer membrane and Western blot chemiluminesence reagents from DuPont (Boston, MA); and sheep anti-mouse IgG–coated dyna beads from Dynal (Oslo, Norway). All chemicals were of analytical reagent grade.
Antibodies.
The goat anti-mouse Ig and the horseradish peroxidase (HRP)-conjugated goat anti-human Ig (Jackson, West Grove, PA), rabbit anti-mouse HRP-conjugated antibody (Dako, Carpentaria, CA), and the streptavidin HRP-conjugated antibody (Amersham, Bucks, UK) were purchased as indicated.
All monoclonal antibodies (MoAbs) were of the IgG class. MOPC21 (Becton Dickinson, San Jose, CA), a murine IgG1 myeloma protein, was used as a control Ig, and SZ2 was purchased from Immunotech (Marseille, France). Gi27 was a generous gift from Dr S. Santoso (Giessen, Germany). AK2, AK3, and FMC25, which are all directed against epitopes on various parts of the human GPIb-IX complex, were raised and characterized in one of our laboratories (M.C.B.) and the preparation of these antibodies has been previously described.8 13
Drug-dependent antibodies.
Sera or plasma from 15 patients (6 men and 9 women; ages 18 years 10 months to 76 years 0 months; mean, 53 years 10 months) with quinine-induced thrombocytopenia were used in this study. The samples were selected at random from patients presenting at this hospital and other hospitals from which samples were forwarded to our laboratory for testing. These patients developed thrombocytopenia when they were receiving quinine and their thrombocytopenia resolved after the cessation of the drug. Other causes of thrombocytopenia were clinically excluded. The diagnosis of quinine-induced thrombocytopenia was confirmed by a positive platelet antibody test.
Cell lines.
Chinese hamster ovary (CHO) cells and mouse L (tk−) cells were obtained from American Type Tissue Collection (Rockville, MD) and maintained as recommended by the supplier.
Synthetic peptides.
The peptide specific for amino acids 283-293 of GPIbα (H-DTEGDKVRATR-NH2), the nonspecific peptide that had the same amino acid sequence in reverse (H-RTARVKDGETD-NH2), and the unrelated nonspecific peptide (H-PTLGDEGDTDLYDYY-NH2) were manufactured by Chiron Technologies (Clayton, Vic, Australia).
Transfection of CHO and L cells with GPIb-IX genes.
CHO DUK− (DHFR−) cells and mouse L (tk−) cells were stably transfected with cDNA encoding the GPIb-IX subunits in various combinations as previously described.14 The cloning of the cDNAs for GPIbα, GPIbβ, and GPIX has been reported previously.15-17 The three cDNAs (each containing the entire coding sequence and the 3′-untranslated region) were cloned separately into the eukaryotic expression vector pDX (a kind gift from Dr K. Berkner, Seattle, WA) in which transcription is driven by the adenovirus major late promoter and the SV40 enhancer. The CHO cells were transfected with the following combinations of GPIb-IX subunit cDNAs: CHO GPIbα + GPIbβ + GPIX, GPIbα + GPIbβ, GPIbα + GPIX, and GPIbβ + GPIX. The L cells were transfected with GPIbα + GPIbβ and GPIbβ + GPIX. Expression of the GPIb-IX subunits in the cell lines was substantiated by Northern blot analysis to detect mRNA and by flow cytometry and enzyme-linked immunosorbent assay (ELISA) to ensure expression of the subunits on the cell surface.
Transient transfections of L αβ cells with GPIX cDNA were also performed by liposome-mediated DNA transfer using a commercial kit (GIBCO-BRL, Gaithersburg, MD). Twelve μL of liposome suspension and 4 μg of GPIX cDNA were separately mixed in 300 μL of serum-free Dulbecco's modified Eagle's medium (DMEM) nutrient mixture F-12 (DMEM/F-12) (Trace, Sydney, Australia). The two suspensions were then combined, mixed gently, and incubated for 30 minutes at room temperature. The mixture was diluted in 1.8 mL of serum-free DMEM/F-12 and added to the cells that had been washed once with the same medium. The cells were exposed to the DNA/liposome mixture for 7 hours under standard culture conditions (37°C, 5% CO2), then 1.6 mL of DMEM/F-12 containing fetal bovine serum (GIBCO-BRL) was added to a final concentration of 10% (vol/vol). The medium was changed 24 hours after the start of the transfection. After 48 hours the cells were detached from the culture dishes with 0.53 mmol/L EDTA/phosphate-buffered saline (PBS) and evaluated by flow cytometry for surface expression of GPIX.
Flow cytometry.
Flow cytometry was performed on a FACStar Plus cytometer (Becton Dickinson) fitted with a 100-mW air-cooled argon ion laser using the 488-nm green line for fluorescence excitation. The cell emission spectra were collected on FL1 (green) using a band pass filter 530 DF 30. Dead cells were excluded using propidium iodide on FL3 using a 700 LP filter. Before analysis the cells were detached from the culture flasks with 0.53 mmol/L EDTA/PBS and resuspended in 1% BSA/PBS. The cells (at a concentration of 2 × 105 per tube) were blocked in 2% BSA/PBS for 10 minutes at room temperature, washed once, pelleted by centrifugation at 2,000g, and resuspended before incubation with MoAb (10 μg/mL) or patient serum (1 in 200 dilution) for 10 minutes at room temperature in the presence or absence of quinine (0.3 mmol/L). In experiments performed in the presence of quinine, the working buffer contained the drug beyond this stage at a concentration of 0.3 mmol/L. The cells were then washed three times in 1% BSA/PBS before being incubated for 10 minutes at room temperature with the relevant fluorescein isothiocyanate (FITC)-conjugated secondary antibody (goat anti-mouse IgG at 1 in 200 dilution or goat anti-human IgG at 1 in 100 dilution). The cells were washed four times before being resuspended in 1% BSA/PBS and were kept in the dark until analysis.
In some experiments the cells were incubated with a proteolytic enzyme before blocking with 2% BSA/PBS. The cells were resuspended in 1% BSA/PBS at 3.5 × 106 cells/mL. The proteolytic enzymes mocarhagin and trypsin (porcine 10× stock, specific activity 1:250; GIBCO-BRL) were added separately to 300 μL of cells at a final concentration of 50 μg/mL. The mocarhagin18was purified in our laboratory. The cells were incubated at 22°C for 10 minutes before the reactions were stopped by the addition of 2 mmoL of PMSF. The cells were divided into six tubes and blocked with 2% BSA/PBS. The labeling of these cells was then continued as described above.
ELISA.
Cells were allowed to grow to confluence before being washed with 1% BSA/PBS and detached from the tissue culture vessel. After resuspension in 1% BSA/PBS the cells were seeded at 5 × 104 cells per well in a 96-well microtiter plate (Corning, Cambridge, MA). The plate was spun at 200g for 10 minutes at 4°C before the supernatant was removed and the cells were allowed to dry overnight at 37°C. The plate was stored at −20°C, with desiccant, in an airtight container until analysis.
After defrosting, the plates were washed twice with 0.05% Tween-20/PBS. The wells were blocked with 200 μL of 2% BSA/PBS for 60 minutes at 37°C and washed three times. The appropriate MoAbs, at a final concentration of 2 μg/mL in 1% BSA/PBS, were added and incubated for 90 minutes at 37°C. The plates were washed four times before the rabbit anti-mouse HRP-conjugated secondary antibody (diluted 1 in 1,000 in 1% BSA/0.05% Tween-20/PBS) was added and incubated for 30 minutes at 37°C. The plate was then washed five times and the ABTS substrate solution was left to develop for 30 minutes in the dark at room temperature. The reaction was stopped with 3% (wt/vol) oxalic acid and the absorbance read on a Titertek photometer (Labsystems, Helsinki, Finland) at 414 nm.
MoAb-specific immobilization of platelet antigens (MAIPA) assay.
The MAIPA assay was performed as previously described with minor modifications.19 Briefly, the cells detached from the culture flasks were centrifuged in test tubes at 2,000g for 2 minutes to give a total of 1.5 to 2 × 106cells per tube. The cells were resuspended in 100 μL PBS/0.2% EDTA buffer with 0.3% BSA/1% FCS (PEBF buffer) and added to 50 μL of patient serum, with or without 5 μL of quinine (final concentration, 0.5 mmol/L), and incubated at 37°C for 30 minutes. The cells were washed once with PEBF buffer. Quinine (0.3 mmol/L) was included in the washing buffer throughout the procedure if the cells were incubated with the drug together with the patient serum at the beginning of the experiment. Twenty μL of MoAb (final concentration, 10 mg/mL) was added and incubated at 37°C for 30 minutes. The cells were pelleted and washed three times with PEBF buffer. The cells were lysed by resuspension in 100 μL 0.01 mol/L Tris-buffered saline (TBS) containing 0.5% Triton X-100 and 0.05% Tween-20 ± 0.3 mmol/L quinine, pH 7.4. The cell lysate was centrifuged at 12,000g for 30 minutes at 4°C. Seventy μL of the supernatant was diluted in 180 μL of TBS buffer. The diluted supernatant (100 μL) was added in duplicate to the wells of a microtiter plate (Immunotech) that had been coated with 100 μL of affinity-purified goat anti-mouse IgG (1/500 in 0.05 mol/L sodium carbonate buffer) by overnight incubation at 4°C, washed, and blocked with 250 μL per well of TBS buffer for 60 minutes at 4°C. The microtiter plate was incubated for 90 minutes at 4°C and washed four times with TBS buffer. Anti-human IgG HRP conjugate (100 μL diluted 1:10,000 in TBS buffer) was added to each well and incubated for 120 minutes at 4°C. After six washes, 100 μL of substrate solution (0.42 mmol/L 3,3′,5,5′-tetramethylbenzidine dihydrochloride in 100 mmol/L sodium acetate/citric acid, pH 6.0, with 1.3 mmol/L hydrogen peroxide) was added. The color reaction was stopped by the addition of 50 μL 2 mol/L sulfuric acid after 15 minutes and read at the dual wavelengths of 450 and 492 nm in a Titertek photometer (Labsystems). The positive control for the assay was normal pooled platelets at a concentration of 20 × 106 plus 50 μL patient serum ± 6.5 μL 7.5 mmol/L quinine (final concentration 0.5 mmol/L).
Biotin labeling of GPIb-IX subunits.
The cells were incubated with 70 μmol/L sulfosuccinimidobiotin for 30 minutes at room temperature in a capped tube, with constant rotation. The free biotin was quenched with a twofold molar excess of glycine for 10 minutes at room temperature, before being washed once. MoAb (AK2, AK3, or SZ2; final concentration 10 μg/mL) was then incubated with the cells for 30 minutes at 4°C, before the cells were washed three times. The cells were lysed, in the presence of protease inhibitors (PMSF 200 mmol/L, bacitracin 10 mg/mL, and benzamidine 200 mmol/L ) by resuspension in 100 μL 0.01 mol/L TBS containing 0.5% Triton X-100, 0.05% Tween-20, and incubation at 4°C for 30 minutes. The cells were then centrifuged at 12,000g for 30 minutes at 4°C. The supernatant (70 μL) was diluted in 430 μL PBS and 1 × 107 goat anti-mouse IgG coated dyna beads were added. After incubation at 4°C for 45 minutes, the dyna beads were washed three times using the MPC-E-1 magnet before they were resuspended in 2 × Laemmli buffer and stored at −80°C until sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Proteolytic digestion of GPIbα.
Cell surfaces labeled with biotin as described above were subjected to proteolytic digestion. Mocarhagin or trypsin was added to 300 μL of cells (3.5 × 106 cells/mL) at a final concentration of 50 μg/mL. The cells were incubated at 22°C for 10 minutes before the reactions were stopped by the addition of 200 mmol/L PMSF. The cells were washed once and the cells and supernatant were transferred to separate tubes. AK3, AK2, or SZ2 were added to the cells and the supernatant samples at a final concentration of 10 μg/mL. After incubation for 30 minutes at 4°C the cells were washed and lysed as described above. The cell lysates and supernatant samples were added to dyna beads (monoclonal sheep anti-mouse IgG) for 45 minutes at 4°C. The dyna beads were separated from the supernatant using the Dynal MPC-E-1 magnet and washed extensively. The samples were stored at −80°C until SDS-PAGE analysis.
SDS-PAGE and detection of proteins.
Samples for SDS-PAGE analysis were treated with 2 μL 0.5 mol/L DTT and boiled for 5 minutes. Free DTT was quenched by the addition of 4 μL 0.5 mol/L iodoacetamide. SDS-PAGE was performed according to the method of Laemmli20 using 4% to 15% Tris-glycine gradient gels (Bio-Rad, Hercules, CA). After completion of electrophoresis the proteins were transferred to PVDF membrane. Membranes were blocked overnight in 5% (wt/vol) skim milk (Diploma, Bonlac Foods Ltd, Melbourne, Australia) and the following morning washed 5 times in PBS/0.05% Tween-20. Streptavidin HRP diluted 1 in 2,000 in 2% BSA/PBS/0.05% Tween-20 was incubated with the membrane for 60 minutes with rocking. After five washes the presence of a signal was detected using the Western blot chemiluminescence reagents according to the manufacturer's instructions.
Peptide inhibition of quinine-dependent antibody binding.
To 10 μL of patient serum (diluted 1 in 200) was added a specific peptide (H-DTEGDKVRATR-NH2) or either a reverse amino acid sequence nonspecific peptide (H-RTARVKDGETD-NH2) or an unrelated nonspecific peptide (H-PTLGDEGDTSLYDYY-NH2) at a final concentration of 200 μmol/L in the presence of quinine (0.3 mmol/L). After incubation at room temperature for 10 minutes this mixture was added to L αβ cells that had been blocked with 2% BSA. The cells were then prepared for flow cytometry as previously described. The optimal concentration of peptide for inhibition was determined using a dose-response curve for the specific peptide.
Statistical analysis.
Statistical analysis was performed using the one-tailed Mann-Whitney U test.
RESULTS
Quinine-dependent antibodies binding to GPIb-IX.
Flow cytometry and MAIPA assays were used to test the binding of sera from 15 patients with quinine-induced thrombocytopenia to mouse L cells that had been stably transfected with either the human GPIbβ and GPIX components of GPIb-IX (L βIX cells) or the human GPIbα and GPIbβ components (L αβ cells). Binding was not observed in the absence of quinine. In the presence of quinine three distinct binding patterns were observed (Table 1). Group 1 (sera from 6 patients) bound to both cell types. Groups 2 (7 patients) and 3 (2 patients) bound only to L βIX cells and L αβ cells, respectively.
Binding of Quinine-Dependent Antibodies to L βIX and L αβ cells
| Patient Sera . | Antibody Binding to L βIX and L αβ Cells . | |||
|---|---|---|---|---|
| L βIX Cells . | L αβ Cells . | |||
| −QN . | +QN . | −QN . | +QN . | |
| Group 1 n = 6 | − | + | − | + |
| Group 2 n = 7 | − | + | − | − |
| Group 3 n = 2 | − | − | − | + |
| Patient Sera . | Antibody Binding to L βIX and L αβ Cells . | |||
|---|---|---|---|---|
| L βIX Cells . | L αβ Cells . | |||
| −QN . | +QN . | −QN . | +QN . | |
| Group 1 n = 6 | − | + | − | + |
| Group 2 n = 7 | − | + | − | − |
| Group 3 n = 2 | − | − | − | + |
Reactions of sera from patients with quinine-induced thrombocytopenia to L cells expressing GPIbβ and GPIX (L βIX cells) or GPIbα and GPIbβ (L αβ cells) in the presence or absence of quinine were assayed by flow cytometry and the MAIPA assay. As shown above, three distinct quinine-dependent antibody binding patterns were observed. A negative control of pooled normal AB sera did not react with either cell type in the presence of quinine (data not shown).
Several group 2 sera were assayed for their ability to bind to L αβ cells before and after transfection with GPIX cDNA. Binding was observed neither in the absence nor in the presence of quinine before transfection of the L αβ cells with GPIX cDNA but was observed in the presence of quinine after transfection with the GPIX cDNA (Fig 1).
Reactivity of sera from group 2 with L αβ cells before and after transfection with GPIX cDNA. L αβ cells were transfected with GPIX cDNA using the GIBCO-BRL lipofectAMINE kit as described in Materials and Methods. The cells were labeled with primary antibody (either anti-GPIX MoAb FMC25 [A and B], or patient sera from group 2 plus quinine [C and D]) followed by a FITC-conjugated secondary antibody and examined by flow cytometry. L αβ cells did not react with anti-GPIX MoAb before transfection with GPIX cDNA (A) but bound anti-GPIX MoAb after GPIX cDNA transfection (B). Patient sera from group 2 did not react with L αβ cells before transfection with GPIX cDNA (C) but were able to bind to L αβ cells expressing GPIX on their surfaces after transfection with GPIX cDNA (D). The graphs are representative of the results observed with three different patients' sera. The solid peak in each graph represents the negative control.
Reactivity of sera from group 2 with L αβ cells before and after transfection with GPIX cDNA. L αβ cells were transfected with GPIX cDNA using the GIBCO-BRL lipofectAMINE kit as described in Materials and Methods. The cells were labeled with primary antibody (either anti-GPIX MoAb FMC25 [A and B], or patient sera from group 2 plus quinine [C and D]) followed by a FITC-conjugated secondary antibody and examined by flow cytometry. L αβ cells did not react with anti-GPIX MoAb before transfection with GPIX cDNA (A) but bound anti-GPIX MoAb after GPIX cDNA transfection (B). Patient sera from group 2 did not react with L αβ cells before transfection with GPIX cDNA (C) but were able to bind to L αβ cells expressing GPIX on their surfaces after transfection with GPIX cDNA (D). The graphs are representative of the results observed with three different patients' sera. The solid peak in each graph represents the negative control.
Patient sera from all three groups were examined for their ability to bind to CHO cells expressing GPIbα alone on the surface, in the presence of quinine. (CHO α cells were initially transfected with GPIbα and GPIbβ but the cells rapidly lost expression of GPIbβ on repeated cell passage.) Sera from groups 1 and 3 bound this cell type but the sera from group 2 did not bind (results not shown).
Altogether, these data indicate that (1) group 1 sera contain at least two antibody populations, one that binds to GPIbα, and another that recognizes either GPIbβ or GPIX; (2) group 2 sera contain an antibody that binds drug dependently to GPIX; and (3) group 3 sera contain an antibody that recognizes a quinine-dependent epitope on GPIbα.
Competitive MAIPA.
Group 1 sera were further analyzed using competitive MAIPA. As shown previously the antibody binding was only observed in the presence of quinine.
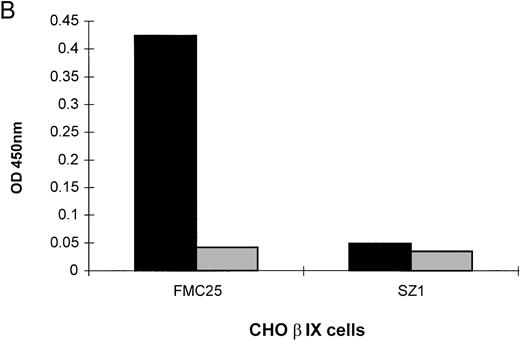
The MAIPA assay is an antigen-capture ELISA which uses a MoAb to capture GPIb-IX and the human drug-dependent antibody then binds to the GP complex at a site distant from the MoAb-binding site. If the human antibody and the murine MoAb-binding sites coincide or are in close proximity to each other, the MoAb cross-blocks the human antibody and a negative result is obtained. Group 1 serum showed quinine-dependent antibody binding to CHO α cells when the non–cross-blocking MoAb, AK2, was used to capture the antigen. Similarly, strong quinine-dependent antibody binding to CHO βIX cells occurred when the non–cross-blocking MoAb, FMC25, captured the GPIbβ-GPIX complex. Figure 2A illustrates the results obtained with one representative patient's serum; similar results were observed with three other patients' sera. These data confirm that group 1 serum contains at least two separate antibodies, one which reacts with GPIbα, and another that reacts with either GPIbβ or GPIX. When the experiment was repeated using the anti-GPIX MoAb, SZ1, binding of the drug-dependent antibody to CHO βIX cells was completely inhibited (Fig 2B), suggesting that the specificity of the second antibody type in group 1 serum is GPIX rather than GPIbβ. In contrast, if SZ1 and CHO αβIX cells were used in the experiment, we detected a mild quinine-dependent reaction which was due to the GPIbα-specific antibody in group 1 serum remaining after the GPIX-specific antibody had been completely cross-blocked by SZ1 (data not shown).
Reactions of group 1 patient sera with CHO cells stably transfected with cDNAs from various parts of the GPIb-IX complex and inhibition of quinine-dependent antibody binding by anti-GPIX MoAbs. MAIPA studies were performed in the presence and absence of quinine with patient serum and a MoAb as indicated in the figure. (A) The results of one representative patient's serum binding to mock-transfected CHO, CHO βIX, and CHO α cells in the presence of quinine. Binding was not observed in the absence of quinine (not shown). In the presence of quinine, binding did not occur with mock-transfected CHO cells but was observed with CHO βIX and CHO α cells. (B) The results of one representative patient's serum binding to CHO βIX cells expressing GPIbβ and GPIX in the presence (▩) or absence (▧) of quinine and a competitive MoAb (FMC25 or SZ1). No binding was observed in the absence of quinine. Competition with FMC25 did not inhibit the quinine-dependent antibody binding. Conversely, competition with SZ1 (specific for GPIX) inhibited binding of the quinine-dependent antibody.
Reactions of group 1 patient sera with CHO cells stably transfected with cDNAs from various parts of the GPIb-IX complex and inhibition of quinine-dependent antibody binding by anti-GPIX MoAbs. MAIPA studies were performed in the presence and absence of quinine with patient serum and a MoAb as indicated in the figure. (A) The results of one representative patient's serum binding to mock-transfected CHO, CHO βIX, and CHO α cells in the presence of quinine. Binding was not observed in the absence of quinine (not shown). In the presence of quinine, binding did not occur with mock-transfected CHO cells but was observed with CHO βIX and CHO α cells. (B) The results of one representative patient's serum binding to CHO βIX cells expressing GPIbβ and GPIX in the presence (▩) or absence (▧) of quinine and a competitive MoAb (FMC25 or SZ1). No binding was observed in the absence of quinine. Competition with FMC25 did not inhibit the quinine-dependent antibody binding. Conversely, competition with SZ1 (specific for GPIX) inhibited binding of the quinine-dependent antibody.
Adsorption of drug-dependent antibody subtypes.
Sera from patients in group 1 were absorbed against three different cell types and the further binding potential of the sera was assessed. When the sera were absorbed with untransfected L cells no difference in the binding pattern was observed. Preadsorption of the same sera with L αβ cells ablated any further binding to the L αβ cells, but the binding to the L βIX cells remained. Conversely, preadsorption of the patient sera with the L βIX cells abrogated further binding to the L βIX cells, but the binding to the L αβ cells was unchanged (Table 2). These data indicate that group 1 sera which reacted with both L αβ and L βIX cells did not contain one antibody (directed against the shared subunit, GPIbβ) but two distinct antibodies, one with specificity for GPIbα and another with specificity for GPIX.
Results of Absorption Study of Group 1 Sera With Untransfected L, L αβ and L βIX Cells
| After Absorbing With: . | Binding of Quinine-Dependent Antibodies to: . | |
|---|---|---|
| L αβ . | L βIX . | |
| L0 Cells | + | + |
| L αβ Cells | − | + |
| L βIX Cells | + | − |
| After Absorbing With: . | Binding of Quinine-Dependent Antibodies to: . | |
|---|---|---|
| L αβ . | L βIX . | |
| L0 Cells | + | + |
| L αβ Cells | − | + |
| L βIX Cells | + | − |
L0 cells are untransfected L cells. L αβ cells are stably transfected with GPIbα cDNA and GPIbβ cDNA and have been confirmed to express the corresponding proteins. L βIX cells are stably transfected with GPIbβ and GPIX cDNA and are expressing GPIbβ and GPIX proteins. Group 1 patient sera were absorbed with L0 cells, L αβ cells, and L βIX cells and then assayed for quinine-dependent antibody binding to L αβ or L βIX cells.
Mapping the antibody-binding site on GPIbα using selective enzymatic cleavages.
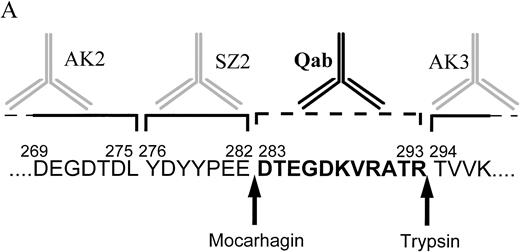
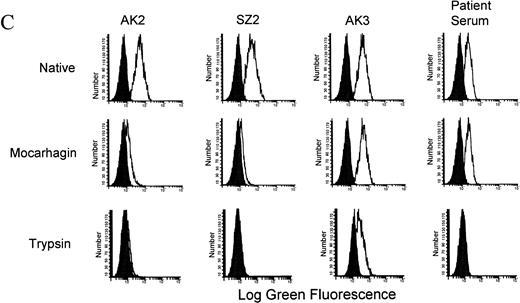
Three enzymes have recently been shown to cleave GPIbα at specific sites.18 Using sequential cleavage with two of these enzymes the location of the patient antibody-binding site on GPIbα was determined (Fig 3A). The degree of digestion of GPIbα, using mocarhagin and trypsin, was determined by labeling the CHO αβIX cell surface proteins with biotin before digestion and extracting the protein components from the cell surfaces and the supernatants before and after cleavage to be run on SDS-PAGE, as described in Materials and Methods. Complete digestion of the GPIbα protein was observed with mocarhagin (Fig 3B), with a 70-kD band immunoprecipitated from the cell lysate by AK3 and a 40-kD band precipitated by AK2 and SZ2 from the supernatant collected after the cleavage process. A similar banding pattern was observed after the digestion with trypsin (results not shown). The GPIX protein was not affected by these enzymes. GPIbβ was also unaffected by the digestions. Flow cytometry was used to analyze the binding of two group 1 serum samples to L αβ cells. The MoAbs AK2, AK3, and SZ2 were used to ensure that specific cleavage had occurred (Fig 3C). After cleavage with mocarhagin, at position 283, AK2 and SZ2 no longer bound, but AK3 and the quinine-dependent antibody binding was unaffected. Cleavage with trypsin, at position 293, resulted in loss of both AK2 and SZ2 binding, but the binding site for AK3 remained intact, although reduced in magnitude. The quinine-dependent antibody binding was inhibited by cleavage with trypsin. These results indicate that the quinine-dependent antibody is binding to GPIbα between the mocarhagin and the trypsin cleavage sites, between amino acids 283 and 293 (Fig3A).
Characterization of the quinine-dependent antibody binding domain on GPIbα. (A) GPIbα amino acid sequence residues 269 to 297. The specific cleavage sites for mocarhagin (a novel cobra venom metalloproteinase) and trypsin are shown. The MoAbs AK2, SZ2, and AK3 are specific for GPIbα. AK2 binds distal to amino acid 275, SZ2 between amino acids 276 to 282, and AK3 proximal to amino acid 294. The quinine-dependent antibody is interacting with GPIbα between amino acids 283 to 293. (B) After labeling the surface proteins of the CHO αβIX cells with biotin, the GPIbIX complex was immunoprecipitated using the MoAb panel. The presence of the GPIbα and GPIX native proteins was shown on the surface of the CHO αβIX cells. Lane 1 shows the components immunoprecipitated using AK3, lane 2 with AK2, and lane 3 with SZ2. After cleavage with mocarhagin, AK3 precipitated a 70-kD GPIbα component and the intact GPIX protein from the surface of the cells (lane 4). AK2 (lane 5) and SZ2 (lane 6) were able to precipitate a 40-kD GPIbα component from the supernatant collected after the cleavage step. (C) Sequential cleavage of GPIbα and checking the ability of the quinine-dependent antibody to continue binding enabled the definition of the domain to which the Ibα-specific antibody was binding. All MoAbs bound to the surface of the untreated L αβ cells. AK2 and SZ2 binding were inhibited after cleavage with mocarhagin. AK3 binding was still present, although at a lower level after cleavage with trypsin. The quinine-dependent antibody binding remained after cleavage with mocarhagin but was inhibited after cleavage with trypsin, indicating that the GPIbα-specific quinine-dependent antibody binding site is located between amino acids 283 and 293.
Characterization of the quinine-dependent antibody binding domain on GPIbα. (A) GPIbα amino acid sequence residues 269 to 297. The specific cleavage sites for mocarhagin (a novel cobra venom metalloproteinase) and trypsin are shown. The MoAbs AK2, SZ2, and AK3 are specific for GPIbα. AK2 binds distal to amino acid 275, SZ2 between amino acids 276 to 282, and AK3 proximal to amino acid 294. The quinine-dependent antibody is interacting with GPIbα between amino acids 283 to 293. (B) After labeling the surface proteins of the CHO αβIX cells with biotin, the GPIbIX complex was immunoprecipitated using the MoAb panel. The presence of the GPIbα and GPIX native proteins was shown on the surface of the CHO αβIX cells. Lane 1 shows the components immunoprecipitated using AK3, lane 2 with AK2, and lane 3 with SZ2. After cleavage with mocarhagin, AK3 precipitated a 70-kD GPIbα component and the intact GPIX protein from the surface of the cells (lane 4). AK2 (lane 5) and SZ2 (lane 6) were able to precipitate a 40-kD GPIbα component from the supernatant collected after the cleavage step. (C) Sequential cleavage of GPIbα and checking the ability of the quinine-dependent antibody to continue binding enabled the definition of the domain to which the Ibα-specific antibody was binding. All MoAbs bound to the surface of the untreated L αβ cells. AK2 and SZ2 binding were inhibited after cleavage with mocarhagin. AK3 binding was still present, although at a lower level after cleavage with trypsin. The quinine-dependent antibody binding remained after cleavage with mocarhagin but was inhibited after cleavage with trypsin, indicating that the GPIbα-specific quinine-dependent antibody binding site is located between amino acids 283 and 293.
Peptide inhibition of quinine-dependent antibody binding.
A dose-response curve for the specific peptide was obtained ranging from 2.5 to 500 μmol/L. Optimal inhibition was observed at 200 μmol/L. Incubation of the patient serum with a peptide specific to the 11 amino acids encompassing the antibody binding site before exposure to the L αβ cells decreased the level of antibody binding by 62.0% ± 8.8% (n = 3). In contrast, incubation of the patient serum with a reverse peptide that was composed of the same amino acid sequence in reverse resulted in 26.6% ± 5.8% decrease in antibody binding (n = 3). Incubation with a peptide homologous to an unrelated region of GPIbα resulted in a 15.5% ± 13.4% decrease in antibody binding. The inhibition with the specific peptide was significantly greater than the inhibition observed with the unrelated peptide (P ≤ .05). The observation of the small decrease in antibody binding in the presence of both unrelated peptides connoted that this decrease was a nonspecific effect, whereas the effect of the specific peptide was twofold to threefold stronger.
DISCUSSION
The aim of this study was to map the binding site(s) of the quinine-dependent antibodies on GPIb-IX. We have previously shown2 that the portion of GPIb-IX that remains associated with the platelet membrane after removal of glycocalcin by proteases and the NH2 terminal of GPIbα are the components of the complex involved in antibody binding. Cell lines transfected with the cDNAs of different GPIb-IX subunits were used to further define the domains to which the quinine-dependent antibodies bind.
Sera from 15 patients who had developed quinine-induced thrombocytopenia were examined. Three distinct binding patterns with the cells were observed. The sera in group 1 were able to bind to both the L αβ and the L βIX cells, indicating these sera contained at least two separate antibody populations with binding sites on different subunits of the GPIb-IX complex. The sera in group 2 were only able to bind to the L βIX cells and sera in group 3 only to the L αβ cells, indicating that each contained a different antibody type.
The sera in group 1 exhibited drug-dependent binding to more than one subunit of the GPIb-IX complex. The MAIPA and flow cytometry results showed that these sera were able to bind to the CHO αβIX, L αβ, CHO βIX, L βIX, and CHO α cells but not to mock-transfected cells in the presence of quinine. These results indicate that these sera contain antibodies specific for at least two of the three subunits of the GPIb-IX complex. The binding to CHO cells expressing GPIbα alone indicates that group 1 sera contain an antibody that reacts specifically to the GPIbα subunit. Because we were unable to generate CHO or L cells that expressed a single subunit of either GPIbβ or GPIX on the surface, alternative experimental approaches were required to characterize the antibody specificity to other subunits besides GPIbα. Our finding that the anti-GPIX MoAb, SZ1,21 could completely abolish binding of group 1 sera to CHO βIX cells indicates that these sera contained antibodies specific for GPIX but no antibody against GPIbβ. Overall, these data indicate that group 1 sera contained two antibody populations, one that reacts with GPIX and another with GPIbα. Consistent with our previous findings,2 we found that the anti-GPIX MoAb, SZ1, strongly inhibited the binding of group 1 sera to CHO αβIX cells (>90% inhibition). Because the quinine-dependent epitope on GPIbα and that on GPIX are not close to each other,2 this MoAb-blocking result indicates that in these sera, the anti-GPIX drug-dependent antibody (rather than the anti-GPIbα antibody) is the predominant one.
Preadsorption of the sera from group 1 with L αβ or L βIX cells inhibited binding when the sera was reexposed to the same cell line. Conversely, on exposure to the other cell line, binding remained. The inability of the L αβ or L βIX cells to completely inhibit antibody binding after preadsorption of the sera further illustrates the presence, in the group 1 sera, of two distinct antibody populations that bind to GPIbα and GPIX and the absence of an antibody that binds to GPIbβ.
The antibody present in the group 2 sera most probably reacted with the GPIX subunit on the L βIX cells because no binding of these sera was detected with the L αβ cells. The observation that binding occurred when the L αβ cells were transfected with GPIX cDNA confirms that this group of sera contained an antibody that bound specifically to GPIX.
The sera from group 3 contained an antibody that bound specifically to GPIbα because sera in this group were able to bind to CHO cells expressing GPIbα alone but were unable to bind to CHO or L cells expressing GPIbβ and GPIX.
The protease sensitivity of GPIbα was used to further define the binding domain for the GPIbα-specific antibody. We have previously defined the specific sites on GPIbα where mocarhagin (a cobra venom metalloproteinase) and trypsin cleave.18 The sequential cleavage of GPIbα with these proteases has enabled us to locate the domain to which the quinine-dependent antibodies are binding. Our previous work has shown that the binding site for the quinine-dependent antibodies is distal to the trypsin cleavage site at amino acid 293 of the GPIbα protein.2 The binding of the drug-dependent antibody was not affected by cleavage of the GPIbα protein with mocarhagin. This study has shown that the binding site is located between the mocarhagin and the trypsin cleavage sites, between amino acids 283 and 293 on the GPIbα protein. The second trypsin-sensitive site on GPIbα was also partially cleaved during this study but the continued presence of the MoAb AK3 binding after this digestion confirmed that complete cleavage at this site did not occur. The cleavage patterns observed by flow cytometry were confirmed by surface-labeling experiments. These data indicated that the quinine-dependent antibody was able to bind to both the 40-kD fragment that results from cleavage at amino acid 293 and the 100-kD glycocalicin fragment that resulted from cleavage close to the surface of the cell. This result is consistent with that of our previous study.2
The peptide inhibition studies have shown that the region encompassing amino acids 283 to 293 of the GPIbα protein is essential for the binding of the quinine-dependent antibody. After incubation of the patient serum with the specific peptide we observed a 62.0% ± 8.8% drop in the level of antibody binding. The unrelated peptides caused an insignificant decrease in the level of antibody binding. This inhibition of antibody binding by the peptide specific for amino acids 283 to 293 indicates that the 11–amino acid region represented by this peptide is involved in the quinine-dependent antibody binding to GPIbα. The incomplete inhibition by the specific peptide possibly implies the epitope is not entirely linear but involves a degree of conformational dependency which is not mirrored by the linear peptide.
We have shown that the quinine-dependent antibodies that bind to GPIb-IX can be defined as two separate populations that bind to independent domains on GPIX and GPIbα, respectively. About 50% of patients only have the antibody which binds to GPIX alone; only about 10% of them have the antibody which reacts with GPIbα alone. About 40% of patients have both antibodies. Detailed characterization of the binding domain on GPIbα has shown that the quinine-dependent antibody is interacting with the GPIbα protein between amino acids 283 and 293. This is the first study to define the quinine-dependent antibody binding sites on GPIb-IX in such detail. Identification of the binding domain for the quinine-dependent antibodies may allow more detailed studies of this and related regions to identify possible genetic abnormalities that predispose individuals to the development of this condition.
The amino acid sequence of importance for drug-dependent antibody binding to platelets in quinine-induced thrombocytopenia may provide an indication of regions of homology that act as the antigenic epitope on other glycoproteins in drug-dependent immune reactions. Knowledge obtained from the analysis of the molecular mechanism of quinine-induced thrombocytopenia may contribute useful insights to the pathogenesis of immune drug-induced damage to other tissues.
ACKNOWLEDGMENT
We thank Dr S. Santoso for generously providing the MoAb Gi27 and Dr K. Berkner for the kind gift of the eukaryotic expression vector pDX. We also thank Sue Evans for assistance with the MAIPA assays and Sasha Tait for assistance with establishing the ELISA assay and the GPIX cDNA transient transfection experiments during this study.
Supported by a grant from the National Health and Medical Research Council of Australia.
Address reprint requests to Beng H. Chong, MBBS, PhD, Department of Haematology, Prince of Wales Hospital, Cnr. High & Avoca St, Randwick, NSW 2031, Australia; e-mail: b.h.chong@unsw.edu.au.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 1. Reactivity of sera from group 2 with L αβ cells before and after transfection with GPIX cDNA. L αβ cells were transfected with GPIX cDNA using the GIBCO-BRL lipofectAMINE kit as described in Materials and Methods. The cells were labeled with primary antibody (either anti-GPIX MoAb FMC25 [A and B], or patient sera from group 2 plus quinine [C and D]) followed by a FITC-conjugated secondary antibody and examined by flow cytometry. L αβ cells did not react with anti-GPIX MoAb before transfection with GPIX cDNA (A) but bound anti-GPIX MoAb after GPIX cDNA transfection (B). Patient sera from group 2 did not react with L αβ cells before transfection with GPIX cDNA (C) but were able to bind to L αβ cells expressing GPIX on their surfaces after transfection with GPIX cDNA (D). The graphs are representative of the results observed with three different patients' sera. The solid peak in each graph represents the negative control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/7/10.1182_blood.v92.7.2366/3/m_blod41916001x.jpeg?Expires=1769154376&Signature=WkU7rngODfaTv7MgU5I5~N2IClW8Km7ipm556-bqQQUJX8wZrFa3B5cQylgtGIaYHusdoZgvfbYQe~N-o~Ahhr3xHyK0RdRO8lkpTWiVTj8Z6Fi6jdgJkKF-3NFUSxji4tWUTR0zriO4vJvwCpRDpAlO6j-56sYaQBgYRq3XqMzW0VoQcLD3OaUr7QEXMIq0UiKzGvXM0mHy8H8uV9fc7-UEh2VqHmwdDx1tHxtclMFqHkA8YcVTKXvzF87VbIV44EGbI9PFVcYL3c5rJd9JBRvXgvE1w3J6wP~T0LIJlADUGrW8vlzLDDN-B0j0YI83U3KOmRpk3DCB5sMtq8G0qA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal