Abstract
Mantle cell lymphomas (MCLs) are characterized by 11q13 chromosomal translocations and cyclin D1 overexpression. The secondary genetic and molecular events involved in the progression of these tumors are not well known. In this study, we have analyzed 45 MCLs (32 typical and 13 blastoid variants) by comparative genomic hybridization (CGH). To identify the possible genes included in the abnormal chromosome regions, selected cases were analyzed for P53, P16INK4a, RB, C-MYC, N-MYC, BCL2, BCL6, CDK4, and BMI-1 gene alterations. The most frequent imbalances detected by CGH were gains of chromosomes 3q (49%), 7p (27%), 8q (22%), 12q (20%), 18q (18%), and 9q34 (16%) and losses of chromosomes 13 (44%), 6q (27%), 1p (24%), 11q14-q23 (22%), 10p14-p15 (18%), 17p (16%), and 9p (16%). High-level DNA amplifications were identified in 11 different regions of the genome, predominantly in 3q27-q29 (13%), 18q23 (9%), and Xq28 (7%). The CGH analysis allowed the identification of regional consensus areas in most of the frequently involved chromosomes. Chromosome gains (P = .02) and losses (P = .01) and DNA amplifications (P = .015) were significantly higher in blastoid variants. The significant differences between blastoid and typical tumors were gains of 3q, 7p, and 12q, and losses of 17p. CGH losses of 17p correlated with P53 gene deletions and mutations. Similarly, gains of 12q and high-level DNA amplifications of 10p12-p13 were associated with CDK4 and BMI-1 gene amplifications, respectively. One of 2 cases with 8q24 amplification showed C-MYC amplification by Southern blot. Alterations in 2p, 3q, 13, and 18q were not associated with N-MYC, BCL6, RB, orBCL2 alterations, respectively, suggesting that other genes may be the targets of these genetic abnormalities in MCLs. Increased number of gains (0 v 1-4 v >4 gains per case) (P = .002), gains of 3q (P = .02), gains of 12q (P = .03), and losses of 9p (P = .003) were significantly associated with a shorter survival of the patients. These results indicate that an increased number of chromosome imbalances are associated with blastoid variants of MCLs and may have prognostic significance.
MANTLE CELL LYMPHOMA (MCL) is a malignant lymphoproliferative disorder derived from naive pregerminal center CD5+ B cells expressing IgM/D.1Morphologically, the typical variant of MCL is composed of a monotonous proliferation of small to medium-sized lymphocytes with irregular nuclei and relatively low proliferative activity.1 Several studies have also recognized a blastoid variant in which the cells may show either rounder nuclei with fine and disperse chromatin resembling lymphoblasts or larger, deeply indented, and pleomorphic nuclei with occasional nucleoli.2,3 These blastoid variants have a higher proliferative activity and a more aggressive biological behavior than the typical variants of the tumor.2-4
Genetically, MCLs are characterized by chromosomal translocations involving the 11q13 region and its molecular counterpart bcl-1 rearrangement, which results in the overexpression of cyclin D1 gene.5,6 The identification of this translocation in virtually all cases of MCL and the constant cyclin D1 overexpression in the tumors indicate that these molecular phenomena are important mechanisms in their pathogenesis.7,8 Experimental studies have shown that cyclin D1 may function as an oncogene in the malignant transformation of different cell types. However, the tumorigenic and transforming properties of cyclin D1 seem to be less effective than other oncogenes.9 On the other hand, cyclin D1 transgenic animals do not develop spontaneous lymphomas, and lymphomagenesis in these animals requires the cooperation with other oncogenes such asC-MYC.10 All of these findings suggest that other mechanisms, in addition to cyclin D1 deregulation, may participate in the development and progression of MCLs.
The secondary genetic and molecular events involved in the pathogenesis of these tumors are not well known. Recent studies indicate that blastoid variants of MCLs harbor more frequent bcl-1 rearrangements at the major translocation cluster locus2 and have a higher incidence of P53 gene mutations11,12 andP16INK4a deletions than typical variants.13,14 Classic cytogenetic studies have identified additional chromosome abnormalities besides the 11q13 translocations in some MCLs.15-19 However, these studies are limited, and no correlations with the morphologic variants or the biological behavior of the tumor have been described. Chromosomal banding techniques are useful for the identification of chromosomal aberrations and imbalances, but they are less reliable for the recognition of potentially amplified regions. On the other hand, these techniques require the analysis of metaphases upon cell culture that may induce a certain subclone selection and underrepresentation of tumor clones. The relatively new technique of comparative genomic hybridization (CGH) allows a rapid analysis of chromosomal imbalances within the tumor genome including mapping of high-level DNA amplifications without the requirement of cell culturing and metaphase preparation.20
The aims of this study were to determine the secondary chromosomal imbalances and high-level DNA amplifications that may play a role in the development and progression of MCLs, to analyze the potential involvement of specific genes located in the altered chromosomal regions in the different variants of MCLs, and to determine the clinical and pathological relevance of these genetic alterations.
MATERIALS AND METHODS
Case selection.
Tumor specimens from 45 MCLs were included in the study. There were 33 males and 12 females. A total of 29 cases were obtained from the Hospital Clı́nic Provincial of Barcelona; 10 cases from the Hospital Clı́nico Universitario, Salamanca; and 6 from the Hospital Virgen de la Salud, Toledo, Spain. A total of 32 cases were classified as typical MCLs and 13 as blastoid variants of MCLs, according to previously described criteria.1-3 All cases were studied at diagnosis and were reviewed and classified by three of us (E.C., M.A.P., and T.F.). The immunophenotype of the tumors was analyzed using immunohistochemistry on tissue sections and/or cell suspensions by flow cytometry. These studies included Ig light and heavy chains, several B-cell (CD19, CD20, CD22, CD45RA, and CD79a) and T-cell (CD2, CD3, CD5, CD7, CD4, CD8, CD45RO, and CD43) markers, CD10, and CD23. Cyclin D1 expression was examined in all cases by Northern blot analysis and/or immunohistochemistry.7 Bcl-1 rearrangement was also examined in all cases by Southern blot analysis or polymerase chain reaction (PCR) according to a previously described method.21 Cytogenetic analysis could be performed in 7 cases. All tumors included in the study had a B-cell phenotype and all, except one blastoid case, coexpressed CD5. The only CD5−tumor had a bcl-1 rearrangement at the MTC locus detected by Southern blot and PCR, and cyclin D1 overexpression by Northern blot and immunohistochemistry. Cyclin D1 overexpression was observed in all tumors. Bcl-1 rearrangement was detected in 20 (44%) tumors. In addition, 4 of the 7 cases examined by cytogenetic analysis had the t(11;14)(q13;q32) translocation.
DNA extraction.
High molecular weight DNA was extracted from 42 lymph nodes and 3 involved peripheral blood with the use of the standard Proteinase K/RNAse treatment and phenol-chloroform extraction. Normal DNA was obtained from 4 male and 1 female healthy blood donors. DNA was diluted to a concentration of 40 to 60 ng/μL, and 1 μL of each sample was analyzed in a 0.8% agarose gel and stained with ethidium bromide to verify its quality and concentration.
CGH.
Normal and tumor DNA were labeled with Spectrum Red-dUTP and Spectrum Green-dUTP by nick translation using a commercial kit (Vysis, Downers Grove, IL). Subsequently, equal amounts of normal and tumor labeled probes (500 ng) and 10 μg of Cot-1 DNA were coprecipitated with the use of ethanol. The precipitated DNA was dissolved in 12 μL of hybridization buffer and denatured at 74°C for 8 minutes. Normal metaphase spreads (Vysis) were denatured for 5 minutes at 74°C and hybridized with the DNA mixture in a moist chamber for 2 to 3 days. Slides were washed according to the protocol supplied by the manufacturer. Chromosomes were counterstained with 4,6-diamino-2-phenylindole (DAPI), resulting in a G banding-like pattern that was used for chromosome identification.
Slides were analyzed with the use of Cytovision Ultra workstation (Applied Imaging, Sunderland, UK). The fluorescent hybridization signals and DAPI-staining patterns were captured. The software performed a calculation of the tumor DNA to normal DNA fluorescent ratios along the length of each chromosome. Ratio values obtained from at least 10 metaphase spreads were averaged, and the resulting profiles were plotted next to the chromosomal ideograms. Ratio values greater than 1.25 and less than 0.75 were considered to represent chromosomal gains and losses, respectively. A high-level DNA amplification was considered when the fluorescence ratio values exceeded 1.5, and, in addition, a distinct band-like hybridization signal of the tumor DNA was seen. Negative control experiments were performed using differentially labeled male versus male DNA and female versus female DNA. In addition, control experiments in which the Red-dUTP and Green-dUTP labels were interchanged between normal and tumor were also performed.
Southern blot analysis.
Genomic DNA (15 μg) was digested with EcoRI, HindIII, and/or BamHI restriction enzymes (BRL, Gaithersburg, MD), separated on 0.8% agarose gels, and transferred to Hybond-N membranes (Amersham, Buckinghamshire, UK). The membranes were prehybridized; hybridized with the P53, P16INK4a,BCL6, BCL2, C-MYC, N-MYC, RB, CDK4, BMI-1, and β-ACTIN probes; and washed as previously described.7
Probes.
The P53 probe was a 2.0-kb EcoRI-BamHI fragment of the p1A65 (pArgSP53) cDNA clone containing the entire coding region of the human P53 gene, which was kindly provided by Dr L.V. Crawford (Imperial Cancer Research Foundation, Cambridge, UK).22 The P16INK4a probe was a fragment of exon 2 obtained by PCR with the use of primers previously described.13 The BCL6 probe was a 1.4-kbEcoRI-Bgl II fragment of the partial cDNA clone ofBCL6 gene, which was kindly provided by Dr B.W. Baron (University of Chicago, Chicago, IL).23 The BCL2probe was a 1.5-kb HindIII fragment of the partial cDNA clone of BCL2 gene, which was kindly provided by Dr J. Boix (University of Lleida, Lleida, Spain).24 TheC-MYC probe was a 1.4-kb Cla I-EcoRI fragment containing the third coding exon, which was kindly provided by Dr R. Dalla Favera (Columbia University, New York, NY). The N-MYCprobe was a 1.0-kb EcoRI-BamHI coding fragment of exon 2 (Oncor, Gaithersburg, MD). The RB probes were Rb0.9 and Rb3.8 representing the 5′ and 3′ portions of RB cDNA, which was kindly provided by Dr R.A. Weinberg (Whitehead Institute, Cambridge, MA).25 The CDK4 probe was a 1.2-kbBamHI/Sma I full cDNA, which was kindly provided by Dr M. Serrano (Centro Nacional Biotecnologia, Madrid, Spain).26 The BMI-1 probe was a 1.5-kb PstI fragment of the partial cDNA, which was kindly provided by Dr M. van Lohuizen (The Netherlands Cancer Institute, Amsterdam, The Netherlands).27 Probes were radiolabeled using a random primer DNA labeling kit (Amersham) with [α-32P]-dCTP. To normalize the DNA loading, the blots were hybridized with a β-ACTIN probe. The signals were quantified using a Fuji Photo Film system (Image Gauge version 2.5.3, Tokyo, Japan).
Statistical analysis.
Differences among the histologic variants and other initial and evolutive characteristics of the patients in terms of the CGH imbalances were compared by the Fisher’s exact test (two-tailed). The differences observed between means of gains, losses or amplifications, and the histologic subtype were compared using the Student’st-test when the data fulfill the criteria for parametric statistics. Nonparametric tests were used when necessary (U-Mann-Whitney). The actuarial survival analysis was performed according to the method described by Kaplan and Meier,28and the curves were compared by the log rank test.29
RESULTS
CGH.
Forty of the 45 patients (89%) showed gains (total, 124) or losses (total, 109) of genetic material (Figs 1 and2). All of the altered cases, except for 5, showed more than one chromosome imbalance. The single alterations identified in these cases were trisomy X (case 35), trisomy 3 (case 17), monosomy 14 (case 41), gain of 18q23 (case 31), and trisomy 20 (case 8). Irrespective of the morphology of the lymphoma, the most frequent imbalances were gains of chromosomes 3q (49%), 7p (27%), 8q (22%), 12q (20%), 18q (18%), and 9q34 (16%) (Table 1). High-level DNA amplifications were identified in 11 different regions of the genome, predominantly in 3q27-q29 (13%), 18q23 (9%), and Xq28 (7%) (Table2). Eight of the 11 (73%) amplifications were localized in chromosomal regions in which known fragile sites have been identified (3q27-q29 and FRA3C, Xq28 and FRAXF, 8q24 and FRA8C, 2p25 and FRA2C, 7p22 and FRA7B, 13q31-q32 and FRA13D, 3q24-q25 and FRA3D, and 17q23-q25 and FRA17B). The most frequent losses were on chromosome 13 (44%), 6q (27%), 1p (24%), 11q14-q23 (22%), 10p14-p15 (18%), 17p (16%), and 9p (16%) (Table3).
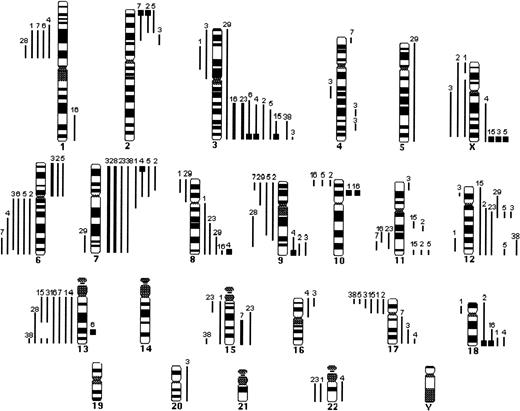
Summary of the CGH data in 32 cases of typical MCL. Lines on the left side of the ideogram indicate loss of chromosomal material; lines on the right side indicate gain of chromosomal material. Thick black bars represent chromosomal gains exceeding 1.5 in a large chromosomal region. High-level DNA amplifications are represented as solid squares. Each line represents a gained or lost region in a single tumor.
Summary of the CGH data in 32 cases of typical MCL. Lines on the left side of the ideogram indicate loss of chromosomal material; lines on the right side indicate gain of chromosomal material. Thick black bars represent chromosomal gains exceeding 1.5 in a large chromosomal region. High-level DNA amplifications are represented as solid squares. Each line represents a gained or lost region in a single tumor.
Summary of the CGH data in 13 cases of blastoid MCL. Lines on the left side of the ideogram indicate loss of chromosomal material; lines on the right side indicate gain of chromosomal material. Thick black bars represent chromosomal gains exceeding 1.5 in a large chromosomal region. High-level DNA amplifications are represented as solid squares. Each line represents a gained or lost region in a single tumor.
Summary of the CGH data in 13 cases of blastoid MCL. Lines on the left side of the ideogram indicate loss of chromosomal material; lines on the right side indicate gain of chromosomal material. Thick black bars represent chromosomal gains exceeding 1.5 in a large chromosomal region. High-level DNA amplifications are represented as solid squares. Each line represents a gained or lost region in a single tumor.
CGH Gains in MCL According to Histologic Subtype
| . | Total Gains . | Histologic Subtype . | P Value . | |||
|---|---|---|---|---|---|---|
| Typical (n = 32) . | Blastoid (n = 13) . | |||||
| n . | (%) . | n . | (%) . | |||
| 3q | 22 | 12 | 37 | 10 | 77 | .02 |
| 7p | 12 | 4 | 12 | 8 | 61 | .002 |
| 8q | 10 | 5 | 16 | 5 | 38 | NS |
| 12q | 9 | 1 | 3 | 8 | 61 | .0003 |
| 18q | 8 | 4 | 12 | 4 | 31 | NS |
| 9q34 | 7 | 4 | 12 | 3 | 23 | NS |
| Xq | 7 | 3 | 9 | 4 | 31 | NS |
| 15q22-q24 | 5 | 3 | 9 | 2 | 15 | NS |
| 16p | 5 | 3 | 9 | 2 | 15 | NS |
| 17q | 5 | 2 | 6 | 3 | 23 | NS |
| 2p | 4 | 0 | — | 4 | 31 | NS |
| 11q25 | 4 | 1 | 3 | 3 | 23 | NS |
| 10p12-p13 | 4 | 2 | 6 | 2 | 15 | NS |
| 22 | 4 | 3 | 9 | 1 | 8 | NS |
| 5p | 3 | 2 | 6 | 1 | 8 | — |
| 6p | 3 | 0 | — | 3 | 23 | — |
| 20 | 2 | 1 | 3 | 1 | 8 | — |
| . | Total Gains . | Histologic Subtype . | P Value . | |||
|---|---|---|---|---|---|---|
| Typical (n = 32) . | Blastoid (n = 13) . | |||||
| n . | (%) . | n . | (%) . | |||
| 3q | 22 | 12 | 37 | 10 | 77 | .02 |
| 7p | 12 | 4 | 12 | 8 | 61 | .002 |
| 8q | 10 | 5 | 16 | 5 | 38 | NS |
| 12q | 9 | 1 | 3 | 8 | 61 | .0003 |
| 18q | 8 | 4 | 12 | 4 | 31 | NS |
| 9q34 | 7 | 4 | 12 | 3 | 23 | NS |
| Xq | 7 | 3 | 9 | 4 | 31 | NS |
| 15q22-q24 | 5 | 3 | 9 | 2 | 15 | NS |
| 16p | 5 | 3 | 9 | 2 | 15 | NS |
| 17q | 5 | 2 | 6 | 3 | 23 | NS |
| 2p | 4 | 0 | — | 4 | 31 | NS |
| 11q25 | 4 | 1 | 3 | 3 | 23 | NS |
| 10p12-p13 | 4 | 2 | 6 | 2 | 15 | NS |
| 22 | 4 | 3 | 9 | 1 | 8 | NS |
| 5p | 3 | 2 | 6 | 1 | 8 | — |
| 6p | 3 | 0 | — | 3 | 23 | — |
| 20 | 2 | 1 | 3 | 1 | 8 | — |
Gains in isolated cases are not represented in the table.
Abbreviation: NS, not significant.
CGH Amplifications in MCL According to Histologic Subtypes
| . | Total Amplifications . | Histologic Subtype . | P Value . | |||
|---|---|---|---|---|---|---|
| Typical (n = 32) . | Blastoid (n = 13) . | |||||
| n . | (%) . | n . | (%) . | |||
| 3q27-q29 | 6 | 3 | 9 | 3 | 23 | NS |
| 18q23 | 4 | 2 | 6 | 2 | 15 | NS |
| Xq28 | 3 | 0 | — | 3 | 23 | — |
| 8q24 | 2 | 1 | 3 | 1 | 8 | — |
| 10p12-p13 | 2 | 0 | — | 2 | 15 | — |
| 2p25 | 2 | 0 | — | 2 | 15 | — |
| 7p22 | 1 | 0 | — | 1 | 8 | — |
| 9q34 | 1 | 0 | — | 1 | 8 | — |
| 13q31-q32 | 1 | 0 | — | 1 | 8 | — |
| 3q24-q26 | 1 | 1 | 3 | 0 | — | — |
| 17q25 | 1 | 1 | 3 | 0 | — | — |
| . | Total Amplifications . | Histologic Subtype . | P Value . | |||
|---|---|---|---|---|---|---|
| Typical (n = 32) . | Blastoid (n = 13) . | |||||
| n . | (%) . | n . | (%) . | |||
| 3q27-q29 | 6 | 3 | 9 | 3 | 23 | NS |
| 18q23 | 4 | 2 | 6 | 2 | 15 | NS |
| Xq28 | 3 | 0 | — | 3 | 23 | — |
| 8q24 | 2 | 1 | 3 | 1 | 8 | — |
| 10p12-p13 | 2 | 0 | — | 2 | 15 | — |
| 2p25 | 2 | 0 | — | 2 | 15 | — |
| 7p22 | 1 | 0 | — | 1 | 8 | — |
| 9q34 | 1 | 0 | — | 1 | 8 | — |
| 13q31-q32 | 1 | 0 | — | 1 | 8 | — |
| 3q24-q26 | 1 | 1 | 3 | 0 | — | — |
| 17q25 | 1 | 1 | 3 | 0 | — | — |
Abbreviation: NS, not significant.
CGH Losses in MCL According to Histologic Subtypes
| . | Total Losses . | Histologic Subtype . | P Value . | |||
|---|---|---|---|---|---|---|
| Typical (n = 32) . | Blastoid (n = 13) . | |||||
| n . | (%) . | n . | (%) . | |||
| 13 | 20 | 10 | 31 | 10 | 77 | NS |
| 6q | 12 | 6 | 19 | 6 | 46 | NS |
| 1p | 11 | 6 | 19 | 5 | 38 | NS |
| 11q14-q23 | 10 | 7 | 22 | 3 | 23 | NS |
| 10p14-p15 | 8 | 5 | 16 | 3 | 23 | NS |
| 17p | 7 | 1 | 3 | 6 | 46 | .0012 |
| 9p | 7 | 3 | 9 | 4 | 31 | NS |
| 15q25-q26 | 4 | 2 | 6 | 2 | 15 | NS |
| Xq11-q27 | 4 | 1 | 3 | 3 | 23 | NS |
| 8p | 3 | 1 | 3 | 2 | 15 | — |
| 15q11-q15 | 3 | 1 | 3 | 2 | 15 | — |
| 3p | 3 | 1 | 3 | 2 | 15 | — |
| 7q | 3 | 2 | 6 | 1 | 8 | — |
| . | Total Losses . | Histologic Subtype . | P Value . | |||
|---|---|---|---|---|---|---|
| Typical (n = 32) . | Blastoid (n = 13) . | |||||
| n . | (%) . | n . | (%) . | |||
| 13 | 20 | 10 | 31 | 10 | 77 | NS |
| 6q | 12 | 6 | 19 | 6 | 46 | NS |
| 1p | 11 | 6 | 19 | 5 | 38 | NS |
| 11q14-q23 | 10 | 7 | 22 | 3 | 23 | NS |
| 10p14-p15 | 8 | 5 | 16 | 3 | 23 | NS |
| 17p | 7 | 1 | 3 | 6 | 46 | .0012 |
| 9p | 7 | 3 | 9 | 4 | 31 | NS |
| 15q25-q26 | 4 | 2 | 6 | 2 | 15 | NS |
| Xq11-q27 | 4 | 1 | 3 | 3 | 23 | NS |
| 8p | 3 | 1 | 3 | 2 | 15 | — |
| 15q11-q15 | 3 | 1 | 3 | 2 | 15 | — |
| 3p | 3 | 1 | 3 | 2 | 15 | — |
| 7q | 3 | 2 | 6 | 1 | 8 | — |
Losses in isolated cases are not represented in the table.
Abbreviation: NS, not significant.
Chromosome imbalances were significantly more frequent in blastoid variants (mean of 10.1 ± 4.89 per case) than in typical tumors (mean of 3.87 ± 3.08 per case; P = .0001). The summary of chromosomal imbalances detected in these patients is shown in Figs 1and 2 and in Tables 1 through 3. Blastoid MCLs had a total of 65 chromosome gains (mean, 5.0 ± 3.27) and 16 high-level DNA amplifications (mean, 1.23 ± 1.25), whereas only 59 gains (mean, 1.84 ± 1.68) and 8 amplifications (mean, 0.25 ± 0.52) were identified in typical cases (P = .02 and P = .015, respectively). Similarly, chromosome losses were also significantly higher in blastoid (mean, 4.3 ± 2.42) than in typical tumors (mean, 1.65 ± 1.77; P = .01). Most chromosome imbalances were similarly distributed in typical and blastoid tumors. However, a number of particular alterations were more frequent in blastoid MCLs. The significant differences between the two variants were gains of 3q, 7p, and 12q and losses of 17p (Tables 1 through 3).
Considering both typical and blastoid MCLs, CGH analysis allowed the delimitation of minimal common regions overrepresented or underrepresented on each of the chromosomes most frequently involved (Figs 1 and 2 and Tables 1 through 3). Twenty-two cases showed gains of 3q, and two consensus regions could be delimited in 3q27-q29 and 3q25 (case 30). The commonly overrepresented region on chromosome 7 was mapped to bands 7p15-p22, with a high-level amplification at 7p22 (case 4). On chromosome 8, 2 cases (cases 4 and 24) with high-level amplifications defined a consensus region in 8q24. For chromosomes 9 and 10, a consensus area of overlap in 9q34 and 10p12-p13, respectively, could be identified. On chromosome 11 and 12, the consensus area was delineated to 11q25 and 12q13, respectively. An amplification (case 6) and two gains (cases 21 and 22) defined the consensus region 13q22-q32. Interestingly, this region was retained in 2 cases (case 3 and 15) with extensive losses of 13q arm (Figs 1 and2). On chromosome 18 and X, the consensus region was 18q23 and Xq28, respectively, with 3 blastoid tumors showing a high-level amplification at Xq28.
The regions frequently involved by loss of genetic material were delineated to 1p21-p22 (cases 11 and 28), 10p14-p15 (cases 2, 5, and 16), 11q21-q22 (cases 14 and 16), and 17p13 (cases 5, 25, and 38). On chromosome 6q, two different areas commonly lost were identified in 6q21-q22 (case 20) and 6q25-q27 (case 12). On chromosome 13, two different areas were also identified: 13q13-q14 (cases 9 and 21) and 13q33-q34 (cases 37 and 38).
Comparison of CGH results with Southern blot analysis.
To identify the possible genes included in abnormal chromosome regions, selected cases were analyzed for P53, P16INK4a, C-MYC, N-MYC, BCL6, BCL2, RB, CDK4, and BMI-1 gene alterations. The results are summarized in Tables 4 and5. The status of the P53 gene was studied by Southern blot and single-stranded conformational polymorphism (SSCP) analysis in the 6 blastoid MCLs with 17p losses. Tumors with an anomalous SSCP pattern were sequenced. Two of these cases showed homozygous deletions of the gene (cases 5 and 15; Table 4 and Fig3A), and the other 4 tumors (cases 1, 2, 3, and 38) had point mutations associated with loss of the remaining allele. Sixteen additional cases with normal chromosome 17 profile were also examined molecularly, and no P53 alterations were observed in any of them. Southern blot analysis confirmed a homozygous deletion of P16INK4a gene in a case with loss of 9p by CGH (case 12). Two cases (cases 2 and 5) with a 25% reduction in the CGH profile of chromosome 9p showed around 20% reduction of theP16INK4a signal by Southern blot. However, CGH did not detect loss of 9p in 2 tumors (cases 4 and 6) in which homozygous deletions of P16INK4a gene were detected by Southern blot. Fifteen additional cases with normal chromosome 9 profile were also examined and noP16INK4a gene alterations were observed (Table 5).
Correlation Between CGH Losses and Gene Deletions/Mutations by Southern Blot
| Chromosomal Region . | Gene Examined . | Cases Studied . | Gene Deletions4-150 . | |
|---|---|---|---|---|
| CGH Loss . | CGH Normal . | |||
| 9p | P16 | 20 | 3/3 | 2/17 |
| 13q | RB | 11 | 0/3 | 0/8 |
| 17p | P53 | 22 | 6/64-151 | 0/16 |
| Chromosomal Region . | Gene Examined . | Cases Studied . | Gene Deletions4-150 . | |
|---|---|---|---|---|
| CGH Loss . | CGH Normal . | |||
| 9p | P16 | 20 | 3/3 | 2/17 |
| 13q | RB | 11 | 0/3 | 0/8 |
| 17p | P53 | 22 | 6/64-151 | 0/16 |
Number of tumors with gene deletion/number of tumors examined by Southern blot. Not all cases examined by CGH could be analyzed molecularly.
Two cases had P53 homozygous deletions and 4 cases had hemyzygous deletions associated with gene mutation.
Correlation Between CGH Gains and High-Level DNA Amplifications and Gene Amplifications by Southern Blot
| Chromosomal Region . | Gene Examined . | Cases Studied . | Gene Amplification/Gain5-150 . | ||
|---|---|---|---|---|---|
| CGH Amplification . | CGH Gain . | CGH Normal . | |||
| 2pter | N-MYC | 12 | 0/2 | 0/1 | 0/9 |
| 3qter | BCL6 | 30 | 0/6 | 0/13 | 0/11 |
| 8qter | C-MYC | 34 | 1/2 | 0/4 | 0/28 |
| 10p12-p13 | BMI-1 | 9 | 2/2 | — | 0/7 |
| 12q13 | CKD4 | 18 | — | 5/6 | 0/12 |
| 18qter | BCL2 | 18 | 0/4 | 0/4 | 0/10 |
| Chromosomal Region . | Gene Examined . | Cases Studied . | Gene Amplification/Gain5-150 . | ||
|---|---|---|---|---|---|
| CGH Amplification . | CGH Gain . | CGH Normal . | |||
| 2pter | N-MYC | 12 | 0/2 | 0/1 | 0/9 |
| 3qter | BCL6 | 30 | 0/6 | 0/13 | 0/11 |
| 8qter | C-MYC | 34 | 1/2 | 0/4 | 0/28 |
| 10p12-p13 | BMI-1 | 9 | 2/2 | — | 0/7 |
| 12q13 | CKD4 | 18 | — | 5/6 | 0/12 |
| 18qter | BCL2 | 18 | 0/4 | 0/4 | 0/10 |
Number of tumors with gene amplification or gains/number of tumors examined by Southern blot. Not all cases examined by CGH could be analyzed molecularly.
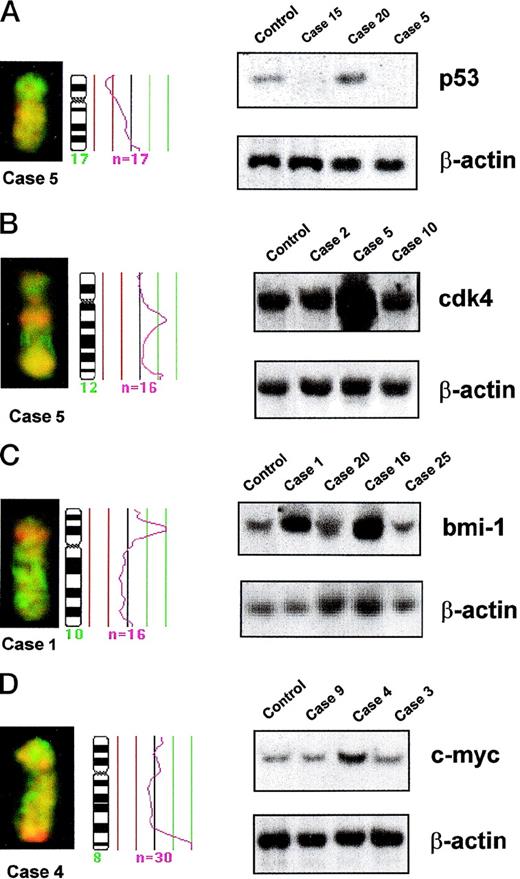
Partial CGH karyotypes, corresponding ratio profiles, and Southern blot analysis of different MCLs. Hybridized tumor DNA was labeled with Spectrum Red-dUTP and control DNA with Spectrum Green-dUTP. On the right side of the ideograms, the average ratios of tumor/normal fluorescence are plotted. The central line indicates a ratio value of 1.0; lines on the left side indicate ratio values of 0.75 and 0.5, respectively; lines on the right side indicate ratio values of 1.25 and 1.5, respectively. n = number of chromosomes analyzed for calculating the respective average ratio profile. β-ACTIN probe was used as loading control in all Southern blots. (A) (Left) Partial CGH karyotype of case 5. A reduction of 17p arm is visible. (Right) Southern blot analysis of DNA of the same case (case 5), another case with a similar chromosome 17 profile (case 15), an additional case with normal chromosome 17 profile (case 20), and DNA from lymphocytes of a healthy control. Both cases with 17p loss by CGH (cases 5 and 15) showed homozygous deletions of P53gene. (B) (Left) Partial CGH karyotype of case 5. A gain of 12q13 is visible. (Right) Southern blot analysis of DNA of the same case (case 5), another case with gain of 12q by CGH (case 2), an additional tumor with normal chromosome 12 profile (case 10), and DNA from lymphocytes of a healthy control. Densitometric evaluation showed an amplification of CDK4 gene in case 5. (C) (Left) Partial CGH karyotype of case 1. An intense, band-like hybridization signal mapping to chromosomal band 10p12-p13 is observed. (Right) Southern blot analysis of DNA of the same case (case 1), another tumor with a similar profile of chromosome 10 (case 16), two additional tumors with normal chromosome 10 profile (cases 20 and 25), and DNA from lymphocytes of a healthy control. Densitometric evaluation showed an amplification ofBMI-1 gene. (D) (Left) Partial CGH karyotype of case 4. An intense, band-like hybridization signal mapping to chromosomal band 8q24 is observed. (Right) Southern blot analysis of DNA of the same case (case 4), two additional tumors with normal chromosome 8 profile (cases 9 and 3), and DNA from lymphocytes of a healthy control. Densitometric evaluation showed an amplification of C-MYC gene.
Partial CGH karyotypes, corresponding ratio profiles, and Southern blot analysis of different MCLs. Hybridized tumor DNA was labeled with Spectrum Red-dUTP and control DNA with Spectrum Green-dUTP. On the right side of the ideograms, the average ratios of tumor/normal fluorescence are plotted. The central line indicates a ratio value of 1.0; lines on the left side indicate ratio values of 0.75 and 0.5, respectively; lines on the right side indicate ratio values of 1.25 and 1.5, respectively. n = number of chromosomes analyzed for calculating the respective average ratio profile. β-ACTIN probe was used as loading control in all Southern blots. (A) (Left) Partial CGH karyotype of case 5. A reduction of 17p arm is visible. (Right) Southern blot analysis of DNA of the same case (case 5), another case with a similar chromosome 17 profile (case 15), an additional case with normal chromosome 17 profile (case 20), and DNA from lymphocytes of a healthy control. Both cases with 17p loss by CGH (cases 5 and 15) showed homozygous deletions of P53gene. (B) (Left) Partial CGH karyotype of case 5. A gain of 12q13 is visible. (Right) Southern blot analysis of DNA of the same case (case 5), another case with gain of 12q by CGH (case 2), an additional tumor with normal chromosome 12 profile (case 10), and DNA from lymphocytes of a healthy control. Densitometric evaluation showed an amplification of CDK4 gene in case 5. (C) (Left) Partial CGH karyotype of case 1. An intense, band-like hybridization signal mapping to chromosomal band 10p12-p13 is observed. (Right) Southern blot analysis of DNA of the same case (case 1), another tumor with a similar profile of chromosome 10 (case 16), two additional tumors with normal chromosome 10 profile (cases 20 and 25), and DNA from lymphocytes of a healthy control. Densitometric evaluation showed an amplification ofBMI-1 gene. (D) (Left) Partial CGH karyotype of case 4. An intense, band-like hybridization signal mapping to chromosomal band 8q24 is observed. (Right) Southern blot analysis of DNA of the same case (case 4), two additional tumors with normal chromosome 8 profile (cases 9 and 3), and DNA from lymphocytes of a healthy control. Densitometric evaluation showed an amplification of C-MYC gene.
Five of the 6 MCL with CGH gains of 12q showed a twofold to eightfold amplification of CDK4 gene, whereas it was in germline configuration in 12 MCL with a normal chromosome 12 CGH profile (Table 5 and Fig 3B). Similarly, Southern blot analysis confirmed a threefold amplification of BMI-1 gene in the two tumors with a high-level DNA amplification of 10p12-p13 (cases 1 and 16), whereas it was in germline configuration in 7 other tumors with normal chromosome 10 CGH profile (Table 5 and Fig 3C). A threefold amplification of the C-MYCgene was detected by Southern blot in one case (case 4), with a high-level DNA amplification at 8qter by CGH (Table 5 and Fig 3D). However, no C-MYC alterations were observed in the other case with 8qter amplification (case 24) or in 4 cases with 8qter gains and 28 cases with normal profiles of chromosome 8.
BCL6 gene was analyzed by Southern blot in 30 MCLs, 19 cases with gains or high-level DNA amplifications involving 3q27, and 11 cases with a normal profile of chromosome 3 in the CGH analysis. No amplifications or rearrangements of BCL6 gene were found in any case. The known HindIII polymorphism was detected in 9% of the tumors, a frequency similar to that described in normal population (13%).30 No N-MYC alterations were found in 3 tumors with 2pter amplification or gains (cases 2, 5, and 7) or in 9 cases with normal chromosome 2. Similarly, BCL2 gene was found in germline configuration in 4 cases with a 18qter high-level DNA amplification, 4 cases with 18q gains, and 10 tumors with a normal profile of chromosome 8. The status of RB gene had been previously examined by Southern blot and immunohistochemistry in 11 and 14 tumors, respectively.31 No alteration of the gene was observed in the Southern blot analysis of 3 cases with loss of chromosome 13 or in 8 tumors with normal chromosome 13. Similar levels of RB protein expression were observed both in 6 cases with loss of chromosome 13 and in 8 tumors with normal chromosome 13 profile.
Clinical significance of CGH imbalances.
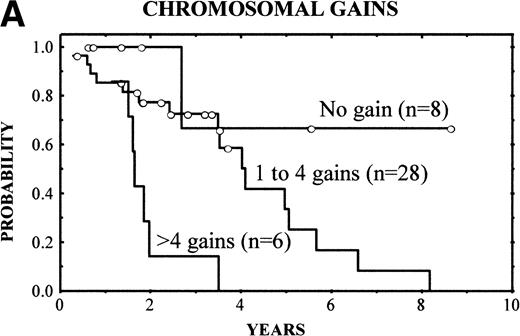
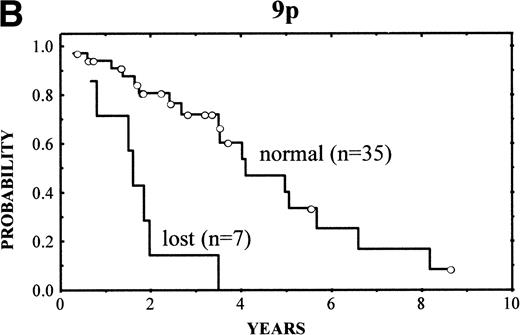
The clinical characteristics of the current series of MCLs were similar to those previously reported4: median age of 62 years (range, 32 to 81 years), male predominance (male/female ratio, 2.7:1), frequent palpable spleen (53%), advanced stage (stage IV, 90%), and extranodal involvement (94%), including bone marrow infiltration in the majority of cases (87%) and high serum LDH levels in 47%. Patients with losses of 9p showed high serum LDH more frequently than the reminders (86% v 36%, respectively; P = .03). No other correlation was found between the clinical and analytical parameters at diagnosis and the above-mentioned genetic alterations. After different treatment approaches (polychemotherapy, 34 cases; monotherapy with alkylating agents, 5 cases; other, 3 cases), the complete response (CR) rate was 17%. No significant differences were found in the CR rate according to the presence of the genetic lesions. Clinical follow-up was available in 42 patients. The overall survival (OS) was 3.5 years. Median OS for patients with typical histology was 4.8 years, whereas it was 1.6 years for those with blastoid variants (P = .005). A number of CGH alterations were associated with a poorer prognosis. The presence of chromosome gains (0 v 1-4v >4 gains per case) was related to a poor overall survival (4-year OS: 68%, 58%, and 0%, respectively; P = 0.02; Fig4A). When the analysis was restricted to the 29 patients with typical variants, the presence of chromosome gains (0 gains v 1-4 gains per case v >4 gains per case) still retained prognostic significance (P = .03). Moreover, gains of 3q (patients with normal 3q v patients with gains of 3q; 4-year OS: 63% v 34%; P = .02), gains of 12q (normal 12q v gains of 12q; 4-year OS: 55% v18%; P = .03), and losses of 9p (normal 9p v loss of 9p; 4-year OS: 61% v 0%; P = .003) (Fig 4B) were associated with a significant shorter survival.
(A) (Left) Survival curves of patients with MCL according to increased number of gains (0 v 1-4 v >4 gains per case;P = .02). (B) (Right) Survival curves of patients with MCL according to losses of 9p (normal 9p v loss of 9p;P = .003).
(A) (Left) Survival curves of patients with MCL according to increased number of gains (0 v 1-4 v >4 gains per case;P = .02). (B) (Right) Survival curves of patients with MCL according to losses of 9p (normal 9p v loss of 9p;P = .003).
DISCUSSION
In the present study, we have identified a high number of chromosomal imbalances and DNA amplifications in MCLs. The most recurrent alterations observed in our series were gains of 3q, 7p, 8q, 12q, 18q, and 9q34 and losses of chromosome 13, 6q, 1p, 11q14-q23, 10p14-p15, 17p, and 9p, as well as amplification in 11 different sites. Previous cytogenetic studies had identified different secondary alterations in MCLs, including gains and trisomies of chromosomes 3, 7, 12, and 18; losses of 6q, 1p, 11q, and 13q12-q14; and monosomies 9, 13, and 17.15-19 Our study confirms these chromosomes as recurrent targets in MCLs, but the frequency of the alterations is higher in the CGH analysis than in conventional cytogenetic studies. In addition, we have observed certain imbalances, such as gains of 3q and 8q and losses of 9p, not previously recognized by classic cytogenetics. A recent CGH study on MCLs showed similar chromosomal alterations in these tumors, although the number of high-level DNA amplifications was lower.32
Most of the individual CGH alterations detected in MCLs have already been observed in other non-Hodgkin’s lymphomas (NHLs). However, the global profile and frequency of these imbalances seem to be relatively characteristic of MCLs. Trisomy 3 and 3q gains have been recently found by CGH in 52% of marginal zone lymphomas. However, alterations in 1p, 7p, 11q, 12q, and 13 were absent or very rare in these tumors.33 Gains of chromosome 12 have been detected in primary mediastinal B-cell lymphomas (31%), primary gastrointestinal large-cell lymphomas (23%), and chronic lymphocytic leukemias (CLL; 16%). However, other genetic alterations in these tumors, such as gains of 9p (50%) and Xq (31%) in mediastinal lymphomas and gains of 11q (25%) in gastrointestinal lymphomas, were different from the abnormalities found in MCLs.34-38 In addition, CLL and gastrointestinal tumors showed preferentially trisomy 12 and the most represented band in mediastinal lymphomas was 12q24, whereas in MCLs the consensus region was localized at 12q13. Similarly, the global CGH alterations in follicular lymphomas,39,40 nodal and extranodal large-cell lymphomas,41-43 and multiple myeloma44 differ from the pattern observed in MCLs.
Imbalances in the long arm of chromosome 13 were particularly frequent in MCLs. Deletions in 13q13-q14 are one of the most frequent structural alterations in CLL.45 Molecular studies have suggested the existence of a novel tumor-suppressor gene in this region, distal toRB gene, probably involved in the pathogenesis of CLL.46 Interestingly, Stilgenbauer et al47 have recently identified that the chromosomal region 13q14, commonly lost in CLL, is also deleted in 70% of MCLs. Our CGH study, delineating a consensus region in 13q13-q14 that is frequently lost in MCLs, is consistent with these molecular findings and supports the idea that deletions of this region may be important in the development of MCL as well as in CLL. Other frequent alterations of the long arm of chromosome 13 were found in our study, particularly deletions of 13q33-q34 and gains of 13q22-q32. The potential target genes in these regions are not known.
Cytogenetic alterations associated with gene amplification such as double minute chromosomes and homogeneously staining regions are relatively rare in NHLs.48 However, CGH studies are detecting an increased number of high-level DNA amplifications in these tumors, suggesting that gene amplification may be more frequent than initially thought.49 In this sense, we found 24 high-level DNA amplifications in 16 MCLs involving 11 different regions. Most of these amplifications have been observed in other NHLs by CGH. However, amplifications in 7p22 and 17q23-q25 have not been previously described. The amplifications at Xq28, 13q31-q32, and 10p12-p13 have only been detected in three MCLs32,49 and a large cell lymphoma.43
Interestingly, we observed a close relationship between high-level DNA amplifications and the location of chromosome fragile sites. In fact, 8 of the 11 amplified regions were localized in chromosomal regions where known fragile sites have been identified. This association between amplified regions and fragile sites has not been recognized in previous CGH studies in NHLs. In a recent review of the literature,50 it can be observed that high-level DNA amplifications in lymphomas have been found in 27 different chromosomal regions. Comparing these amplification sites with the location of fragile sites, 19 (70%) of the amplified regions were indeed associated with known fragile sites, suggesting that this relationship may be a general phenomenon. In vitro studies have recently shown that activation of fragile sites play an important role in the amplification of chromosomal units by initiating breakage-fusion-bridge cycles and determining the size of the amplified region. The amplicons at 11q13 and 12q13-q14, which include known human oncogenes, are also associated with fragile sites.51 Similarly, our findings in this CGH study and the review of the CGH literature in NHLs expand these observations and support the hypothesis that fragile sites may be implicated in the amplification of certain chromosomal regions during tumor progression.
Molecular studies were performed to correlate the CGH analysis with the potential alterations of different genes. 17p losses were strongly associated with P53 gene alterations in our series. Inactivation of P53 gene by mutations and hemizygous deletions is a well-known mechanism in blastoid MCLs and other NHLs and is associated with 17p abnormalities.11,52 However,P53 homozygous deletions, as in the 2 cases observed in this study, are extremely rare in NHLs, although they have been described in solid tumors.52,53 9p losses were associated withP16INK4a homozygous deletion in 1 case and 20% reduction in the Southern signal in 2 additional cases. These findings are consistent with the previous findings ofP16INK4a deletions in MCLs that may be either homozygous or hemizygous and may only be present in a subpopulation of tumor cells.13 14
The other genes analyzed in this study have not been previously examined in MCLs. Interestingly, we have observed CDK4 gene amplification in tumors with 12q gains. CDK4 has been mapped to 12q13, a chromosomal region frequently amplified in different tumors, particularly gliomas and sarcomas.50,54,55 This chromosomal band also contains GLI and MDM2 genes that may also be coamplified with CDK4. Amplification of these genes has been recently observed in diffuse large B-cell lymphomas,56 and they were associated with advanced stage disease. Our results in MCLs suggest that CDK4 amplification may also be involved in the pathogenesis of these tumors. BMI-1 is an oncogene that participates in murine lymphomagenesis, probably cooperating with c-myc.27 57BMI-1 mRNA expression has been detected in a human Burkitt’s lymphoma cell line. BMI-1 has been mapped to 10p13, a chromosomal region that was found to be amplified in 2 blastoid MCL in our study. Southern blot analysis of these cases showed that BMI-1 was amplified in these cases, but it was in germline configuration in other 7 tumors with normal chromosome 10p profile. These findings suggest that BMI-1 may be a target of this amplification and it may also play a role in these tumors.
Transgenic animal models have indicated that C-MYC activation cooperates with cyclin D1 in lymphomagenesis.10 However, the role of C-MYC in human MCLs is not known. In this study, we have detected a C-MYC amplification in 1 case with a high-level DNA amplification at 8qter, but it was in germline configuration in other 10 tumors, including 4 cases with gains of 8qter. No alterations of N-MYC, BCL6, RB, and BCL2 genes were detected in different number of tumors, including cases with chromosomal alterations in 2p, 3q, 13q, and 18q, suggesting that these genes do not play a relevant role in the pathogenesis of MCLs and that other genes may be the targets of these genetic abnormalities in these lymphomas.
Blastoid MCLs have a more aggressive behavior than typical variants.4 We have now demonstrated that blastoid MCLs have a higher number of chromosomal imbalances and amplifications than typical variants. In addition, specific alterations, including gains of 3q, 7p, and 12q and losses of 17p, were significantly more frequent in blastoid tumors, suggesting that these alterations could be important events in the progression of MCLs. Monni et al32 had also observed a higher number of changes in blastoid variants. However, no significant differences could be detected between blastoid and typical cases, probably because of the small number of cases included in their study.32 The association between 17p and 9p losses and blastoid MCLs is concordant with previous molecular findings ofP53 and P16INK4a gene alterations in aggressive MCLs and other NHLs.11-14,58,59Deletions in 6q and gains of chromosome 7 and 12 have been associated with aggressive histologies in NHLs.16,43,60 Interestingly, gains of 11q and 12q have also been recently associated with aggressive behavior in gastrointestinal large-cell lymphomas.34 These findings support the idea that, in contrast with the association between primary genetic alterations and specific lymphomas, certain secondary genetic events involved in aggressive variants may be similar in different types of lymphomas.16
In this study, we have also examined the clinical significance of CGH alterations in MCLs. In agreement with other cytogenetic studies in NHLs,34,60 the complexity of the genetic alterations, and particularly the number of gains, were significantly associated with a shortened median survival. Interestingly, the prognostic significance of the chromosome gains was also found when the analysis was restricted to the patients with typical variants. Furthermore, gains of 3q and 12q and losses of 9p were associated with poor prognosis. Structural and numerical alterations on chromosome 3q have been associated with more aggressive subtypes of NHLs.61,62 Although BCL6 is located in 3q27, our results indicate that this gene does not seem to play an important role in MCLs. The prognostic significance of 9p losses is concordant with the association betweenP16INK4a deletion and a worse prognosis in NHLs.63
In conclusion, we have demonstrated that MCLs have a high number of chromosomal alterations and DNA amplifications. The pattern of chromosomal aberrations in these tumors seems to be relatively different from the profile detected in other lymphomas. High-level DNA amplifications were closely associated with fragile sites, which support the idea that these structures may participate in chromosomal amplifications. The significant differences in some alterations between blastoid and typical variants suggest that they may be involved in the pathogenesis of these aggressive variants. In addition, our findings indicate that certain imbalances detected by CGH may be of prognostic significance in MCLs.
ACKNOWLEDGMENT
The authors thank Iracema Nayach and Nerea Peiró for their excellent technical assistance and Eva Cid for her linguistic advice.
Supported by Grant No. SAF 99/20 from CICYT, Grant No. SAF 96/177, Maratón-TV3 Cáncer, Asociación Española contra el Cáncer, and Generalitat de Catalunya SGR52/96. S.B., M.P., and L.H. were fellows supported by Spanish Ministerio de Educación y Cultura (S.B.), Maratón-TV3 Cáncer (M.P.), and Fundació Rius i Virgili (L.H.).
S.B. and M.R. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Elı́as Campo, MD, Laboratory of Pathology, Hospital Clı́nic, Villarroel 170, 08036-Barcelona, Spain; e-mail: campo@medicina.ub.es.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal