Abstract
BCL3 encodes a protein with close homology to IκB proteins and interacts with p50 NF-κB homodimers. However, the regulation and transcriptional activity of BCL3 remain ill-defined. We observed here that interleukin-9 (IL-9) and IL-4, but not IL-2 or IL-3, transcriptionally upregulated BCL3 expression in T cells and mast cells. BCL3 induction by IL-9 was detected as soon as 4 hours after stimulation and appeared to be dependent on the Jak/STAT pathway. IL-9 stimulation was associated with an increase in p50 homodimers DNA binding activity, which was mimicked by stableBCL3 expression. This contrasts with tumor necrosis factor (TNF)-dependent NF-κB activation, which occurs earlier, involves p65/p50 dimers, and is dependent on IκB degradation. Moreover, IL-9 stimulation or BCL3 transient transfection similarly inhibited NF-κB–mediated transcription in response to TNF. Taken together, our observations show a new regulatory pathway for the NF-κB transcription factors through STAT-dependent upregulation ofBCL3 gene expression.
THE BCL3 GENE WAS originally identified at the junction in the t(14;19) chromosome translocation in certain cases of human B-cell chronic lymphocytic leukemias.1 In leukemic cells with this translocation, expression of this gene is constitutive, pointing to a role forBCL3 in leukemogenesis,1 although there is no experimental evidence as yet for a role of BCL3 in transformation. BCL3 mRNA is detected in different tissues, especially in spleen and other lymphoid organs.2BCL3 null mutant mice develop normally but exhibit severe defects in cellular and humoral immune responses to pathogens and show altered spleen micro-architecture associated with a loss of splenic B cells.3 4 Although these observations show that BCL3 is involved in the regulation of immune responses, its precise role is still unknown.
BCL3 belongs to the IκB family of proteins, featuring 7 tandem ankyrin repeats that are similar to those found in p105/NF-κB15 and other IκB members: p100/NF-κB2, IκBα, IκBβ, and IκBε (for review see Baldwin6and Baeuerle and Baltimore7). Apart from this conserved domain, BCL3 also features a Pro-rich amino-terminal domain and a Pro- and Ser-rich carboxy-terminal domain.1
Like other IκB proteins, BCL3 interacts with NF-κB transcription factors via its ankyrin repeats. NF-κB consists of dimers of subunits that contain a conserved Rel homology domain required for dimerization, interaction with IκB, and DNA binding. The NF-κB family can be divided into two groups: (1) c-Rel, p65/RelA, and RelB, which contain a well-defined transactivation domain; and (2) p50 and p52, which are generated by proteolytic processing from their precursors, p105/NF-κB1 and p100/NF-κB2, respectively. The major form of NF-κB, in most cases, is a heterodimer of the p50 and p65 subunits. The classical pathway leading to NF-κB activation has been extensively reviewed.8 9 p65/p50 dimers are sequestrated in the cytoplasm by interaction with IκB proteins and translocated into the nucleus in response to various extracellular stimuli that induce IκB phosphorylation and degradation. This results in κB-dependent transcription of a variety of genes implicated in stress and immune responses.
Contrasting with IκBα and IκBβ, which specifically interact with dimers containing p65 or c-Rel,10,11 BCL3 specifically interacts with p50 or p52 homodimers.12,13 Moreover, BCL3 is predominantly a nuclear protein,2,14 whereas IκB is mainly a cytoplasmic protein.14 15
Whether BCL3 acts as a bona fide IκB molecule has been disputed. Initial reports indicated that BCL3 can inhibit p50 homodimers DNA binding.12,16 However, Fujita et al17 showed that BCL3 can form a ternary complex on DNA in association with p50 homodimers. Recently, it was clearly demonstrated that BCL3 favors p50 dimer formation by recruiting p50 monomers from the cytoplasmic pool of p105/p50 dimers and enhances nuclear translocation and DNA binding of p50 dimers.18
The precise role of p50 homodimers is still a matter of controversy: some investigators have reported that they are unable to activate transcription,12,19 whereas others have reported that p50 homodimers can activate a promoter containing κB sites.20,21 In this context, BCL3 was proposed to be an activator of transcription, either by removing p50 homodimers from κB sites and allowing p65/p50 transactivating complexes to target DNA12 or by coactivation through p50 homodimers.17 Other observations suggest that BCL3 inhibits κB-dependent transcription.22
The regulation of BCL3 expression has been poorly investigated so far. This gene was shown to be induced by mitogenic stimuli in B and T cells.1,23 The role of cytokines in this process has not been studied yet. Interleukin-9 (IL-9) is a multifunctional cytokine secreted by activated T cells24 and exerts its effects on a variety of cell types, especially in the immune system (mast cells, B and T lymphocytes, hematopoietic progenitors, and eosinophils).25 Interestingly, IL-9 was described as an antiapoptotic factor for thymic lymphomas,26 and transgenic mice expressing the IL-9 protein at a high level are more susceptible to lymphoma development than control littermates.27 More recently, a role for IL-9 has been suggested in allergy and asthma.28
We report here that IL-9 induces the expression of BCL3 mRNA and protein and enhances the DNA binding of p50 homodimers. Moreover, BCL3 induction by IL-9 appears to be STAT-dependent and leads to repression of tumor necrosis factor (TNF)-induced NF-κB transactivation. Our results point towards a previously undocumented pathway leading to NF-κB regulation by cytokines independently of IκB degradation.
MATERIALS AND METHODS
Cell culture, cell transfections, and cytokines.
T-helper cell clones TS2 and TS329 were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS), 50 μmol/L 2-mercaptoethanol, 0.55 mmol/L L-arginine, 0.24 mmol/L L-asparagine, and 1.25 mmol/L L-glutamine. These factor-dependent cell lines were able to grow in the presence of either IL-2, IL-4, or IL-9 without antigen and feeder cells.24 29 ST2K9 (a gift from Dr E. Schmitt, Gütenberg University, Mainz, Germany) was cultured in the same medium. BW5147, a murine lymphoma cell line (ATCC, Rockville, MD), and MC9p3, an autonomous variant of the MC9 mast cell line (a gift from Dr C. Petit-Frère, Institut Henri Beaufour, Les Ullys, France), were cultured in Iscove-Dulbecco’s medium supplemented with 10% FCS, 50 μmol/L 2-mercaptoethanol, 0.55 mmol/L L-arginine, 0.24 mmol/L L-asparagine, and 1.25 mmol/L L-glutamine. L138 (a gift from Dr L. Hültner, GSF, München, Germany) and MC9, two mast cell lines, were cultured in the same medium in the presence of IL-3 or IL-9. Mouse bone marrow-derived mast cells (BMMC) were obtained by culturing bone marrow from Balb/c mice for 4 weeks in enriched medium (RPMI-1640 containing 0.55 mmol/L L-arginine, 0.24 mmol/L L-asparagine, 1.25 mmol/L L-glutamine, 50 μmol/L 2-mercaptoethanol, and 20% FCS) supplemented with either recombinant murine IL-3 (20 U/mL) or a combination of IL-3 (20 U/mL) and IL-9 (200 U/mL). Fluorescence-activated cell sorting (FACS) analysis of this population showed an homogenous staining by biotinylated IgE and no staining with anti-Mac1, anti-Mac2, anti-Mac3, and anti-Thy1 antibodies.
For stable transfection of a BCL3 expression plasmid, theBCL3 cDNA was cloned in the pEF-BOS plasmid30 in which a puromycin resistance gene has been inserted for selection of transfectants. Cells were transfected by electroporation (1,500 μF, 74 ohm, and 290 V for MC9p3; 1,500 μF, 74 ohm, and 230 V for BW5147) with 40 μg of sterile DNA. Clones and pools of transfected cells were selected using puromycin (1.8 μg/mL), and BCL3 expression was checked by Western blot analysis with an affinity-purified rabbit polyclonal antibody directed against the C-terminal part of BCL3 (Santa Cruz-sc#185; Santa Cruz Laboratories, Santa Cruz, CA; 1 μg/mL). BW5147 cells expressing wild-type or mutated hIL-9R were obtained as previously described31 and cultured in the presence of 1.5 μg/mL puromycin. Two additional hIL-9R mutants, mut6 (activating STAT1 and 3) and mut7 (activating STAT5), were similarly transfected in BW5147 (Demoulin et al, manuscript in preparation). In the mut6 hIL-9R, mutation of leucine 368 into arginine reduced STAT5 activation to less than 5% of the activation observed with the wild-type hIL-9R, without affecting the activation of other STATs. In the mut7 hIL-9R, replacement of glutamine 370 by a leucine abolished STAT3 and STAT1 activation, without affecting that of STAT5.
Recombinant human IL-9 (2 × 107 U/mg), mouse IL-4 (3.8 × 106 U/mg), mouse IL-6 (109 U/mg), and mouse IL-9 (5 × 107 U/mg) were produced in the baculovirus system in our laboratory and purified as previously described.32 Human recombinant IL-2 (3.5 × 106 U/mg) was provided by Cetus (Chiron Corp, Amsterdam, The Netherlands) and mouse recombinant IL-3 (2 × 107 U/mg) was a gift of Dr J.Y. Bonnefoy (Glaxo, Geneva, Switzerland). Human recombinant TNFα was supplied by Peprotech (Rocky Hill, NJ). Actinomycin D (ICN, Irvine, CA), protein synthesis inhibitor cycloheximide (Sigma, St Louis, MO), and proteasome inhibitor MG-132 (Biomol, Plymouth Meeting, PA) were used at 5, 10, and 50 μmol/L, respectively.
Subtractive hybridization.
Total RNA was prepared from TS2 cells stimulated with IL-2 (200 U/mL) or mIL-9 (200 U/mL) for 48 hours, using guanidium isothiocyanate lysis and CsCl gradient centrifugation.33 Polyadenylated RNA was purified from total RNA with oligo(dT) cellulose columns (Pharmacia, Uppsala, Sweden). Double-stranded cDNA was generated from 5 μg polyA+ RNA using an oligo(dT) primer and the SuperScript Choice System for cDNA synthesis, according to the manufacturer’s recommendations (GIBCO BRL, Grand Island, NY). Representational difference analysis was performed as described.34
After 3 rounds of subtraction, final difference products were digested with Dpn II and cloned into the BamHI site of pTZ19R. Double-stranded plasmid DNA was prepared and sequenced with a Thermo-sequenase Sequencing kit (Amersham, Arlington Heights, IL). Sequence comparisons with the GenBank and EMBL databases were performed with the BLAST search program (NCBI, Bethesda, MD). Oligo(dT)-primed cDNA libraries generated from IL-9–stimulated TS2 cells were screened with the mouse BCL3 Dpn II fragment.
Preparation of mRNA and Northern blot.
Total RNA was prepared using guanidium isothiocyanate lysis and CsCl gradient centrifugation from TS2, TS3, ST2K9, L138, MC9, and BMMC. These IL-9–responsive cell lines were cultured in medium containing saturating concentrations of the indicated cytokines as follows: 10 days in the presence of 100 U/mL IL-2 or 200 U/mL IL-9 (or IL-4) for TS2, TS3, and ST2K9 cells; 10 days in the presence of 200 U/mL IL-3 or 200 U/mL IL-9 for L138 and MC9 cells; and 28 days in the presence of 20 U/mL IL-3 or 20 U/mL IL-3 and 200 U/mL IL-9 for BMMC. BW5147 was cultured for 2 days with or without 500 U/mL IL-9 or IL-4 or 10,000 U/mL IL-6. Total RNA was extracted from BW5147 cells transfected with mutants of the human IL-9 receptor and stimulated for 24 hours with 500 U/mL human or murine IL-9 using Trizol reagent (GIBCO BRL), according to the manufacturer’s guide.
Ten micrograms of total RNA was fractionated by electrophoresis in a 1.3% agarose gel containing 2.2 mol/L formaldehyde and were transferred onto a Hybond-C Extra nitrocellulose membrane (Amersham). cDNAs were labeled using the Rediprime DNA labeling kit from Amersham. Hybridizations and washes were performed as described.35The BCL3 probe was a 1.8-kb cDNA containing the complete coding sequence. After autoradiography, all blots were reprobed with a chickenβ-actin probe to control for even loading of RNA.
Reverse transcription-polymerase chain reaction (RT-PCR) analysis and Southern blotting.
Reverse transcription was performed on 5 μg Trizol-purified total RNA with an oligo(dT) primer. cDNA corresponding to 20 ng of total RNA was amplified for 25 cycles by PCR with specific primers for murineBCL3 as follows: sense 5′-GCGCAGCGGCTGCGACGT-3′ (from position 204 of the cDNA sequence; GenBank accession no.AF067774) and antisense 5′-CATCCGTCTCAGCTGCTTCCT-3′ (from position 1394 of the cDNA sequence); and for β-actin: sense 5′-ATGGATGACGATATCGCTGC-3′ and antisense 5′-GCTGGAAGGTGGACAGTGAG-3′ (20 cycles). The post-PCR products were analyzed in ethidium bromide-stained 1% agarose gel. Specific amplification was confirmed after blotting (Zeta-Probe membrane; Bio-Rad, Hercules, CA) by hybridization with internal radioactive probes. The internal probe was 5′-GTCCACGACATCATGCTACTG-3′ for β-actin and 5′-TGTGGTGATGACAGCCAGGTG-3′ for the mouse BCL3. Radioactive signals were quantified by Phosphorimager (Molecular Dynamics, Sunnyvale, CA) and theBCL3/actin ratios were calculated.
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts were prepared as described,36 with minor modifications. Briefly, cells (1.5 × 107) were stimulated for the indicated periods with IL-9 (100 U/mL) or TNFα (10 ng/mL), washed with phosphate-buffered saline (PBS), and resuspended in 1 mL ice-cold hypotonic buffer A for 15 minutes (10 mmol/L HEPES buffer, pH 7.5, containing 10 mmol/L KCl, 1 mmol/L MgCl2, 5% glycerol, 0.5 mmol/L EDTA, 0.1 mmol/L EGTA, 0.5 mmol/L dithiothreitol, 1 mmol/L Pefabloc [Boehringer Mannheim, Mannheim, Germany], 10 μg/mL aprotinin, 1 mmol/L Na3VO4, and 5 mmol/L NaF). The cells were lyzed by adding 65 μL NP-40 10% and vortexing. Nuclei were pelleted (30 seconds at 14,000 rpm) and extracted for 30 minutes in 100 μL hypertonic buffer B (buffer A supplemented with HEPES [20 mmol/L], glycerol [20%], and NaCl [420 mmol/L]). Nuclear debris were removed through 2 minutes of centrifugation. Analysis of DNA binding activity was performed as described37 using32P-labeled oligonucleotide probes corresponding to (1) a palindromic κB motif38: upper strand, 5′-GATCCAACGGCAGGGGAATTCCCCTCTCCTTA-3′, and bottom strand, 5′-GATCTAAGGAGAGGGGAATTCCCCTGCCGTTG-3′; and (2) an Sp1 DNA binding motif39: upper strand, 5′-ATTCGATCGGGGCGGGGCGAGC-3′, and bottom strand, 5′-ATGCTCGCCCCGCCCCGATCGA-3′.
Supershifts were performed by adding antibodies to the incubating mixture of nuclear extracts and labeled DNA probe. Four micrograms of anti-p50 antibody (sc#1192-X; Santa Cruz) and 2 μg of anti-p65 antibody (sc#372-X; Santa Cruz) were used per lane.
Western blot.
IκBα phosphorylation was assayed using the PhosphoPlus IκBα(Ser32) Antibody Kit (New England Biolabs, Beverley, MA). Briefly, cells (2 × 106) were stimulated with IL-9 (100 U/mL) or TNFα (10 ng/mL) for 5, 10, or 30 minutes and lyzed in 100 μL SDS Sample Buffer (Bio-Rad). Proteins were fractionated on precast Novex sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 8%) and electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham). Membranes were blocked in 5% nonfat dry milk, washed, and probed with diluted (1/1,000) affinity-purified rabbit polyclonal antibodies (directed against IκBα or the phosphorylated Ser32 of this protein) and with horseradish peroxidase-linked antirabbit antibody (1/2,000). An ECL detection kit (Phototope-HRP Western Detection Kit; New England Biolabs) was used for expression of chemiluminescence.
For BCL3 detection, BW5147 cells (2 × 107) were stimulated or not stimulated with IL-9 (100 U/mL) for different periods of time. Total cell extracts were prepared by lysis in RIPA buffer (PBS, pH 7.4, 1% NP40, 0.5% sodium deoxycholate, and 0.1% SDS). Fifty micrograms of proteins was loaded with 1 vol of SDS Sample Buffer (Bio-Rad) on a precast Novex SDS-PAGE polyacrylamide gel (8%). After transfer, the PVDF membrane (Amersham) was blocked in 5% nonfat dry milk, washed, and probed with an affinity-purified rabbit polyclonal antibody directed against the C-terminal part of BCL3 (sc#185; Santa Cruz; 2 μg/mL) and with horseradish peroxidase-linked antirabbit antibody (1/5,000; Transduction Laboratories, Lexington, KY). An ECL detection kit (Phototope-HRP Western Detection Kit; New England Biolabs) was used for expression of chemiluminescence.
Luciferase activity assay.
The reporter plasmid (pGL3-TK-κBPD) was obtained by subcloning two palindromic NF-κB binding sites (5′ GGGGAATTCCCC 3′) and the TK promoter into pGL3 basic (Promega, Madison, WI) upstream of the firefly luciferase coding sequence. As an internal control, we used the pRL-TK vector (Promega) containing theRenilla luciferase gene under the control of the TK promoter. For BW5147 cells, 10 million cells were coelectroporated with 40 μg pGL3-TK-κBPD and 1 μg pRL-TK (230 V, 74 Ω, and 1,500 μF). The pool of transfected cells was divided into four equal fractions, and each fraction was stimulated either with mIL-9 (100 U/mL), mIL-4 (500 U/mL), hTNFα (10 ng/mL), or a combination of TNFα and IL-9 or IL-4 or was left unstimulated. After 8 or 48 hours, cells were pelleted and lyzed. Both luciferase activities were monitored with the Dual-Luciferase Reporter Assay System kit (Promega) according to the manufacturer’s instructions. For COS cells, 5 × 105cells per well were seeded in 6-well plates 24 hours before transfection with Lipofectamin (GIBCO BRL). Cells were transfected with 250 ng pGL3-TK-κBPD, 250 ng pRL-TK, and either 1 μg of theBCL3 cDNA in the pEF-BOS plasmid or 1 μg of the empty vector. After 24 hours, cells were stimulated or not stimulated with TNFα (10 ng/mL). Luciferase activities were monitored 8 hours later as described above.
RESULTS
IL-9 induces BCL3 gene expression in mouse T lymphocytes and mast cells.
To find a specific gene expression pattern that could explain the effects of IL-9 on mouse T lymphocytes, we performed a representational difference analysis of gene expression on TS2 cells. These T-helper cells were cultured either with IL-2 or IL-9 for 10 days. Poly-A+ RNA was extracted and the respective oligo(dT)-primed cDNAs were prepared, digested with Dpn II, and then used to create both amplicons. After three rounds of subtractive hybridization, the third difference product was cloned and 57 clones were sequenced. Five independent IL-9–specific cDNA sequences were identified and one of them was found to match the sequence ofBCL3.
Full-length cDNA clones obtained from a TS2 cDNA library were sequenced and found to differ from the published BCL3 gene23in 23 nucleotides, resulting in 11 amino acid changes. Because the same amino acid changes were found in a cDNA library from another unrelated cell line (BW5147 from AKR mice) and because we have never identified any cDNA that matches the published sequence, we assume that these clones correspond to the bona fide BCL3 gene and not to a new BCL3-related gene (GenBank accession no. AF067774).
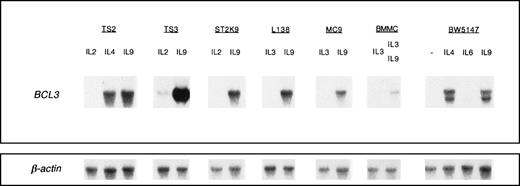
To confirm BCL3 mRNA expression upon IL-9 stimulation, we performed a Northern blot hybridization with RNA from T-helper cell clones that can grow in the presence of either IL-2, IL-4, or IL-9 (TS2, TS3, or ST2K9). As shown in Fig 1, a strong BCL3 signal was detected in all T-cell clones stimulated with IL-9 but not when the cells were cultured in the presence of IL-2. Interestingly, both IL-9 and IL-4 induced the BCL3 mRNA in TS2 cells as well as in the BW5147 thymic lymphoma. We observed a similarBCL3 gene induction by IL-9 in primary BMMC and in mast cell lines, such as MC9 and L138, that can grow either with IL-3 or IL-9.
IL-9 induces BCL3 expression. The indicated IL-9–responsive cells were cultured in medium containing saturating concentrations of the indicated cytokines: 10 days in the presence of 100 U/mL IL-2, 200 U/mL IL-4, or 200 U/mL IL-9 for T-helper cell clones (TS2, TS3, and ST2K9) and 10 days in the presence of 200 U/mL IL-3 or 200 U/mL IL-9 for mast cell lines (L138 and MC9). BMMC were cultured in the presence of 20 U/mL IL-3 or a combination of IL-3 and IL-9 (200 U/mL). The mouse lymphoma line BW5147 was stimulated for 2 days with 500 U/mL IL-9 or IL-4 or 10,000 U/mL IL-6. After electrophoresis of 10 μg of total RNA and transfer to nitrocellulose, filters were hybridized with a 32P-labeled mouse BCL3 cDNA probe. Hybridization with a β-actin probe confirmed that comparable amounts of RNA had been loaded in each lane.
IL-9 induces BCL3 expression. The indicated IL-9–responsive cells were cultured in medium containing saturating concentrations of the indicated cytokines: 10 days in the presence of 100 U/mL IL-2, 200 U/mL IL-4, or 200 U/mL IL-9 for T-helper cell clones (TS2, TS3, and ST2K9) and 10 days in the presence of 200 U/mL IL-3 or 200 U/mL IL-9 for mast cell lines (L138 and MC9). BMMC were cultured in the presence of 20 U/mL IL-3 or a combination of IL-3 and IL-9 (200 U/mL). The mouse lymphoma line BW5147 was stimulated for 2 days with 500 U/mL IL-9 or IL-4 or 10,000 U/mL IL-6. After electrophoresis of 10 μg of total RNA and transfer to nitrocellulose, filters were hybridized with a 32P-labeled mouse BCL3 cDNA probe. Hybridization with a β-actin probe confirmed that comparable amounts of RNA had been loaded in each lane.
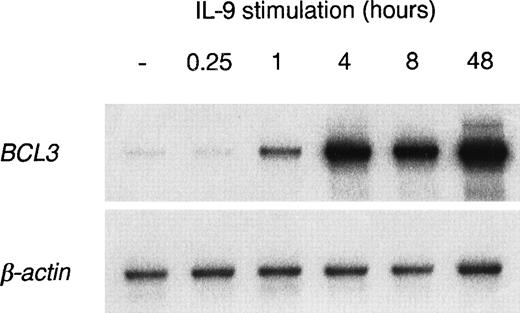
The kinetics of BCL3 expression in BW5147 cells were studied by RT-PCR after IL-9 stimulation for various periods of time (15 minutes to 48 hours). As shown in Fig 2, theBCL3 message is upregulated within 1 hour of IL-9 stimulation, whereas maximal expression is reached upon 4 to 8 hours of stimulation.
Kinetics of BCL3 upregulation by IL-9. BW5147 cells were cultured in the absence or in the presence of 100 U/mL IL-9 for the indicated periods of time. Total RNA was extracted and RT-PCR amplification was performed with oligonucleotides specific forBCL3 or β-actin. A Southern blot of the PCR products was performed and filters were hybridized with the BCL3 orβ-actin cDNA probes.
Kinetics of BCL3 upregulation by IL-9. BW5147 cells were cultured in the absence or in the presence of 100 U/mL IL-9 for the indicated periods of time. Total RNA was extracted and RT-PCR amplification was performed with oligonucleotides specific forBCL3 or β-actin. A Southern blot of the PCR products was performed and filters were hybridized with the BCL3 orβ-actin cDNA probes.
IL-9 induces delayed, IκBα-independent NF-κB binding.
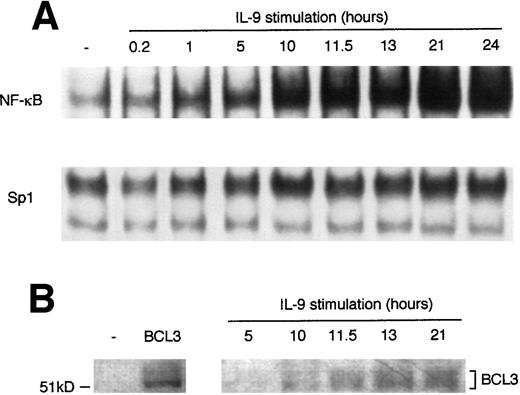
Because BCL3 has been reported to interact with NF-κB dimers,12 40 we studied NF-κB DNA binding in IL-9–stimulated BW5147 lymphoma. Cells were stimulated with IL-9 for periods of time varying from 12 minutes to 24 hours. Nuclear extracts were prepared and analyzed in a band shift assay with a32P-labeled palindromic NF-κB probe or a Sp1 probe as a control. As shown in Fig 3A, at least 10 hours of IL-9 stimulation are required to observe a significant increase in NF-κB binding, and maximum levels of binding are reached after 21 hours of stimulation. Using Sp1 binding as a control, NF-κB binding was quantified with a PhosphorImager and we found an average fold induction of 2.8- ± 0.3-fold from 7 independent experiments with 24 hours of IL-9 stimulation (P < .0001;t-test). The plateau of NF-κB binding was stable upon long-term stimulation by IL-9 (at least 4 days; data not shown). By Western blot analysis, the kinetics of upregulation of the BCL3 protein correlated with NF-κB DNA binding activity (Fig 3B). A similar increase in NF-κB binding in response to IL-9 was observed in other cell lines in which IL-9 also upregulates BCL3 expression, such as the TS2 T-helper clone and the MC9 mast cell line (data not shown).
IL-9 increases NF-κB DNA binding. (A) EMSA analysis of IL-9–stimulated BW5147 cells. Cells were cultured in the absence or in the presence of 100 U/mL IL-9. Nuclear extracts were prepared after various stimulation times and a mobility shift assay was performed using a 32P-labeled palindromic κB site. An Sp1 DNA binding oligonucleotide was used as a control for the loading of similar protein quantities in each lane. (B) Western blot analysis of BCL3 induction by IL-9. The left panel shows BCL3 expression in BW5147 cells transfected with a control vector (−) or the BCL3 cDNA cloned into the pEF-BOS plasmid (BCL3) as a positive control. In the right panel, cells were cultured as in (A). Total extracts were prepared and analyzed by Western blot with anti-BCL3 antibodies as described in Materials and Methods.
IL-9 increases NF-κB DNA binding. (A) EMSA analysis of IL-9–stimulated BW5147 cells. Cells were cultured in the absence or in the presence of 100 U/mL IL-9. Nuclear extracts were prepared after various stimulation times and a mobility shift assay was performed using a 32P-labeled palindromic κB site. An Sp1 DNA binding oligonucleotide was used as a control for the loading of similar protein quantities in each lane. (B) Western blot analysis of BCL3 induction by IL-9. The left panel shows BCL3 expression in BW5147 cells transfected with a control vector (−) or the BCL3 cDNA cloned into the pEF-BOS plasmid (BCL3) as a positive control. In the right panel, cells were cultured as in (A). Total extracts were prepared and analyzed by Western blot with anti-BCL3 antibodies as described in Materials and Methods.
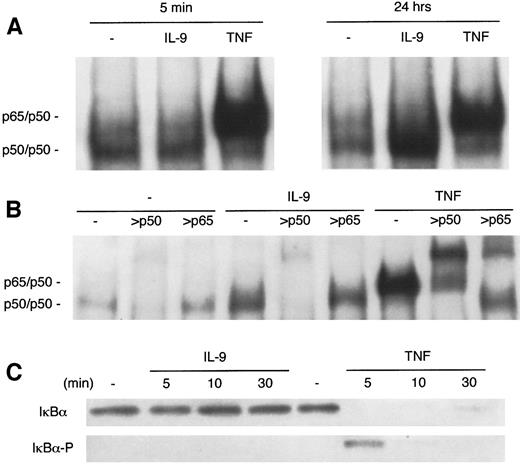
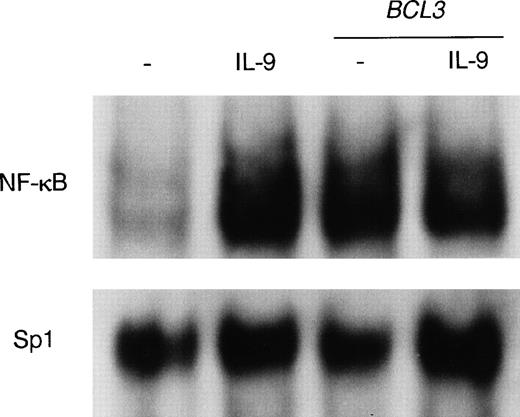
To compare the activity of IL-9 with that of a classical activator of NF-κB, BW5147 cells were stimulated for 5 minutes or 24 hours with either IL-9 or TNFα, and a band shift assay was performed with the palindromic NF-κB site. As shown in Fig4A, 5 minutes of TNFα stimulation was sufficient to result in a clear NF-κB band shift, whereas IL-9–mediated activation is optimal only after 24 hours. In addition to this difference in the kinetics of activation, Fig 4A shows that NF-κB complexes induced by TNF and IL-9 exhibited distinct eletrophoretic mobilities. IL-9–induced complexes migrated faster than those induced by TNF. Moreover, antibodies against the p65/RelA NF-κB subunit supershifted TNF-induced complexes but not IL-9–induced DNA binding (Fig 4B). By contrast, the latter complex is supershifted by anti-p50. Because anti-p52 antibodies did not affect IL-9–induced complexes (data not shown), the IL-9–induced NF-κB–binding complexes are likely composed of p50 homodimers.
Differences in NF-κB binding induction by IL-9 and TNF. (A) Delayed induction by IL-9 of a fast-migrating NF-κB complex. BW5147 cells were stimulated in the absence or in the presence of IL-9 (100 U/mL) or TNF (10 ng/mL) for 5 minutes or 24 hours. Nuclear extracts were prepared and a mobility shift assay was performed using a32P-labeled palindromic κB site. The two types of NF-κB dimers binding to the probe (p65/p50 and p50 homodimers) are indicated. (B) NF-κB complexes induced by IL-9 contain only p50 subunits. BW5147 cells were stimulated in the absence or in the presence of IL-9 (100 U/mL) or TNF (10 ng/mL) for 24 hours. Nuclear extracts were prepared and a mobility shift assay was performed using a32P-labeled palindromic κB site in the presence of anti-p50 (4 μg per lane) or anti-p65 (2 μg per lane) antibodies. The two types of NF-κB dimers binding to the probe (p65/p50 and p50 homodimers) are indicated. (C) IL-9 does not induce the phosphorylation and degradation of IκB. BW5147 cells were stimulated with 100 U/mL IL-9 or 10 ng/mL TNF for 5, 10, or 30 minutes or were left unstimulated. Total cell extracts were loaded on a gel and Western blotted. The membrane was hybridized with two antibodies: one directed against total IκB and another specific for phosphorylated IκB (Ser32).
Differences in NF-κB binding induction by IL-9 and TNF. (A) Delayed induction by IL-9 of a fast-migrating NF-κB complex. BW5147 cells were stimulated in the absence or in the presence of IL-9 (100 U/mL) or TNF (10 ng/mL) for 5 minutes or 24 hours. Nuclear extracts were prepared and a mobility shift assay was performed using a32P-labeled palindromic κB site. The two types of NF-κB dimers binding to the probe (p65/p50 and p50 homodimers) are indicated. (B) NF-κB complexes induced by IL-9 contain only p50 subunits. BW5147 cells were stimulated in the absence or in the presence of IL-9 (100 U/mL) or TNF (10 ng/mL) for 24 hours. Nuclear extracts were prepared and a mobility shift assay was performed using a32P-labeled palindromic κB site in the presence of anti-p50 (4 μg per lane) or anti-p65 (2 μg per lane) antibodies. The two types of NF-κB dimers binding to the probe (p65/p50 and p50 homodimers) are indicated. (C) IL-9 does not induce the phosphorylation and degradation of IκB. BW5147 cells were stimulated with 100 U/mL IL-9 or 10 ng/mL TNF for 5, 10, or 30 minutes or were left unstimulated. Total cell extracts were loaded on a gel and Western blotted. The membrane was hybridized with two antibodies: one directed against total IκB and another specific for phosphorylated IκB (Ser32).
All known inducers of NF-κB cause phosphorylation-dependent degradation of IκBα, which allows NF-κB dimers to migrate freely into the nucleus.9 We analyzed the phosphorylation and expression of IκBα by performing a Western blot with two different antibodies: one recognizing total IκBα protein and one specific for the Ser32-phosphorylated form of IκBα. As shown in Fig4C, 5 minutes of TNFα stimulation resulted in a transient IκBα phosphorylation in BW5147 cells. The protein was then degraded and began reappearing after 30 minutes of TNFα stimulation. Unlike TNFα, IL-9 did not induce a phosphorylation/degradation of IκBα, indicating that IL-9 does not trigger the classical IκB-dependent pathway to activate NF-κB.
Because the unusual kinetics of NF-κB binding and the nature of the dimers activated by IL-9 pointed to a role for BCL3 in this process, we transfected the MC9 mast cell line with the BCL3 cDNA under the control of the EF1α promoter to obtain a strong constitutive expression of BCL3. When compared with control MC9 cells,BCL3-expressing transfectants showed an increased nuclear NF-κB activity that is similar to the effect of IL-9 on control cells. In addition, IL-9 failed to further increase NF-κB binding inBCL3-expressing transfectants (Fig5). In 5 independent experiments, the IL-9–dependent NF-κB induction was quantified as a 2.9- ± 0.3-fold increase in control MC9 cells (P = .0001; t-test), compared with a 1.1- ± 0.4-fold increase in BCL3 transfectants (P = .6;t-test).
Similar NF-κB DNA binding induced by IL-9 and BCL3. MC9 mast cells transfected with either a control vector or the pEF-BOS plasmid containing the BCL3 cDNA were stimulated with 100 U/mL IL-9 for more than 24 hours or were left unstimulated. Shown is a representative EMSA performed with the palindromic NF-κB probe. An Sp1 DNA binding oligonucleotide was used as a control for the loading of similar protein quantities in each lane.
Similar NF-κB DNA binding induced by IL-9 and BCL3. MC9 mast cells transfected with either a control vector or the pEF-BOS plasmid containing the BCL3 cDNA were stimulated with 100 U/mL IL-9 for more than 24 hours or were left unstimulated. Shown is a representative EMSA performed with the palindromic NF-κB probe. An Sp1 DNA binding oligonucleotide was used as a control for the loading of similar protein quantities in each lane.
IL-9 inhibits TNF-induced NF-κB–dependent transcription.
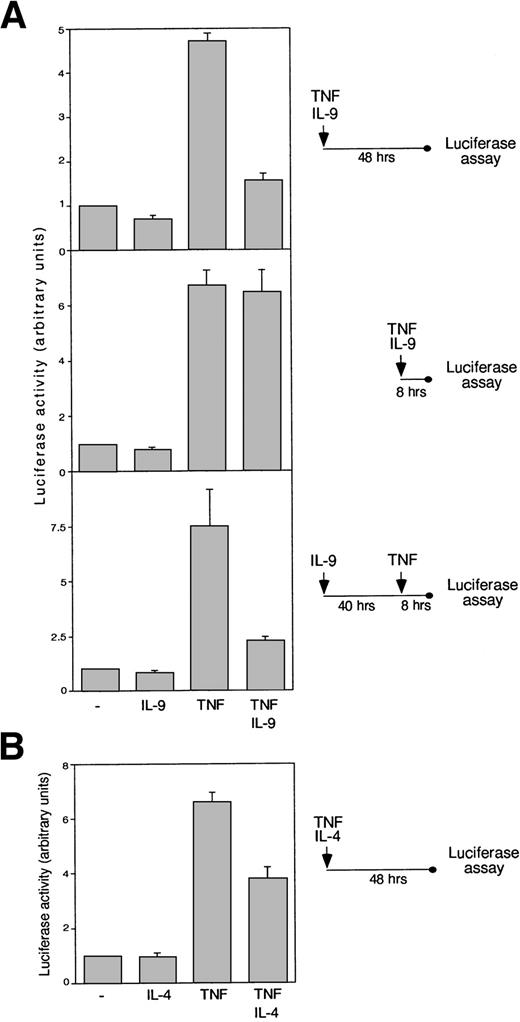
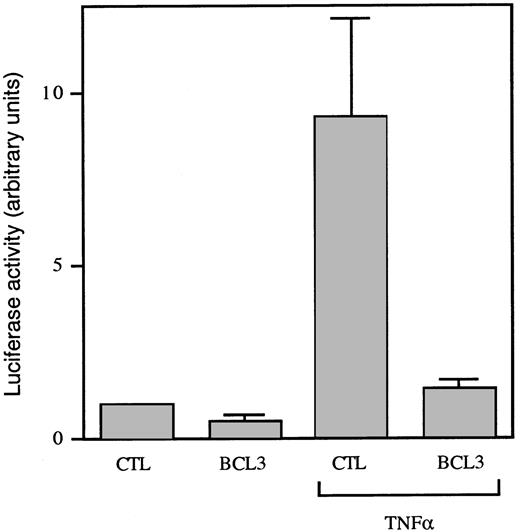
The role of BCL3 in NF-κB–mediated transcription is still controversial, because both stimulatory and inhibitory activities of BCL3 have been reported.12,13,17,22,41 42 To analyze the effect of IL-9 on NF-κB transcriptional regulation, two palindromic NF-κB binding sites were cloned upstream of the TK promoter followed by a luciferase reporter gene to obtain the pGL3-TK-κBPD construct. As shown in Fig 6A, IL-9 did not significantly affect the basal luciferase expression driven by this promoter in BW5147 cells. However, 48 hours of IL-9 stimulation drastically inhibited TNF-induced transcriptional activity (Fig 6A, upper panel). Importantly, 8 hours after TNFα stimulation, IL-9 could only inhibit TNF-induced NF-κB transcriptional activity when cells were preincubated with IL-9, supporting the role of BCL3induction in this process (Fig 6A, middle and lower panels). As expected from the observation that IL-4 induces BCL3 expression in BW5147 (Fig 1), this cytokine also partially inhibited the activity of TNF under the same experimental conditions (Fig 6B). To directly examine the effect of BCL3 on TNF-mediated NF-κB activation, we transiently transfected COS cells with the pGL3-TK-κBPD reporter plasmid and a BCL3 expression construct. As shown in Fig 7, BCL3 expression completely inhibited TNF-induced luciferase activity.
IL-9 and IL-4 repress TNF-induced NF-κB–dependent transcription. (A) BW5147 cells were transfected with the pGL3-TK-κBPD plasmid and pRL-TK as an internal control and were stimulated with IL-9 (100 U/mL) or TNF (10 ng/mL) or were left unstimulated. In the upper panel, cells were cultured with or without cytokines for 48 hours before luciferase assay. In the middle panel, cells were stimulated only for 8 hours. In the bottom panel, cells were incubated or not incubated with IL-9 for 48 hours, and TNF was added for the last 8 hours before luciferase assay. The results are expressed as arbitrary units of luciferase activity, with 1 U being defined as the luciferase activity in unstimulated cells. (B) BW5147 cells were transfected with pGL3-TK-κBPD plasmid and pRL-TK as an internal control and stimulated with IL-4 (500 U/mL) with or without TNF (10 ng/mL) or were left unstimulated. Luciferase was monitored after 48 hours.
IL-9 and IL-4 repress TNF-induced NF-κB–dependent transcription. (A) BW5147 cells were transfected with the pGL3-TK-κBPD plasmid and pRL-TK as an internal control and were stimulated with IL-9 (100 U/mL) or TNF (10 ng/mL) or were left unstimulated. In the upper panel, cells were cultured with or without cytokines for 48 hours before luciferase assay. In the middle panel, cells were stimulated only for 8 hours. In the bottom panel, cells were incubated or not incubated with IL-9 for 48 hours, and TNF was added for the last 8 hours before luciferase assay. The results are expressed as arbitrary units of luciferase activity, with 1 U being defined as the luciferase activity in unstimulated cells. (B) BW5147 cells were transfected with pGL3-TK-κBPD plasmid and pRL-TK as an internal control and stimulated with IL-4 (500 U/mL) with or without TNF (10 ng/mL) or were left unstimulated. Luciferase was monitored after 48 hours.
BCL3 represses TNF-induced NF-κB–dependent transcription. COS cells were transiently transfected with pGL3-TK-κBPD, pRL-TK and either the BCL3 cDNA in the pEF-BOS plasmid or the empty vector. After 24 hours, cells were stimulated or not stimulated with TNF (10 ng/mL). Luciferase activities were monitored 8 hours later. The results correspond to the mean ± SD of 4 individual transfections and are expressed as arbitrary units of luciferase activity, with 1 U being defined as the luciferase activity in unstimulated cells.
BCL3 represses TNF-induced NF-κB–dependent transcription. COS cells were transiently transfected with pGL3-TK-κBPD, pRL-TK and either the BCL3 cDNA in the pEF-BOS plasmid or the empty vector. After 24 hours, cells were stimulated or not stimulated with TNF (10 ng/mL). Luciferase activities were monitored 8 hours later. The results correspond to the mean ± SD of 4 individual transfections and are expressed as arbitrary units of luciferase activity, with 1 U being defined as the luciferase activity in unstimulated cells.
Role of STAT factors in BCL3 and NF-κB induction by IL-9.
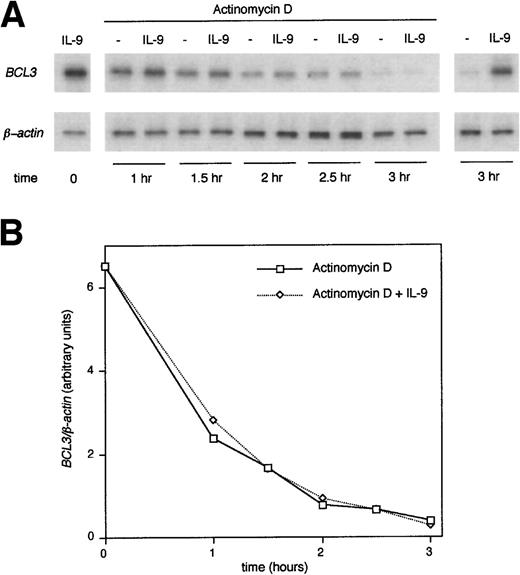
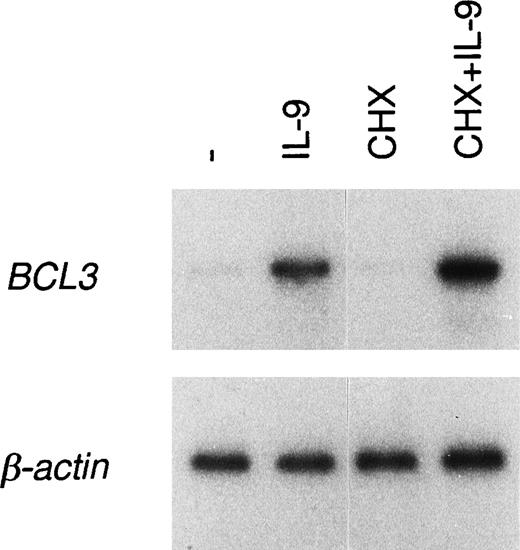
The increase in BCL3 mRNA induced by IL-9 could result either from a transcriptional activation or from stabilization of theBCL3 message. To address this question, cells preincubated in the presence of IL-9 were washed and further cultured for up to 3 hours with or without IL-9 and actinomycin D, an inhibitor of RNA synthesis. After 3 hours, the BCL3 message was barely detectable without IL-9, whereas a strong expression was maintained in the presence of IL-9. By contrast, with actinomycin D, this effect of IL-9 was completely abolished, indicating that transcription is required for this activity (Fig 8). To determine whether IL-9 directly upregulates BCL3 expression or whether this process requires protein synthesis, BW5147 cells were stimulated with IL-9 in the presence of cycloheximide. As shown in Fig 9, cycloheximide did not affectBCL3 expression, as assessed by RT-PCR analysis, indicating that new protein synthesis is not required for this IL-9 activity.
BCL3 upregulation by IL-9 is dependent on RNA transcription. BW5147 cells were preincubated with 100 U/mL IL-9 for 24 hours, extensively washed, and restimulated for 1, 1.5, 2, 2.5, and 3 hours with or without IL-9 and actinomycin D (5 μg/mL) as indicated in the figure. Total RNA was extracted and RT-PCR amplification was performed with oligonucleotides specific for BCL3 orβ-actin. A Southern blot of the PCR products was performed and filters were hybridized with a BCL3 or β-actinoligonucleotide. (A) shows the autoradiography of the blot. (B) shows the intensity of the BCL3 signals from the same blot quantified by PhosphorImager analysis and standardized to theβ-actin level.
BCL3 upregulation by IL-9 is dependent on RNA transcription. BW5147 cells were preincubated with 100 U/mL IL-9 for 24 hours, extensively washed, and restimulated for 1, 1.5, 2, 2.5, and 3 hours with or without IL-9 and actinomycin D (5 μg/mL) as indicated in the figure. Total RNA was extracted and RT-PCR amplification was performed with oligonucleotides specific for BCL3 orβ-actin. A Southern blot of the PCR products was performed and filters were hybridized with a BCL3 or β-actinoligonucleotide. (A) shows the autoradiography of the blot. (B) shows the intensity of the BCL3 signals from the same blot quantified by PhosphorImager analysis and standardized to theβ-actin level.
New protein synthesis is not required for BCL3upregulation by IL-9. BW5147 cells were stimulated with 100 U/mL IL-9 and with or without the combination of 10 μg/mL cycloheximide and 50 μmol/L MG-132 (a proteasome inhibitor) for 4.5 hours, or the cells were left unstimulated. Total RNA was extracted and RT-PCR amplification was performed with oligonucleotides specific forBCL3 or β-actin. A Southern blot of the PCR products was performed and filters were hybridized with the BCL3 orβ-actin cDNA probes.
New protein synthesis is not required for BCL3upregulation by IL-9. BW5147 cells were stimulated with 100 U/mL IL-9 and with or without the combination of 10 μg/mL cycloheximide and 50 μmol/L MG-132 (a proteasome inhibitor) for 4.5 hours, or the cells were left unstimulated. Total RNA was extracted and RT-PCR amplification was performed with oligonucleotides specific forBCL3 or β-actin. A Southern blot of the PCR products was performed and filters were hybridized with the BCL3 orβ-actin cDNA probes.
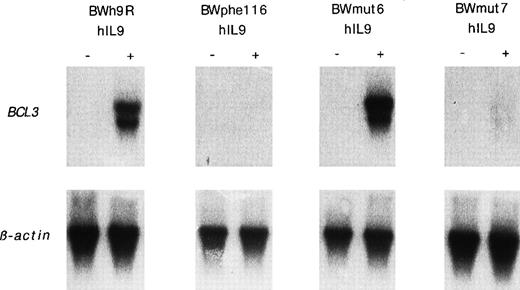
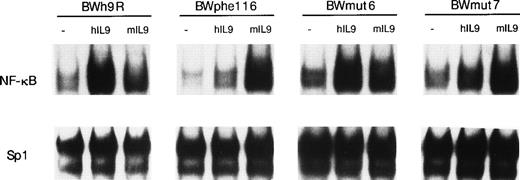
To analyze further the mechanisms of BCL3 upregulation in response to IL-9, we took advantage of BW5147 transfectant cells expressing mutated forms of the human IL-9 receptor (hIL-9R). When cells were transfected with the wild-type hIL-9R, hIL-9 induced a similar BCL3 expression to that induced by mIL-9 in the parental cells (Fig 10). By contrast, hIL-9 was inactive in cells expressing a mutated form of hIL-9R (phe116) in which tyrosine residue 116 was replaced by a phenylalanine, resulting in the inability of the receptor to activate STAT transcription factors.31 To determine the respective role of the different STAT proteins activated by IL-9 (STAT1, 3, and 5), other hIL-9R mutants that specifically activate STAT5 (mut7) or STAT1 and 3 (mut6) were stably transfected in BW5147. As shown in Fig 10, STAT5 is not necessary for BCL3 expression, because this gene is still induced in BWmut6 cells (in which STAT5 is not activated in response to hIL-9). In addition, STAT5 is not sufficient forBCL3 expression, because this gene is not significantly upregulated in BWmut7 (in which STAT5 is the only STAT activated by hIL-9). As expected, the NF-κB activation measured by EMSA in BW5147 transfected with the different hIL-9R mutants matched BCL3expression (Fig 11). The mean NF-κB DNA binding induction in response to IL-9 were 3.9- ± 0.5-fold (P = .01; t-test) for BWh9R, 1.3- ± 0.2-fold (P = .19; t-test) for BWphe116, 5.1- ± 0.6-fold (P = .016; t-test) for BWmut6, and 1.5- ± 0.2-fold (P = .26; t-test) for BWmut7.
BCL3 upregulation by IL-9 in cells expressing mutated hIL-9R. BW5147 cells transfected with the wild-type human IL-9R (BWh9R) or mutants of this receptor partially (BWmut6 and BWmut7) or totally (BWphe116) defective in STAT activation (see text) were stimulated with 500 U/mL human IL-9 for 24 hours. After electrophoresis of 5 μg of total RNA and transfer to nitrocellulose, filters were hybridized with a 32P-labeled mouse BCL3 cDNA probe. Hybridization with a β-actin probe confirmed the even loading of RNA in each lane.
BCL3 upregulation by IL-9 in cells expressing mutated hIL-9R. BW5147 cells transfected with the wild-type human IL-9R (BWh9R) or mutants of this receptor partially (BWmut6 and BWmut7) or totally (BWphe116) defective in STAT activation (see text) were stimulated with 500 U/mL human IL-9 for 24 hours. After electrophoresis of 5 μg of total RNA and transfer to nitrocellulose, filters were hybridized with a 32P-labeled mouse BCL3 cDNA probe. Hybridization with a β-actin probe confirmed the even loading of RNA in each lane.
NF-κB DNA binding induced by IL-9 in cells expressing mutated hIL-9R. BW5147 cells transfected with the wild-type human IL-9R (BWh9R) or mutants of this receptor partially (BWmut6 and BWmut7) or totally (BWphe116) defective in STAT activation (see text) were stimulated with 100 U/mL human IL-9 for 24 hours or with the equivalent amount of mIL-9 as a positive control. Nuclear extracts were prepared and a mobility shift assay was performed using a32P-labeled palindromic κB site. An Sp1 DNA binding site was used as a control for the even loading of proteins in each lane.
NF-κB DNA binding induced by IL-9 in cells expressing mutated hIL-9R. BW5147 cells transfected with the wild-type human IL-9R (BWh9R) or mutants of this receptor partially (BWmut6 and BWmut7) or totally (BWphe116) defective in STAT activation (see text) were stimulated with 100 U/mL human IL-9 for 24 hours or with the equivalent amount of mIL-9 as a positive control. Nuclear extracts were prepared and a mobility shift assay was performed using a32P-labeled palindromic κB site. An Sp1 DNA binding site was used as a control for the even loading of proteins in each lane.
DISCUSSION
Although BCL3 was cloned several years ago as a putative proto-oncogene associated with human B-cell leukemias, its function in the immune system began to be unravelled only recently, with the analysis of knock-out mice.3 4 These mice display severe defects in antigen-specific immune responses, which clearly points to a role for BCL3 in the regulation of the immune system. However, so far, no clear picture of how BCL3 is regulated can be drawn up. In this report, we identify IL-9 as the first physiological stimulus triggering BCL3 expression in T cells and in mast cells. In addition, we show that IL-4 can also induce BCL3 expression in BW5147 lymphoma cells and T-helper cell clones.
The mechanisms underlying this induction were studied using IL-9R mutants defective in STAT activation. STAT1 and STAT3 were found to be particularly important for BCL3 expression in BW5147 stimulated with IL-9. In line with this observation, STAT1 and STAT3 are also activated by IL-4 in these cells (our unpublished observation). By contrast, interferon γ activated only STAT1 and did not induce BCL3 upregulation in BW5147 (data not shown), suggesting that STAT3 is the most important (if not the only) factor involved in BCL3 upregulation by IL-9 and IL-4. IL-6, which activates STAT3 weakly in BW5147 (our unpublished observation), induced a faint BCL3 expression that is detectable by more sensitive techniques, such as RT-PCR (data not shown). The fact that STAT proteins are transcription factors and that BCL3 upregulation by IL-9 required RNA transcription but not protein synthesis indicates that this process is directly regulated at the transcriptional level.
As expected from several reports,17,18 43BCL3expression was followed by an increase in the DNA binding of p50 homodimers. The role of BCL3 in our system is supported by the following observations: (1) anti-p50, but not anti-p65 antibodies supershifted NF-κB dimers that were activated by IL-9; (2) IL-9 did not induce IκB phosphorylation and degradation; (3) BCL3 protein expression correlated with the late NF-κB DNA binding induced by IL-9; (4) cells transfected with the BCL3 cDNA showed a constitutive DNA binding activity of p50 homodimers that is similar to that induced by IL-9 and is not modified further by IL-9 stimulation; and (5) in cells transfected with mutants of the IL-9R, we found a good correlation between the level of BCL3 expression and the DNA binding of p50 homodimers.
The function of BCL3 as a regulator of NF-κB–dependent transcription is still a matter of debate. Based on transient transfection experiments using NF-κB and BCL3 expression plasmids, some reports suggest that BCL3 promotes κB-dependent transcription,12,17 and others suggest that BCL3 inhibits κB-dependent transcription.22 We found here that IL-9 repressed TNF-dependent transcriptional activation through two palindromic κB sites, and a similar effect was observed with IL-4. This repression requires preincubation in the presence of IL-9 for at least 24 hours, in agreement with the kinetics of BCL3induction. The role of BCL3 in this process is further supported by the observation that BCL3 transient transfection similarly inhibited TNF-dependent NF-κB activity.
Interestingly, the IL-9 effect on NF-κB–mediated transcription was dependent on the nature of NF-κB binding sites cloned in the reporter construct. For example, no repression was observed with a reporter plasmid containing NF-κB sites from the TNF promoter (data not shown). Such differential activity may result from the fact that p50 homodimers could more efficiently compete with p65/p50 heterodimers for a palindromic sequence than for other types of NF-κB sites. Taken together, these observations suggest that IL-9 may specifically downregulate a particular set of genes induced by NF-κB. Further experiments will have to identify these genes and to determine their role in IL-9 activities in vivo.
In particular, the role of BCL3 in IL-9–induced tumors needs further investigation. However, BCL3 is unlikely to play a role in the antiapoptotic activity of IL-9, because BCL3 transfection does not protect T-cell lymphomas against corticoid-induced apoptosis (data not shown) and because this activity of IL-9 could be mediated by both STAT5- and STAT3-activating mutants of the IL-9 receptor (Demoulin et al, manuscript in preparation).
In conclusion, the data reported here shed some light on BCL3regulation and point to a link between the Jak-STAT signal transduction pathway and the NF-κB system. The identification of genes that are regulated by cytokines such as IL-9 and IL-4 through this process could lead to a better understanding of the mechanisms underlying the role of BCL3 and these cytokines in immune regulation.
ACKNOWLEDGMENT
The authors thank Drs S. Nagata, A. Burgess, and C. Wildmann for their generous donations of reagents and Dr L. van Fiets and V. Bours for helpful discussions.
Supported in part by the Belgian Federal Service for Scientific, Technical and Cultural Affairs and the Opération Télévie. M.R. and J.L. are scientific associates (Télévie), J.-B.D. is a research assistant, and J.-C.R. is a research associate with the Fonds National de la Recherche Scientifique, Belgium.
Address rerpint requests to Jean-Christophe Renauld, MD, Ludwig Institute for Cancer Research, Avenue Hippocrate, 74, B-1200 Brussels, Belgium; e-mail: renauld@licr.ucl.ac.be.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal