Abstract
A homozygous cross-reacting material negative factor XII-deficient patient with 3% antigen and activity levels of factor XII was screened for the identification of a mutation at the genomic level. Low-ionic strength single-stranded conformation polymorphism (SSCP) analysis and sequence analysis showed that the proband’s gene for factor XII had an A→G substitution at nucleotide position 7832 in exon 3, resulting in a Tyr34 to Cys substitution in the NH2-terminal type II domain of factor XII. We designated this mutation as factor XII Tenri. Mutagenic polymerase chain reaction (PCR), followed by KpnI digestion, showed a homozygous mutation in the proband’s gene and heterozygous mutations in his parents and sister. Immunoprecipitation and Western blot analyses of plasma samples from the factor XII Tenri family indicated that the proband had a trace amount of variant factor XII with an apparent molecular mass of 115 kD, which was converted to the normal 80-kD form after reduction, suggesting that factor XII Tenri was secreted as a disulfide-linked heterodimer with a ≈35-kD protein, which we identified as 1-microglobulin by immunoblotting. Pulse-chase experiments using baby hamster kidney (BHK) cells showed that Tenri-type factor XII was extensively degraded intracellularly, but the addition of cystine resulted in increased secretion of the mutant. Using membrane-permeable inhibitors, we observed that the degradation occurred in the pre-Golgi, nonlysosomal compartment and a proteasome appeared to play a major role in this process. On the basis of these in vitro results, we speculate that the majority of the factor XII Tenri is degraded intracellularly through a quality control mechanism in the endoplasmic reticulum (ER), and a small amount of factor XII Tenri that formed a disulfide-linked heterodimer with 1-microglobulin is secreted into the blood stream.
HUMAN COAGULATION factor XII (Hageman factor) is a single chain glycoprotein (Mr 80,000) that circulates in blood as an inactive zymogen at concentrations of 29 to 40 μg/mL.1,2 Human factor XII is composed of 596 amino acid residues with N-linked and O-linked carbohydrate chains.3-5 The domain organization of factor XII is analogous to those of several fibrinolytic proteins, including tissue-type plasminogen activator and urokinase. Factor XII binds to negatively-charged molecules such as kaolin, ellagic acid, dextran sulfate, sulfatide, and endotoxin.1,2 The binding is required not only for the expression of factor XIIa proteolytic activity, but also for the cleavage of factor XII by kallikrein. The human factor XII gene is located on the chromosomal band 5q33-qter, and 12 kb of the entire genomic sequence with 14 exons and 13 introns have been analyzed.6 Factor XII is converted to a two-chain serine protease (α-factor XIIa) with an NH2-terminal heavy chain (Mr 50,000) and a COOH-terminal light chain (Mr 28,000) that is capable of initiating the intrinsic pathway of blood coagulation, kinin production, and fibrinolysis.1,2 The heavy chain of factor XIIa contains five different domains including a type I and a type II domain of fibronectin, two epidermal growth factor (EGF)-like domains, a kringle domain, and a proline-rich domain, whereas the light chain consists of a typical trypsin-like serine protease domain.3-5 On activation, further cleavages take place in the heavy chain, resulting in the production of β-factor XIIa consisting of the nonapeptide (Asn335-Arg343) and disulfide-linked 28-kD protease domain.7
Hereditary factor XII deficiency is clinically asymptomatic, but can be detected by an in vitro test due to a prolonged activated partial thromboplastin time (APTT). Typically, homozygous or compound heterozygous carriers exhibit almost no (<1%) factor XII activity as compared with normal subjects, whereas heterozygotes display intermediate activity. Most of these subjects also lack immunologically detectable factor XII and are referred to as cross-reacting material (CRM) negative.8,9 The majority of CRM negative factor XII deficiency cases are known to be caused by mutations such as a change in the splice sites or a deletion of portions of the coding sequence resulting in a frameshift.10 Hageman trait, the best known factor XII deficiency, is associated with an aberrant TaqI restriction site in intron B, a putative regulatory site.11,12 In contrast, factor XII Washington D.C., a CRM positive factor XII deficiency, contains a Cys571 to Ser replacement, resulting in the disruption of a disulfide-linkage between Cys540-Cys571 near the active site Ser544.13 Factor XII Locarno, another CRM positive factor XII deficiency, is induced by an amino acid substitution of Arg353 by Pro, resulting in a loss of the kallikrein cleavage site at Arg353-Val354.14 Recently, Schloesser et al15 investigated 31 unrelated factor XII-deficient patients and identified two missense mutations, Arg398→Gln and Leu395→Met, as well as two CRM positive factor XII deficiencies caused by amino acid substitutions of Asp442→Asn and Gly570→Arg. Kanaji et al16 reported an abnormal factor XII in which Arg126 has been replaced by Pro, and a common HgaI polymorphism (46 C/T) in the 5′-untranslated region is associated with low translational efficiency resulting in a decreased antigen level of factor XII.17
We report here a Japanese family with CRM negative factor XII deficiency, which we designated as factor XII Tenri. We demonstrated that the proband, who showed 3% antigen and activity levels of factor XII, had an amino acid substitution of Tyr34→Cys in the type II domain, and a small amount of the patient’s factor XII found in plasma was a heterodimeric complex with α1-microglobulin. Moreover, pulse-chase analyses, using stably-transfected wild-type and Tenri-type factor XII cDNAs in baby hamster kidney (BHK) cells, showed that most of the synthesized mutant factor XII was degraded by a proteasome through the quality control mechanism in the endoplasmic reticulum (ER).
MATERIALS AND METHODS
Subject.
A 29-year-old Japanese man was found to have a prolonged APTT (58.2 seconds) before a reconstruction operation of his nasal septum, although he had no history of abnormal bleeding. Both his antigen and activity levels of factor XII were determined as 3% of normal. Hemostatic laboratory findings were as follows (% values represent relative activity unless otherwise stated): prothrombin time, 11.3 seconds; fibrinogen, 2.2 mg/mL; factors XI, 105%; X, 100%; IX, 98%; VIII, 82%; VII, 98%; and V, 72%; prothrombin, 85%; antithrombin, 97%; and α2-plasmin inhibitor, 98%. From the laboratory investigation and the patient’s pedigree (Fig 1), we diagnosed the proband as CRM negative homozygous factor XII deficiency, which we named factor XII Tenri.
Enzymes, antibodies, and reagents.
Restriction endonucleases and DNA modifying enzymes were purchased from Takara Shuzo (Kyoto, Japan), Toyobo (Osaka, Japan), and New England Biolabs (Beverly, MA). Thermo Sequenase cycle sequencing kit and Sculptor in vitro mutagenesis system were the products of Amersham Life Science (Tokyo, Japan). GeneAmp PCR reagent kit, AmpliWax PCR Gem 100, and AmpliTaq DNA polymerase were obtained from Takara Shuzo. EXPRE35S35S (mixture of 72%L-[35S]Met and 18%L-[35S]Cys) was purchased from NEN-DuPont Japan (Tokyo, Japan). Chloroquine and brefeldin A were obtained from Sigma Chemicals (St Louis, MO). Leupeptin, [L-3-trans-ethoxycarbonyloxiran-2-carbonyl]-L-leucine(3-methylbutyl)amide (E64d), carbobenzoxy-Leu-Leu-lucinal (LLL), and carbobenzoxy-Leu-leucinal (LL) were the products of Peptide Institute (Osaka, Japan). Lactacystin was a generous gift from Dr Satoshi Omura (The Kitasato Institute, Tokyo, Japan). Rabbit and goat antihuman factor XII antisera were purchased from Calbiochem (San Diego, CA) and Nordic Immunology (Tilburg, The Netherlands), respectively. Rabbit antihuman α1-microglobulin antibody was obtained from Cosmobio (Tokyo, Japan). Oligonucleotides were synthesized by an Applied Biosystems 394 DNA/RNA synthesizer (Applied Biosystems, Foster City, CA). All other reagents were of the highest quality commercially available.
Polymerase chain reaction (PCR).
Leukocytes from peripheral blood samples were prepared using Lymphoprep (Daiichi Chemicals, Tokyo, Japan), and the leukocyte DNA was obtained by proteinase K digestion and phenol extraction, followed by ethanol precipitation. All of the exons and flanking regions for the factor XII gene were amplified by a DNA Thermal Cycler (Perkin-Elmer Cetus Instruments, Norwalk, CT). Primers used were designed as listed in Table 1. To prevent a nonspecific annealing of primers to genomic DNA, a solution containing 200 μmol/L deoxyribonucleoside 5′-triphosphates (dNTPs) mixture and 100 pmol each of 5′- and 3′-primers was sealed by AmpliWax. Then, 10 μL of 10x PCR buffer, 1 μg of leukocyte DNA, and 1 μL of AmpliTaq DNA polymerase were overlaid. The hot-start PCR reaction was performed for 30 cycles as: denaturation at 94°C for 1 minute, annealing at 63°C for 1 minute, and polymerization at 72°C for 2 minutes.
Screening of mutation site and sequence analysis.
To detect the mutation site in the factor XII Tenri gene, low-ionic strength single-stranded conformation polymorphism (LIS-SSCP) analysis was performed.18 Briefly, 1 μL of PCR product was mixed with 20 μL of LIS solution (10% sucrose/0.01% Bromophenol Blue/0.01% xylene cyanol). After heat denaturation at 97°C for 2 minutes, 4 to 10 μL of sample was applied to a 10% polyacrylamide gel and electrophoresed in 45 mmol/L Tris/45 mmol/L boric acid/1 mmol/L EDTA, pH 8.0 at 22°C. Single-strand DNA fragments were detected by silver staining.
For sequence analyses, PCR fragments were purified by Centricon 100 (Amicon, Beverly, MA) and digested by restriction enzymes at the sites designated in 5′- and 3′-primers, and subsequently ligated to pUC118. At least six independent plasmids were sequenced using Thermo Sequenase cycle sequencing kit and a DSQ-1 automated DNA sequencer (Shimadzu, Kyoto, Japan).
Mutagenic PCR and KpnI digestion.
To confirm the assigned mutation site in factor XII Tenri gene, a mutagenic primer, exon III-33M was designed as 5′-AGCCCTGCCACTTCCCCTTCCGGT-3′ (mutagenic G is underlined) to create a KpnI recognition site in the PCR product from the normal factor XII gene. PCR amplification by primers exon III-33M and exon III-32A (Table 1) was performed as described above. The PCR products were purified by Centricon 100, digested with 6 U of KpnI at 37°C for 2 hours, and then electrophoresed in a 3% agarose gel.
Western blotting.
To examine the secretion of Tenri-type factor XII in the proband’s plasma, immunoprecipitation followed by Western blotting was performed. Briefly, 450 μL of plasma from either the Tenri family or normal individuals was reacted with rabbit antihuman factor XII antiserum and then with protein A-agarose (Sigma). The immunoprecipitates were electrophoresed in an 8% sodium dodecyl sulfate (SDS) gel,19 and transferred to a polyvinylidene difluoride membrane (Millipore, Japan, Tokyo). The membrane was blocked overnight with 1% bovine serum albumin in 25 mmol/L Tris-HCl, pH 7.4/0.15 mol/L NaCl. Factor XII antigen was detected by incubating the membrane with biotinylated anti-factor XII antibody followed by the avidin-peroxidase detection system (Vector Laboratories, Burlingame, CA) as described.20 To examine the heterodimer complex of factor XII with α1-microglobulin, the membrane was reacted with biotinylated rabbit anti-α1–microglobulin antibody and detected by the same method as described above.
Site-directed mutagenesis and construction of the expression vectors.
A full-length (1.6 kb) cDNA for human factor XII,4generously provided by Dr Ross T.A. MacGillivray (University of British Columbia, Vancouver, British Columbia, Cananda), was ligated to theXbaI site of pUC118. Single-strand DNA was prepared by the addition of helper phage M13K07 and kanamycin. To prepare the Tenri-type factor XII cDNA, site-directed mutagenesis was performed using a Sculptor in vitro mutagenesis kit and a mutagenic oligonucleotide of 5′-TTCCAGTGCCACCGGCAGCT-3′ (mutagenic G is underlined) according to the manufacturer’s instructions. The mutation was confirmed by sequence analysis.
Wild-type and Tenri-type factor XII cDNAs in pUC118, thus prepared, were transferred to XbaI site of the pcD2-SRα expression vector21 and purified by CsCl gradient ultracentrifugation. BHK cells, supplied from Japan Cancer Research Resources Bank (Tokyo, Japan), were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Filtron, Brooklyn, Australia) and antibiotic-antimycotic liquid (GIBCO BRL Life Technology, Grand Island, NY). A total of 7 μg of expression vector was transfected into 2 × 105 cells by the calcium phosphate coprecipitation method.21 To obtain stable transfectants, cells were selected by the addition of 400 μg/mL G418 to the medium. A pool of stably-transfected BHK cells was used in subsequent experiments.
Pulse-chase, immunoprecipitation, and gel electrophoresis.
To examine the secretion rate of Tenri-type factor XII, pulse-chase experiments were performed as described previously.22,23Briefly, stably-transfected BHK cells (5 × 105 cells) were starved for 30 minutes with DMEM lacking Met and Cys (Sigma)/10% dialyzed fetal bovine serum, and subsequently labeled with 100 μCi/mL of EXPRE35S35S for 30 minutes. Cells were chased with DMEM/10% fetal bovine serum/ 2 mmol/L Met, and cystine (0.5 mmol/L) was added to the chase media in selected experiments. After incubation for the indicated times, conditioned media were harvested and cells were lysed by 1% NP-40/0.1% SDS solution.22 23 The 35S-labeled factor XII was immunoprecipitated using goat antihuman factor XII antiserum and Staphylosorb (Mercian, Tokyo, Japan). After washing, immunoadsorbed proteins were dissociated by heating at 85°C for 5 minutes, and then electrophoresed. The radioactivities in dried SDS-gels were measured quantitatively by a Fujix BAS2000 Bio-Imaging Analyzer system (Fuji Photo Film, Tokyo, Japan).
RESULTS
Identification of the mutation site in factor XII Tenri gene.
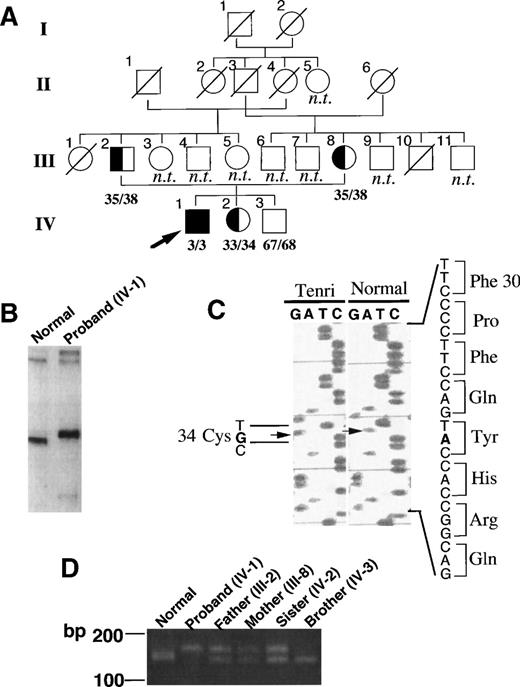
Figure 1A shows a pedigree of the factor XII Tenri family. Both factor XII antigen and activity levels in the proband were quantitated as 3% of normal, suggesting homozygous CRM negative factor XII deficiency as described in Materials and Methods. The proband’s parents, who are married consanguineously, in addition to his sister, also showed reduced antigen and activity levels (≈35%) of factor XII, suggesting heterozygous deficiency. The proband’s brother showed 67% antigen and 68% activity levels, suggesting normality. None of the family members had a bleeding tendency or a history of thromboembolism. To elucidate the molecular basis of factor XII Tenri, we first amplified all 14 exons and flanking regions of factor XII gene from the proband by PCR using the primers listed in Table 1, then analyzed them by the LIS-SSCP method.18 As shown in Fig 1B, a PCR fragment derived from exon 3 of the proband exhibited a slower migration than that of a normal subject. No aberrant migrations on LIS-SSCP analysis were observed for the other PCR fragments (data not shown). Sequence analysis of exon 3 from the proband showed a substitution of A to G at nucleotide position 7832 (Fig 1C). All eight sequenced clones had the identical mutation, suggesting homozygous mutation at this site. This mutation induced an amino acid substitution of Tyr34 (TAC) to Cys (TGC) in the type II domain of factor XII. Sequence analysis of all other exons and flanking regions indicated no mutations. An aberrant TaqI restriction site in intron B, which is well-known as a cause for Hageman trait, was not detected in factor XII Tenri gene (data not shown).
Identification of the mutation site in factor XII Tenri. (A) Pedigree of the factor XII Tenri family. Affected subjects with reduced factor XII levels are shadowed and the proband is indicated by an arrow. Deceased family members and unexplored subjects are indicated by ○/ or □/ and n.t., respectively. Antigen/activity levels of factor XII are shown. (B) LIS-SSCP analysis of exon 3 of factor XII gene. PCR fragments of exon 3 derived from a normal subject (left) and the proband (right) were analyzed by LIS-SSCP as described in Materials and Methods. (C) Nucleotide sequence showing the A to G substitution in exon 3 of the factor XII Tenri gene. This substitution mutates Tyr34 to Cys in the type II domain of factor XII. (D) PCR-Kpn I digestion analysis of the factor XII Tenri family. For each member, exon 3 amplified by PCR as described in Materials and Methods was digested with KpnI and subsequently analyzed by agarose gel electrophoresis. PCR product from normal allele showed a cleaved fragment of 141 bp, whereas that from Tenri-type allele showed an uncleaved band of 163 bp.
Identification of the mutation site in factor XII Tenri. (A) Pedigree of the factor XII Tenri family. Affected subjects with reduced factor XII levels are shadowed and the proband is indicated by an arrow. Deceased family members and unexplored subjects are indicated by ○/ or □/ and n.t., respectively. Antigen/activity levels of factor XII are shown. (B) LIS-SSCP analysis of exon 3 of factor XII gene. PCR fragments of exon 3 derived from a normal subject (left) and the proband (right) were analyzed by LIS-SSCP as described in Materials and Methods. (C) Nucleotide sequence showing the A to G substitution in exon 3 of the factor XII Tenri gene. This substitution mutates Tyr34 to Cys in the type II domain of factor XII. (D) PCR-Kpn I digestion analysis of the factor XII Tenri family. For each member, exon 3 amplified by PCR as described in Materials and Methods was digested with KpnI and subsequently analyzed by agarose gel electrophoresis. PCR product from normal allele showed a cleaved fragment of 141 bp, whereas that from Tenri-type allele showed an uncleaved band of 163 bp.
PCR Primers Used for Amplification of Exons in the Factor XII Gene
| Exon . | Primer No. . | Sequence (5′-3′) . | Nucleotide Numbering* . |
|---|---|---|---|
| Exon I | 11 | CAA GGA AGT TGC TCC ACT TGG CT | −102 to −79 |
| 12A | AAC AAT CCT GGG ACA ATC CTG GTT C | 119 to 144 | |
| Exon II | 21 | GAG AGG AAG CTT CAG ACT AGC AAC AGA | 4659 to 4685 |
| 22A | ATA CGG ATC CCC ACC CAA GGG TTC C | 4824 to 4848 | |
| Exon III | 31 | TCA GAG AGT GTG TTG TCC CTG CAG TTC | 7766 to 7792 |
| 32A | AGC AGG GAT CCT TGG ACA GAA GGA CA | 7943 to 7968 | |
| 33M† | AGC CCT GCC ACT TCC CCT TCC GGT | 7808 to 7831 | |
| Exon IV | 41 | CCT GGG ATC CAT GTA CCC TGC CTG T | 7921 to 7945 |
| 42A | CCA GAA TTC CAG GTG TGT GGG GTC TG | 8185 to 8210 | |
| Exon V and VI | 51 | AAC CAA GCT TGG AAA CTT GGA GTA GCA A | 8299 to 8326 |
| 62A | TCC TCC CTG GCC GGA TCC CAA CCA | 8807 to 8830 | |
| Exon VII | 71 | GAG CAG ATG GTT GGG ATC CGG CCA GGG A | 8800 to 8827 |
| 72A | TCT CAT AAG CTTTCC GCA CTC TCC CTC CTC | 9106 to 9135 | |
| Exon VIII | 81 | TGC GGA AAG CTT ATG AGA GGG AGG CA | 9118 to 9143 |
| 82A | TCG TTG TCC GGG ATC CTG TAG CCA C | 9432 to 9458 | |
| Exon IX | 91 | TCC GGG ATC CCG GCG CTC TAA CGG CG | 9397 to 9422 |
| 92A | GGT CAG GGA ATT CGG CTG CTC CCG CTT | 9778 to 9804 | |
| Exon X | 101 | TAG GAA TTC GGG GGG GAA GGA GGA GCC GA | 9664 to 9692 |
| 102A | GTG GAA AGA AGC TTG GGG CGG GAG AAG GTA | 10088 to 10117 | |
| Exon XI | 111 | AGG AAG CTT GAA CAC GGG ATT GGG GTT | 10139 to 10165 |
| 112A | TCC GGA TCCTCC TGA AGG CGC AAC AG | 10462 to 10487 | |
| Exon XII | 121 | CCC GTA AGC TTC CAG CAC GAC CTG GGT | 10354 to 10380 |
| 122A | GGA ACG ATA CCG AAT TCG CGG GCT TCT T | 10665 to 10692 | |
| Exon XIII | 131 | GAG AAG CTT TGT CCA TCG TCC GGG CGG C | 11086 to 11113 |
| 132A | TGT GGA CCT GAG AAT TCT GCC TGA CGG CCT | 11341 to 11370 | |
| Exon XIV | 141 | GTG CCA GGT AAG CTT TTA GCC CGG TTG | 11301 to 11327 |
| 142A | ACA CAA TCT TGA ATT CCA TGC CCC AGC CA | 11615 to 11643 |
| Exon . | Primer No. . | Sequence (5′-3′) . | Nucleotide Numbering* . |
|---|---|---|---|
| Exon I | 11 | CAA GGA AGT TGC TCC ACT TGG CT | −102 to −79 |
| 12A | AAC AAT CCT GGG ACA ATC CTG GTT C | 119 to 144 | |
| Exon II | 21 | GAG AGG AAG CTT CAG ACT AGC AAC AGA | 4659 to 4685 |
| 22A | ATA CGG ATC CCC ACC CAA GGG TTC C | 4824 to 4848 | |
| Exon III | 31 | TCA GAG AGT GTG TTG TCC CTG CAG TTC | 7766 to 7792 |
| 32A | AGC AGG GAT CCT TGG ACA GAA GGA CA | 7943 to 7968 | |
| 33M† | AGC CCT GCC ACT TCC CCT TCC GGT | 7808 to 7831 | |
| Exon IV | 41 | CCT GGG ATC CAT GTA CCC TGC CTG T | 7921 to 7945 |
| 42A | CCA GAA TTC CAG GTG TGT GGG GTC TG | 8185 to 8210 | |
| Exon V and VI | 51 | AAC CAA GCT TGG AAA CTT GGA GTA GCA A | 8299 to 8326 |
| 62A | TCC TCC CTG GCC GGA TCC CAA CCA | 8807 to 8830 | |
| Exon VII | 71 | GAG CAG ATG GTT GGG ATC CGG CCA GGG A | 8800 to 8827 |
| 72A | TCT CAT AAG CTTTCC GCA CTC TCC CTC CTC | 9106 to 9135 | |
| Exon VIII | 81 | TGC GGA AAG CTT ATG AGA GGG AGG CA | 9118 to 9143 |
| 82A | TCG TTG TCC GGG ATC CTG TAG CCA C | 9432 to 9458 | |
| Exon IX | 91 | TCC GGG ATC CCG GCG CTC TAA CGG CG | 9397 to 9422 |
| 92A | GGT CAG GGA ATT CGG CTG CTC CCG CTT | 9778 to 9804 | |
| Exon X | 101 | TAG GAA TTC GGG GGG GAA GGA GGA GCC GA | 9664 to 9692 |
| 102A | GTG GAA AGA AGC TTG GGG CGG GAG AAG GTA | 10088 to 10117 | |
| Exon XI | 111 | AGG AAG CTT GAA CAC GGG ATT GGG GTT | 10139 to 10165 |
| 112A | TCC GGA TCCTCC TGA AGG CGC AAC AG | 10462 to 10487 | |
| Exon XII | 121 | CCC GTA AGC TTC CAG CAC GAC CTG GGT | 10354 to 10380 |
| 122A | GGA ACG ATA CCG AAT TCG CGG GCT TCT T | 10665 to 10692 | |
| Exon XIII | 131 | GAG AAG CTT TGT CCA TCG TCC GGG CGG C | 11086 to 11113 |
| 132A | TGT GGA CCT GAG AAT TCT GCC TGA CGG CCT | 11341 to 11370 | |
| Exon XIV | 141 | GTG CCA GGT AAG CTT TTA GCC CGG TTG | 11301 to 11327 |
| 142A | ACA CAA TCT TGA ATT CCA TGC CCC AGC CA | 11615 to 11643 |
“A” in the primer number denotes an antisense strand oligonucleotide. Nucleotide sequences designed for the digestion by restriction enzymes are underlined.
Nucleotide numberings are based on Cool and MacGillivray.6
Primer used for mutagenic PCR.
To confirm the mutation site in the genes from the members of the factor XII Tenri family, we performed mutagenic PCR followed byKpnI digestion analyses (Fig 1D). The mutagenic primer was designed to change the codon (CAG) for Gln33 to CGG to create a KpnI site (GGTACC) in the normal allele, but not in the Tenri-type allele. After a 2-hour digestion by KpnI, PCR products derived from the younger brother and normal individuals showed a cleaved fragment of 141 bp, while the PCR fragment from the proband was uncleaved and remained as a 163-bp fragment. PCR products from the parents and sister of the proband showed two fragments of 163 bp and 141 bp, suggesting heterozygosity. These results were consistent with the antigen and activity levels of factor XII in the Tenri family.
A portion of the secreted Tenri-type factor XII is complexed with α1-microglobulin.
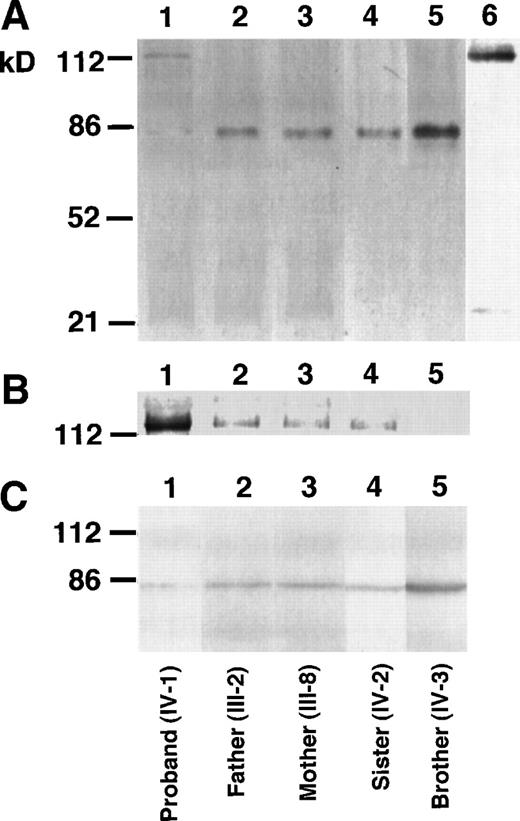
The proband of factor XII Tenri showed low, but detectable (≈3%), antigen and activity levels of factor XII in plasma. This suggests that a small amount of variant factor XII having Cys34 is secreted into the blood stream. To demonstrate Tenri-type factor XII in plasma, we performed immunoprecipitation followed by Western blotting using anti-factor XII antibody. Under nonreducing conditions, immunoblots of factor XII derived from the proband’s plasma showed a 115-kD band, in addition to a faint 80-kD band (Fig 2A, lane 1). In contrast, plasma samples from his parents, sister, and brother exhibited a major 80-kD band that was the same size as normal factor XII (Fig 2A, lanes 2 through 5). To elucidate the presence of a 115-kD variant factor XII among family members, overexposure of the membrane was performed (Fig 2B), which showed that immunoblots of parents and sister, ie, heterozygous deficients, also had a very faint 115-kD band. This suggested that the 115-kD band was derived from the factor XII Tenri allele. After reduction of the SDS sample with 2-mercaptoethanol, all family members showed only a single band of 80-kD (Fig 2C), indicating that the variant factor XII was complexed with a 35-kD protein through a mixed disulfide linkage. We suspected the protein to be α1-microglobulin because α1-microglobulin has a molecular mass of 33 kD and is known to form a disulfide bond to some abnormal coagulation factors with a free Cys residue introduced into the molecule.24 25To confirm this possibility, the same preparation in lane 1 of Fig 2A was blotted using anti-α1–microglobulin antibody. Under these conditions, the 115-kD band was detected by this antibody (Fig 2A, lane 6). Furthermore, we could not detect a 115-kD band by anti-α1–microglobulin antibody in normal plasma. Factor XII dimer (≈160 kD) or albumin-bound form (≈145 kD) of the factor XII Tenri was not observed in the proband’s plasma (data not shown).
Western blot analysis of the factor XII Tenri family. Factor XII in plasmas of the proband (lane 1), his father (lane 2), mother (lane 3), sister (lane 4), and brother (lane 5) were immunoprecipitated by anti-factor XII antibody. After heat dissociation, immunoprecipitates were electrophoresed on 8% SDS-polyacrylamide gels under nonreducing (A and B) or reducing (C) conditions. Electrophoresed proteins were transferred to polyvinylidene difluoride membranes and then blotted by the biotinylated anti-factor XII antibody followed by avidin-peroxidase. (B) is the overexposure of the high molecular mass region of (A). The sample in lane 6 in (A) was the same as in lane 1, but was blotted by the biotinylated anti-1–microglobulin antibody.
Western blot analysis of the factor XII Tenri family. Factor XII in plasmas of the proband (lane 1), his father (lane 2), mother (lane 3), sister (lane 4), and brother (lane 5) were immunoprecipitated by anti-factor XII antibody. After heat dissociation, immunoprecipitates were electrophoresed on 8% SDS-polyacrylamide gels under nonreducing (A and B) or reducing (C) conditions. Electrophoresed proteins were transferred to polyvinylidene difluoride membranes and then blotted by the biotinylated anti-factor XII antibody followed by avidin-peroxidase. (B) is the overexposure of the high molecular mass region of (A). The sample in lane 6 in (A) was the same as in lane 1, but was blotted by the biotinylated anti-1–microglobulin antibody.
Cellular basis for the secretion defect of factor XII Tenri.
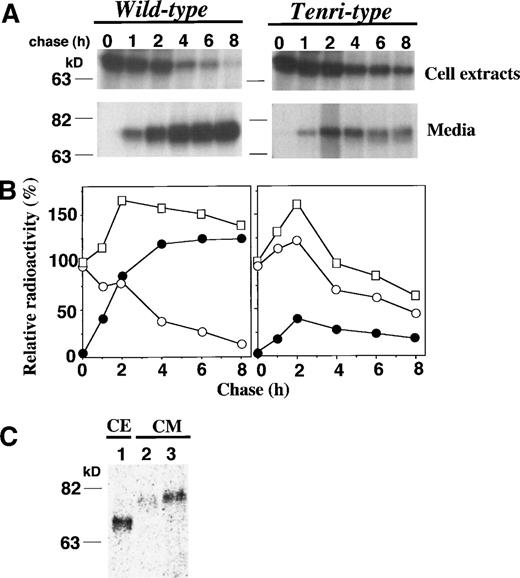
To elucidate the molecular basis whereby substitution of Tyr34→Cys causes a reduction of factor XII antigen level in plasma, we performed pulse-chase experiments of the Tenri-type factor XII using stably-transfected BHK cells and compared the results with that of wild-type factor XII. Nascent wild-type and Tenri-type factor XIIs were detected as a single band of 76 kD in the cell extracts (Fig 3A). The intracellular Tenri-type factor XII was sensitive to endoglycosidase H, suggesting the presence of high mannose-type oligosaccharides attached to the polypeptide in the pre-Golgi compartment (data not shown). By kinetic analysis of the wild-type factor XII, almost all of the pulse-labeled radioactivity was recovered from the media after a 4-hour chase, and the total amount of radioactivity was maintained during the chase period (Fig 3B, left panel). In contrast, Tenri-type mutant showed impaired secretion and a significant reduction (≈50%) in the total amount of radioactivity was observed during an 8-hour chase, suggesting intracellular degradation of Tenri-type factor XII (Fig 3B, right panel).
Pulse-chase analyses of wild-type and Tenri-type factor XIIs in stably-transfected BHK cells. (A) Stably-transfected BHK cells were pulse-labeled for 30 minutes with 100 μCi/mL EXPRE35S35S and chased for 0, 1, 2, 4, 6, and 8 hours. Labeled factor XIIs from cell extracts and from culture media were immunoprecipitated and analyzed on 8% SDS-polyacrylamide gel electrophoresis. (B) In kinetic analyses, the amount of radioactivity in the pulse-labeled cell extracts was taken as 100%, and relative radioactivities of intracellular and secreted fractions are shown by ○ and •, respectively. The sum of the radioactivities of both fractions at each time is shown by □. (C) BHK cells expressing Tenri-type factor XII were pulse-labeled for 30 minutes with 100 μCi/mL EXPRE35S35S (lane 1) and chased in DMEM for 8 hours in the absence (lane 2) or presence (lane 3) of excess amounts of cystine. Factor XII was immunoprecipitated from the pulse-labeled cell extracts (lane 1) and chased media (lanes 2 and 3), and then electrophoresed on an 8% SDS-polyacrylamide gel.
Pulse-chase analyses of wild-type and Tenri-type factor XIIs in stably-transfected BHK cells. (A) Stably-transfected BHK cells were pulse-labeled for 30 minutes with 100 μCi/mL EXPRE35S35S and chased for 0, 1, 2, 4, 6, and 8 hours. Labeled factor XIIs from cell extracts and from culture media were immunoprecipitated and analyzed on 8% SDS-polyacrylamide gel electrophoresis. (B) In kinetic analyses, the amount of radioactivity in the pulse-labeled cell extracts was taken as 100%, and relative radioactivities of intracellular and secreted fractions are shown by ○ and •, respectively. The sum of the radioactivities of both fractions at each time is shown by □. (C) BHK cells expressing Tenri-type factor XII were pulse-labeled for 30 minutes with 100 μCi/mL EXPRE35S35S (lane 1) and chased in DMEM for 8 hours in the absence (lane 2) or presence (lane 3) of excess amounts of cystine. Factor XII was immunoprecipitated from the pulse-labeled cell extracts (lane 1) and chased media (lanes 2 and 3), and then electrophoresed on an 8% SDS-polyacrylamide gel.
As shown in Fig 2, we found that an α1-microglobulin–bound form of factor XII Tenri was secreted in the proband’s plasma. Although BHK cells do not synthesize α1-microglobulin constitutively, we speculated that a mixed-disulfide formation through the free sulfhydryl group of Cys34 in Tenri-type factor XII might enhance the secretion. To test this hypothesis, pulse-chase experiments were performed in the absence or presence of excess amounts of cystine (Fig 3C). Compared with its secretion in the absence of cystine (lane 2), a larger amount of the mutant was secreted in the presence of 0.5 mmol/L cystine (lane 3). Thus, we conclude that the addition of excess amounts of cystine, presumably resulting in the mixed-disulfide formation, resulted in the increased secretion of Tenri-type factor XII from BHK cells.
Proteasome-mediated degradation of factor XII Tenri.
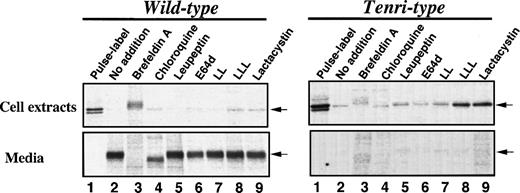
To localize the site and identify the protease(s) responsible for the intracellular degradation of factor XII Tenri, we next examined the effects of various inhibitory reagents on the degradation of Tenri-type factor XII expressed in stably-transfected BHK cells (Fig 4). Taking the amount of radioactivity of factor XII in the cell extracts immediately after pulse-labeling as 100%, ≈97% of wild-type factor XII was detected in the 16-hour–chased media (Fig 4, left panel, lane 2 v 1). In contrast, >85% of the Tenri-type factor XII disappeared during the 16-hour chase (Fig 4, right panel, lane 2 v 1). In the presence of 5 μg/mL brefeldin A, which blocks intracellular transport by redistribution of Golgi proteins into the ER,26 wild-type factor XII accumulated in the cells and ≈60% of the radioactivity was recovered from the cell extracts after a 16-hour chase (Fig 4, left panel, lane 3). However, in the case of Tenri-type factor XII, 76% of the total radioactivity disappeared during a 16-hour chase in the presence of brefeldin A, suggesting that the Tenri-type factor XII was selectively degraded in a pre-Golgi compartment (Fig 4, right panel, lane 3). In the presence of brefeldin A, the intracellular Tenri-type factor XII exhibited a slower migration, which may be due to the processing of carbohydrate chains by the incorporated Golgi enzymes as reported.27 Among the lysosomotropic agents, 100 μmol/L chloroquine (Fig 4, lane 4), 30 mmol/L NH4Cl (data not shown), and 100 μmol/L leupeptin (lane 5), a lysosomal protease inhibitor of microbial origin,28 had no effect on the degradation of Tenri-type factor XII. These results suggest that the degradation of Tenri-type factor XII occurs in a nonlysosomal compartment. In the presence of chloroquine, secreted wild-type factor XII showed faster migration than others (Fig 4, left panel, lane 4). This is probably due to prevention of terminal saialic acid transfer as described previously.29 Among the membrane-permeable protease inhibitors we examined, 100 μmol/L E64d (Fig 4, lane 6) and 20 μmol/L LL (Fig 4, lane 7), which are inhibitors for intracellular cysteine protease calpain,30 showed no inhibitory effects on the degradation of Tenri-type factor XII or secretion of wild-type factor XII. However, 20 μmol/L LLL (Fig 4, lane 8) and 20 μmol/L lactacystin (Fig 4, lane 9), two chemically different potent proteasome inhibitors,31 32 strongly inhibited the degradation of Tenri-type factor XII. As a result, Tenri-type factor XII was retained in the cells, while these proteasome inhibitors essentially showed no effect on the secretion of wild-type factor XII (Fig 4, left panel, lanes 8 and 9). Other peptidyl-aldehyde inhibitors of proteasome such as carbobenzoxy-l-leucyl-l-leucyl-l-norvalinal and carbobenzoxy-l-isoleucyl-γ-t-butyl-l-glutamyl-l-alanyl-l-leucinal showed similar inhibitory effects as LLL and lactacystin on the intracellular degradation of Tenri-type factor XII (data not shown). These results suggest that a proteasome plays a role in the intracellular degradation of the Tenri-type factor XII.
Effects of various inhibitors on the secretion and degradation of wild-type and Tenri-type factor XIIs. Stably transfected BHK cells expressing wild-type (left panel) or Tenri-type (right panel) factor XII were pulse-labeled for 30 minutes with 100 μCi/mL EXPRE35S35S and then chased for 16 hours in the presence of various inhibitors. Factor XII in the cell extracts and culture media were subjected to immunoprecipitation followed by SDS-PAGE analysis. Lane 1, sample from pulse-labeled cells; lane 2, sample from 16-hour chased cells without inhibitors; lanes 3 through 9, samples from 16-hour chased cells in the presence of inhibitors; lane 3, 5 μg/mL brefeldin A; lane 4, 100 μmol/L chloroquine; lane 5, 100 μg/mL leupeptin; lane 6, 100 μmol/L E64d; lane 7, 20 μmol/L LL; lane 8, 20 μmol/L LLL; and lane 9, 20 μmol/L lactacystin. Positions of factor XII are indicated by arrows.
Effects of various inhibitors on the secretion and degradation of wild-type and Tenri-type factor XIIs. Stably transfected BHK cells expressing wild-type (left panel) or Tenri-type (right panel) factor XII were pulse-labeled for 30 minutes with 100 μCi/mL EXPRE35S35S and then chased for 16 hours in the presence of various inhibitors. Factor XII in the cell extracts and culture media were subjected to immunoprecipitation followed by SDS-PAGE analysis. Lane 1, sample from pulse-labeled cells; lane 2, sample from 16-hour chased cells without inhibitors; lanes 3 through 9, samples from 16-hour chased cells in the presence of inhibitors; lane 3, 5 μg/mL brefeldin A; lane 4, 100 μmol/L chloroquine; lane 5, 100 μg/mL leupeptin; lane 6, 100 μmol/L E64d; lane 7, 20 μmol/L LL; lane 8, 20 μmol/L LLL; and lane 9, 20 μmol/L lactacystin. Positions of factor XII are indicated by arrows.
DISCUSSION
In this study, we show that a novel type of missense mutation (exon 3, 7832A→G), leading to the substitution of Tyr34 to Cys in the NH2-terminal type II domain of the factor XII molecule, is the genetic basis for a CRM negative factor XII deficiency (Fig 1). Hereditary factor XII deficients in the factor XII Tenri family are clinically asymptomatic, but were found by chance due to the prolongation of the APTT by laboratory tests before surgery.8,9 CRM negative factor XII deficiency is mainly associated with an additional TaqI restriction site in intron B, which is known as the Hageman trait.11,12 Recently, Schloesser et al10 identified a mutation that was induced by the presence of a T→C transition 224 bp upstream of exon 3 and shown to be associated with a mutation in the 5′-flanking region of the factor XII gene (exon 1, -8G→C). Two missense mutations resulting in CRM negative factor XII deficiency were also reported by Schloesser et al15; ie, Leu395→Met and Arg398→Gln in exon 10, but the mechanism whereby the mutations lead to factor XII deficiency is not known. Thus, to our knowledge, our data is the first report on the cellular analysis of a CRM negative factor XII deficiency induced by an amino acid substitution.
A small amount (≈3%) of mutant factor XII was secreted into the proband’s plasma. We showed by immunobloting that the majority of the secreted Tenri-type factor XII was complexed with α1-microglobulin (Fig 2). α1-Microglobulin, a lipocalin superfamily protein, is known to be synthesized in the liver as an NH2-terminal portion of the light chain precursor of inter-α–trypsin inhibitor.33,34 A furin-like protease is responsible for the cleavage of the dibasic amino acids in the central portion, resulting in the production of two functionally different proteins.34 Human α1-microglobulin is a glycoprotein with an apparent molecular mass of 33 kD and is detected in various body fluids as a monomer or as a complexed form with IgA. The protein has three Cys residues, two of which are involved in the formation of an intramolecular disulfide bridge, while the other is covalently bound to IgA.33 Previously, Wojcik et al24 reported that a mutant factor IX, factor IX Zutphen, had a Cys18→Arg mutation resulting in the disruption of Cys18-Cys23 linkage. This mutant factor IX, which exhibited ≈65% antigen and <4% activity levels, was identified as a heterodimer with α1-microglobulin in patient’s plasma. In a subsequent report,25 these investigators extensively examined the intermolecular complex between α1-microglobulin and mutants of protein C and prothrombin. The protein C mutants with Arg-1→Cys, Arg9→Cys, or Ser12→Cys were shown to be present in plasma as a complex with α1-microglobulin, whereas a prothrombin mutant with Tyr44→Cys was not. They concluded that unpaired Cys residues in the propeptide or in the NH2-terminal half of the Gla domain of vitamin K-dependent proteins formed a complex with α1-microglobulin.25 In the present report, we show that not only vitamin K-dependent proteins, but the Tyr34→Cys mutation in type II domain of factor XII, also caused complex formation with α1-microglobulin. In addition to α1-microglobulin binding, a variety of modifications to the newly-introduced Cys are reported in clotting factors. For example, albumin was bound to fibrinogen Milano VII (γ chain Ser358→Cys)35 and fibrinogen Fukuoka II (Bβ chain Gly15→Cys)36, and free Cys was attached in cases of fibrinogen Osaka II (γ chain Arg275→Cys)37 and antithrombin Toyama (Arg47→Cys)38,39. It is interesting to note that a CRM positive factor XII deficiency, factor XII Washington D.C., has an amino acid substitution of Cys571→Ser, which resulted in a generation of free Cys540.13 Unlike Tenri-type factor XII, however, factor XII Washington D.C. was efficiently secreted into the blood stream.
In pulse-chase experiments, Tenri-type factor XII was poorly secreted from stably-transfected BHK cells and the total amounts of radioactivity decreased to ≈50% during an 8-hour chase (Figs 3A and B) and to ≈15% during a 16-hour chase (Fig 4). Thus, an extensive intracellular degradation of Tenri-type factor XII was suggested by these experiments. Although endogenous α1-microglobulin expression was negligible in BHK cells, the addition of cystine, which presumably enhanced a mixed disulfide-linkage formation of α1-microglobulin with Tenri-type factor XII, improved secretion of the mutant, suggesting resistance to intracellular degradation as a result of blocking the free sulfhydryl group (Fig 3C). The degradation occurred in pre-Golgi, nonlysosomal compartments, and a proteasome appeared to play a critical role in this degradation (Fig 4). This type of degradation is referred to as “ER-associated degradation” or “quality control in the ER” (see reviews in Kopito40 and Kim and Arvan41). Currently, various nascent proteins that fail to fold or to oligomerize correctly are known to be degraded through this mechanism. We have shown that protein C synthesized in the presence of warfarin22 and type I (secretion defect) antithrombin deficiency mutants42 were degraded by the quality control in the ER. Tenri-type factor XII presumably folded incorrectly due to the introduction of Cys34 and was retained in the ER associating with molecular chaperones, eg, calnexin, calreticulin, and Ig binding protein (BiP).43 Finally, misfolded Tenri-type factor XII was degraded by a proteasome presumably after the retrograde transport from the ER to the cytosol through a translocon.40 41 It should be noted that a trace amount of α1-microglobulin–bound form of factor XII Tenri escaped the quality control mechanism and was secreted into the blood stream. The difference in the association of ER chaperones with Tenri-type factor XII, with or without binding to α1-microglobulin, remains to be investigated.
ACKNOWLEDGMENT
We thank Drs Satoshi Omura and Ross T.A. MacGillivray for supplying us with lactacystin and the factor XII cDNA, respectively. We also thank Dr Sadao Wakabayashi and Kazuya Hara of our department for helpful discussion and technical assistance, respectively.
Supported by a Grant-in-Aid for Scientific Research on Priority Areas (Intracellular Proteolysis) from the Ministry of Education, Science, and Culture of Japan (to T. K.).
S.K. and F.T. contributed equally to this study.
Address reprint request to Takehiko Koide, DSc, Department of Life Science, Faculty of Science, Himeji Institute of Technology, Harima Science Garden City, Hyogo 678-1297, Japan; e-mail: koide@sci.himeji-tech.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal