Abstract
The receptor for hyaluronan (HA)-mediated motility (RHAMM) controls motility by malignant cells in myeloma and is abnormally expressed on the surface of most malignant B and plasma cells in blood or bone marrow (BM) of patients with multiple myeloma (MM). RHAMM cDNA was cloned and sequenced from the malignant B and plasma cells comprising the myeloma B lineage hierarchy. Three distinct RHAMM gene products, RHAMMFL, RHAMM−48, and RHAMM−147, were cloned from MM B and plasma cells. RHAMMFL was 99% homologous to the published sequence of RHAMM. RHAMM−48 and RHAMM−147 variants align with RHAMMFL, but are characterized by sequence deletions of 48 bp (16 amino acids [aa]) and 147 bp (49 aa), respectively. The relative frequency of these RHAMM transcripts in MM plasma cells was determined by cloning of reverse-transcriptase polymerase chain reaction (RT-PCR) products amplified from MM plasma cells. Of 115 randomly picked clones, 49% were RHAMMFL, 47% were RHAMM−48, and 4% were RHAMM−147. All of the detected RHAMM variants contain exon 4, which is alternatively spliced in murine RHAMM, and had only a single copy of the exon 8 repeat sequence detected in murine RHAMM. RT-PCR analysis of sorted blood or BM cells from 22 MM patients showed that overexpression of RHAMM variants is characteristic of MM B cells and BM plasma cells in all patients tested. RHAMM also appeared to be overexpressed in B lymphoma and B-chronic lymphocytic leukemia (CLL) cells. In B cells from normal donors, RHAMMFL was only weakly detectable in resting B cells from five of eight normal donors or in chronically activated B cells from three patients with Crohn’s disease. RHAMM−48 was detectable in B cells from one of eight normal donors, but was undetectable in B cells of three donors with Crohn’s disease. RHAMM−147 was undetectable in normal and Crohn’s disease B cells. In situ RT-PCR was used to determine the number of individual cells with aggregate RHAMM transcripts. For six patients, 29% of BM plasma cells and 12% of MM B cells had detectable RHAMM transcripts, while for five normal donors, only 1.2% of B cells expressed RHAMM transcripts. This work suggests that RHAMMFL, RHAMM−48, and RHAMM−147 splice variants are overexpressed in MM and other B lymphocyte malignancies relative to resting or in vivo–activated B cells, raising the possibility that RHAMM and its variants may contribute to the malignant process in B-cell malignancies such as lymphoma, CLL, and MM.
MULTIPLE MYELOMA (MM) is an incurable cancer characterized by the presence of monoclonal Ig in the blood, lytic bone lesions, and monoclonal plasma cells in the bone marrow (BM). Median survival is approximately 3 years. Compared with its incidence, MM accounts for a disproportionate number of the deaths from hematologic malignancies. Although many patients achieve initial clinical remission, nearly all relapse and become refractory to treatment.1,2 The molecular basis of the malignancy is unknown, and no consistent oncogenic or genetic abnormality characterizes all myeloma patients. Although clinically MM is viewed as a disease restricted to the BM, molecular studies confirm it to be a systemic malignancy that includes peripheral blood. Many groups, including ours, have identified clonotypic IgH VDJ among peripheral blood mononuclear cells (PBMC) of MM patients,3-11 or purified B cells from MM PBMC.6,12-14 We have identified and verified the IgH VDJ clonal sequences that characterize each MM patient, and have shown that clonotypic B cells are frequent in the circulation of myeloma patients (0.16 × 109/L blood).12-14 These B cells are late-stage, drug-resistant cells with stem-cell–like properties.6,12-18 The majority of B cells in MM PBMC are surface Ig+ (sIg), express pan-B cell markers (CD19, CD20, CD24) and by a variety of measures appear to be in an activated late stage of B-cell differentiation.13,16,19,20 They express the stem-cell marker CD34,12 and a large constellation of receptors involved in adhesion to endothelium or extracellular matrix (ECM) and in motility.13,17,20-23 Morphologically and functionally, these B cells are distinct from plasma cells.17,24,25 Both MM B and plasma cells are characterized by their expression of the receptor for hyaluronan (HA)-mediated motility (RHAMM), but only circulating B cells are motile.17,22 24 The heterogeneity in phenotype and morphology seen among these clonotypic B cells, including resting B cells, lymphoblasts, and more differentiated phenotypes, is suggestive of sequentially related cell types resulting from continuous stimulation.
Interactions between HA and RHAMM are required for motile behavior of a wide variety of cells, including sperm, fibroblasts, astrocytes, microglia, and white blood cells.17,22,26-30Binding of HA by RHAMM triggers signal transduction, dissolution of focal adhesions, and motility, and is involved in cell cycle control.24,26,31-34 RHAMM is a glycophosphatidylinositol (GPI)-anchored receptor, lacking both a signal sequence and a transmembrane domain, that is alternatively spliced to yield soluble, cytoplasmic, and surface receptor isoforms.26,27 RHAMM mediates motility, is itself oncogenic when overexpressed, and is essential for ras-mediated transformation.27Activated normal lymphocytes transiently express a low level of RHAMM.22,28 In MM, RHAMM on B and leukemic plasma cells binds HA and mediates HA-dependent motility.17 Although nonmotile BM-anchored MM plasma cells also express RHAMM, its function on these cells is as yet unknown. The relatively abundant RHAMM on most B and plasma cells in MM, as well as its apparently constitutive functional properties,17 suggests it may be deregulated or overexpressed, and thus implicated in the malignant process.
In this study, we have characterized the RHAMM expression of malignant B-lineage cells in myeloma by sequencing and mapping the expression patterns of RHAMM transcripts in MM B and plasma cells from patients as compared with malignant B cells in lymphoma and chronic lymphocytic leukemia (CLL) and B cells from normal donors or donors with nonmalignant but activated B cells (Crohn’s disease). We find that three distinct RHAMM transcripts are detectable ex vivo in MM B and plasma cells, lymphoma cells, and CLL cells, two of which are novel deletion splice variants. All three forms of RHAMM are detectably overexpressed in malignant B-lineage cells as compared with resting or activated normal B cells.
MATERIALS AND METHODS
Patients.
Peripheral blood was obtained from 101 patients with MM, three patients with CLL, eight healthy normal volunteers, and three patients with Crohn’s disease after informed consent; BM was obtained from 11 patients with MM. Malignant lymphoma nodes from three patients were pathology specimens obtained after surgery.
Antibodies and reagents.
B4-FITC (CD19) was from Coulter (Hialeah, FL). FMC63 (CD19) was from Dr H. Zola (Adelaide Women’s and Children’s Hospital, Australia)35 36 and was conjugated to fluorescein isothiocyanate (FITC). Anti-CD38 (Leu-17–phycoerythrin [PE]) was purchased from Becton Dickinson (San Jose, CA). Goat antimouse Ig conjugated to FITC or PE and antihuman Ig (goat antihuman Ig [H+L]-FITC) was from Southern Biotechnology (Birmingham, AL). RHAMM monoclonal antibody (mAb) 3T3.5 was from Dr Eva Turley (Hospital for Sick Children University of Toronto, Toronto, Canada).
Tissue and cell preparations.
All cell preparations were used immediately ex vivo. Peripheral blood or BM cells from MM and CLL patients and normal donors or donors with Crohn’s disease were collected into heparinized vacutainer tubes, centrifuged over a Ficoll-hypaque gradient, washed, and resuspended in Trizol (GIBCO-BRL/Life Technologies, Burlington, Ontario, Canada). Lymphoma nodes were minced to prepare single-cell suspensions, pelleted, and resuspended in Trizol. Only nodes with greater than 50% B cells among lymph node mononuclear cells were used for this study. Our previous work indicated that only B cells in nodes from lymphoma patients expressed surface RHAMM, and that RHAMM was not expressed by normal B or T cells from lymph node.22 PBMC from B-CLL samples included greater than 98% B cells.
Immunofluorescence and sorting.
For phenotypic analysis, MM PBMC or BM cells, or normal PBMC, were stained in two-color immunofluorescence with mAb to CD19 (direct conjugate) and to RHAMM or an IgG1 isotype-matched control (indirect staining) followed by washing, blocking with mouse gamma-globulin, and detection with a goat antimouse Ig fluorescent conjugate, as previously described.6,17 MM B cells are CD19+CD20med/hi,6 and B cells from normal donors are CD19+CD20hi. BM plasma cells in MM are CD19loCD20−/lo. Identical staining patterns were obtained with either B4 or FMC63. Samples were analyzed by FACSsort (Becton Dickinson), collecting 10,000 to 20,000 events gated to exclude RBCs and dead cells. Files were analyzed by gating for CD19+ and plotting the staining of RHAMM or IgG1. Staining was considered positive only if it exceeded that of the isotype control. All B cells from myeloma PBMC (termed MM B cells) express CD19 and IgH mRNA.12 MM B cells express IgH transcripts identical to those of autologous BM plasma cells, confirming their membership within the malignant clone in MM.12-14
For sorting, mononuclear cells were collected from the interface, washed twice in cold phosphate-buffered saline (PBS), resuspended in PBS/10% fetal calf serum (FCS) at 2 × 106 cells/mL, stained with anti-CD19, or anti-CD38 plus antihuman Ig at 4°C for 30 to 60 minutes, and washed twice with cold PBS containing 0.06 g/L EDTA to inhibit cell aggregation. Mononuclear cells were sorted on a Coulter Elite flow cytometer. B cells from normal donors and patients with Crohn’s disease were sorted as CD19+ cells gated for low forward and side scatter to exclude monocytes. B cells from MM PBMC were sorted as CD19+ cells with no gates set on scatter beyond those excluding RBCs and dead cells, as previously described.12-14 For MM PBMC, unlike normal or Crohn’s PBMC, nearly all cells within the monocytoid scatter gates are B cells, as confirmed by their by their IgH and CD19 transcripts, as well as by their phenotypic profile.12-14 BM plasma cells were sorted as CD38hi, Ig+ cells; nearly all of the plasma cells within these gates expressed the clonal marker sequence unique to each MM patient,12-14 confirming selection of malignant cells. Sort gates were set to include cells staining brighter than isotype controls. Reanalysis of sorted samples gave a purity of 97% or better. Cells were sorted into PBS plus 1% FCS, washed, resuspended in Trizol, and stored at −80°C.
Reverse-transcriptase polymerase chain reaction.
RNA was extracted using Trizol, resuspended in diethyl pyrocarbonate (DEPC)-treated water, and stored at −80°C. Next, total RNA (500 ng or 1 μg) was denatured at 70°C for 10 minutes, annealed to 0.5 μmol/L oligo dT15, and reverse-transcribed with 0.5 mmol/L deoxyribonucleoside triphosphates (dNTPs), 0.01 mol/L dithiothreitol (DTT), 200 U Superscript (GIBCO/BRL), and 1× reverse transcriptase (RT) buffer (GIBCO BRL) in a volume of 20 μL at 42°C for 60 minutes, and heat-inactivated at 99°C for 3 minutes. The cDNA template (1 to 4 μL) was amplified in a 50-μL polymerase chain reaction (PCR) mix (Boehringer Mannheim, Laval, Quebec, Canada; BMH) containing 1× PCR buffer, 0.2 mmol/L dNTPs, 0.4 μmol/L upstream and downstream primers, and 1U Taq DNA polymerase. The PCR cycling parameters were denaturation for 5 minutes at 94°C, followed by 35 cycles of denaturation for 1 minute at 94°C, annealing for 30 seconds, and extension at 72°C for 2 minutes, with a final extension period of 7 minutes at 72°C. Primer annealing temperature (TA) was calculated as TA= 4 × (G + C) + 2 × (A + T) − 5.
The primers used to amplify RHAMM were as follows: (1) 5′ GGCCGTCAACATGTCCTTTCCTA, 3′ TTGGGCTATTTTCCCTTGAGACTC; (2) 5′ CAGGTCACCCAAAGGAGTCTCG, 3′ CAAGCTCATCCAGTGTTTGC; (3) 5′ GCAAACACTGGATGAGCTTG, 3′ TTGCCTTCTTTTAATGGGGTC; (4) 5′ AGGAGGAACAAGCTGAAAGG, 3′ TTCCTGAGCTGCACCATGTT; (5) 5′ GGCCGTCAACATGTCCTTTCCTA, 3′ ACAGCAACATCAATAACAACAAGA; (6) 5′ GAGAATTCTAAGCTTGGAGTTG, 3′ CAAGCTCATCCAGTGTTTGC; (7) 5′ TCCTAGAAGAAAAGCTGAAAGGGAA, 3′ CTTGGCCGCTTTTTCCTGTAATGA; and (8) 5′ GTTTCTGGAGCTGGCCGTC, 3′ ACTGGTCCTTTCAATACTTCTAAAGT.
Cloning and sequencing.
The PCR products were ligated into the PCR TM II or PCR TM2.1 vector and transformed into One ShotTM competent cells with the TA cloning kit according to the manufacturer’s instructions (Invitrogen, San Diego, CA). Single bacterial colonies were introduced into a 25-μL PCR reaction mix containing 1× PCR buffer, 0.2 mol/L dNTPs, 0.4 μmol/L upstream and downstream primers, and 0.5 U Taq DNA polymerase. The PCR cycling parameters were bacterial lysis for 10 minutes at 94°C, followed by 25 cycles of denaturation for 1 minute at 94°C, annealing at TA for 30 seconds, and extension at 72°C for 1 minute, with a final extension period of 7 minutes at 72°C. The amplified products were analyzed by agarose gel electrophoresis. The cloned inserts were sequenced with the ABI PRISM TM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer, Applied Biosystems, Mississauga, Ontario, Canada) with Ampli Taq DNA polymerase on a Perkin Elmer 373 A DNA Sequencer.
Amplification and detection of RHAMM isoforms.
Total RNA (500 ng or 1 μg) was prepared and reverse-transcribed as described earlier. To detect RHAMMFL and RHAMM−48, 5 μL of cDNA was amplified with RHAMM primer set 1 (described earlier).
To ensure mRNA integrity, 1 μL of cDNA was amplified with histone primers: 5′ CCACTGAACTTCTGATTCGC, 3′ GCGTGCTAGCTGGATGTCTT; or 5 μL of cDNA was amplified with CD19 primers: 5′ TACTATGGCACTGGCTGCTG, 3′ CACGTTCCCGTACTGGTTCT.
The amplified products were analyzed on a 1% agarose, ethidium bromide, 50 mmol/L Tris, 45 mmol/L boric acid, 0.5 mmol/L EDTA, pH 8.3 (0.5× TBE) gel.
To detect RHAMM−147, 5 μL of cDNA was amplified with RHAMM primer set 3 (described earlier), electrophoresed on a 1% agarose gel, and transferred overnight onto Hybond-N nylon membrane (Amersham Canada Ltd, Oakville, Ontario, Canada). The membrane was prehybridized in 5× SSC, 0.1%N-lauroylsarcosine, 0.2% sodium dodecyl sulfate (SDS), and 2% blocking reagent (BMH) for 1 hour at 65°C and then hybridized to a 20-ng/mL digoxigenin (DIG)-labeled RHAMM probe overnight at 65°C in prehybridization solution. The probe template was amplified from BM plasma-cell cDNA with RHAMM primer set 9: 5′ GAAAAGTAGTGCTGCTCATACC, 3′ GGTCTGCAGATCTAGAAGCATC; and labeled with DIG-11-dUTP with a DIG DNA-labeling kit (BMH) according to the manufacturer’s instructions. After hybridization, the membrane was washed once in 2× SSC, 0.1% SDS for 10 minutes at room temperature, twice in 0.1× SSC, 0.1% SDS for 15 minutes at 65°C, blocked with 2% blocking reagent for 30 minutes at room temperature, incubated with 1:5,000 anti-DIG Fab alkaline phosphatase/2% blocking solution for 30 minutes at room temperature, washed twice in 0.1 mol/L maleic acid, 0.1 mol/L NaCl, 0.3% Tween-20 for 15 minutes at room temperature and equilibrated in 0.1 mol/L Tris-HCl, 0.15 mol/L NaCl pH 9. Amplified RHAMM was detected by incubating the membrane with 4.5 μg/mL 4-Nitro blue tetrazolium chloride (NBT), 1.7 μg/mL 5-bromo-4-chloro-3-indolyl-phosphate (BCIP; BMH) in 0.1 mol/L Tris-HCl, 0.15 mol/L NaCl, 0.05 mol/L MgCl2 pH 9 at room temperature and monitoring color development or by overlaying the membrane with Lumi-phos 530 chemiluminescence substrate sheet (Schleicher & Schuell, Keene, NH) and exposure to Hyperfilm-ECL (Amersham).
Comparison of primer efficiency.
The open-reading frame (ORF) of each cloned RHAMM isoform was restriction-digested from the PCR TM II vector with EcoRV plusKpn I, electrophoresed on a low-melting-point agarose gel, and purified with β-Agarase (New England Biolabs, Beverly, MA) according to the manufacturer’s instructions. RHAMMFLand RHAMM−48 (100 pg, 10 pg, 1 pg) were amplified with primer set 1, and the products analyzed by electrophoresis on an ethidium bromide–stained agarose gel. RHAMMFL and RHAMM−147 (100 pg, 10 pg, 1 pg) were amplified with primer set 3, and the products analyzed by electrophoresis, transfer to a nylon membrane, hybridization to a DIG-labeled RHAMM probe, and chemiluminescent detection as described earlier.
In situ RT-PCR.
In situ RT-PCR was as previously described.12 13 Briefly, normal B cells, MM B cells, and BM plasma cells were purified and stained as described earlier. The cells were fixed in 10% formalin/PBS overnight and sorted onto In Situ PCR glass slides (Perkin Elmer) at 10,000 cells/spot. Each slide had three spots that included a negative control, which was not reverse-transcribed to confirm digestion of genomic DNA, a positive control with intact genomic DNA, and a test spot. The slides were air-dried, permeabilized with 2 mg/mL pepsin/0.01N HCl (BMH) at times optimal for each cell type, normal B cells for 45 minutes, MM B cells for 40 minutes, and MM BM plasma cells for 35 minutes, followed by a 1-minute wash in DEPC-treated water and then 1 minute in 95% ethanol. The negative control and test spot were digested with 1,000 U/mL DNAse I (RNAse-free; BMH) overnight at 37°C followed by a water and ethanol wash. The test sample was reverse transcribed with Superscript according to the manufacturer’s recommendations followed by a water and ethanol wash. Amplification was performed on the negative, positive and test spots with an In Situ Core Kit (Perkin Elmer) with direct incorporation of DIG-11-dUTP (BMH). Cycling parameters were denaturation for 5 minutes at 94°C, followed by 25 cycles of denaturation for 1.5 minutes at 94°C, annealing for 1.5 minutes at 65°C, and extension for 2 minutes at 72°C, with a final extension period of 7 minutes at 72°C. The primers used to amplify RHAMM were primer set 10: 5′ ATCACAAAGATTTAAACAACAAAAAGAAT, 3′ CTTCCATCTTTTCCAACTCAGTTTC.
The slides were washed in 2× SSC (20× SSC = 3 mol/L sodium chloride, 0.3 mol/L sodium citrate, pH 7) for 5 minutes at room temperature, blocked with 0.1× SCC/0.2% bovine serum albumin (BSA) for 5 minutes at 45°C, and equilibrated for 10 minutes in 0.1 mol/L Tris-HCl, 0.15 mol/L NaCl pH 7.5. Amplified DNA was detected by incubation with 1:200 anti-DIG Fab alkaline phosphatase (BMH) for 30 minutes at room temperature. The slides were washed in 0.1 mol/L Tris-HCl, 0.15 mol/L NaCl, 0.05 mol/L MgCl2 pH 9.5 for 5 minutes, and incubated with 0.2 mg/mL NBT, 0.1 mg/mL BCIP at room temperature for up to 1 hour. Color development was monitored under the microscope. The reaction was stopped by washing in water.
RESULTS
RHAMM is expressed by members of the B-lineage hierarchy in myeloma.
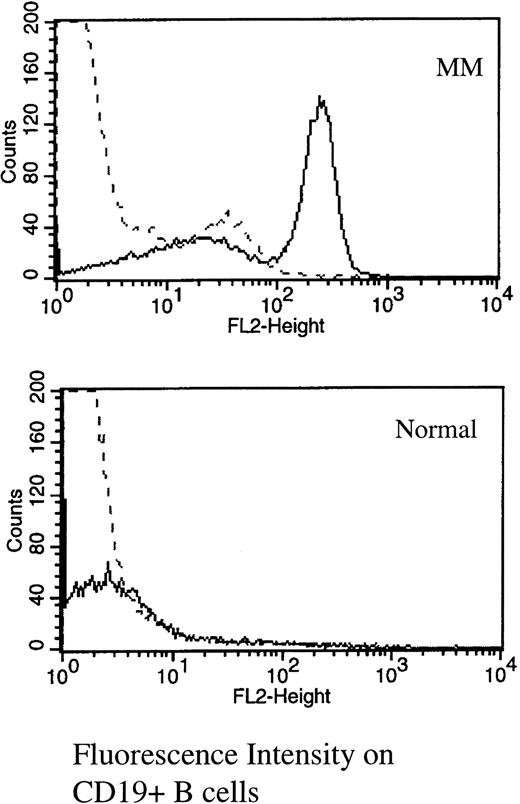
Multicolor immunofluorescence was used to determine the expression of RHAMM on MM B and plasma cells (Table 1 and Fig 1). The majority of MM plasma cells resident in the BM express RHAMM (mean, 66% ± 9%). The MM clone also includes circulating B cells expressing IgH transcripts identical to those of autologous BM plasma cells.12-14 Of circulating CD19+ MM B cells, 56% ± 5% expressed RHAMM (Table 1and Fig 1). In contrast, of B cells from healthy donors, only 3.5% expressed RHAMM, possibly representing the minority of normal circulating B cells in the early stages of activation in vivo.22
Expression of RHAMM on B-Lineage Cells in MM
| Cell Type (no. of patients) . | % Expressing RHAMM . | ||
|---|---|---|---|
| 25th Percentile . | Median . | 75th Percentile . | |
| MM | |||
| BM plasma cells (15)* | 41 | 74† | 93 |
| Blood B cells (90) | 28 | 57† | 92 |
| Normal donors | |||
| B cells (12) | 1 | 3.5 | 9 |
| Cell Type (no. of patients) . | % Expressing RHAMM . | ||
|---|---|---|---|
| 25th Percentile . | Median . | 75th Percentile . | |
| MM | |||
| BM plasma cells (15)* | 41 | 74† | 93 |
| Blood B cells (90) | 28 | 57† | 92 |
| Normal donors | |||
| B cells (12) | 1 | 3.5 | 9 |
PBMC or BMC were stained in two color immunofluorescence as indicated in Materials and Methods. Files were gated for B or plasma cells and the expression of RHAMM plotted as a histogram (as in Fig1), in comparison to identically gated isotype-matched control staining.
From Turley et al.22
P < .001 as compared with normal B cells.
RHAMM is expressed on the majority of MM blood B cells, but is absent from most normal B cells. PBMC from MM or normal donors were stained with B4-FITC (CD19) and RHAMM or an isotype-matched IgG1 control (indirect immunofluorescence with a second-stage goat antimouse Ig-PE). Files were gated for B cells and the expression of RHAMM plotted as a histogram. (—) RHAMM staining; (—-) isotype control staining.
RHAMM is expressed on the majority of MM blood B cells, but is absent from most normal B cells. PBMC from MM or normal donors were stained with B4-FITC (CD19) and RHAMM or an isotype-matched IgG1 control (indirect immunofluorescence with a second-stage goat antimouse Ig-PE). Files were gated for B cells and the expression of RHAMM plotted as a histogram. (—) RHAMM staining; (—-) isotype control staining.
Sequence analysis of RHAMM gene products.
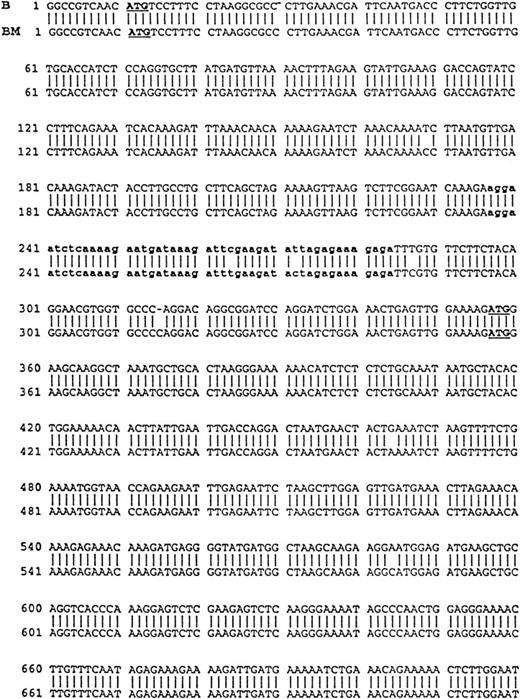
Using cDNA pools from the BM plasma cells of two MM patients and from the B cells of six MM patients, RHAMM cDNA fragments were amplified by RT-PCR with RHAMM-specific primers (Fig 2). The amplified cDNA fragments were cloned and colonies screened by PCR with the appropriate RHAMM primers to identify those containing potential RHAMM cDNA inserts. Each cloned insert was sequenced and identified as a RHAMM cDNA fragment by alignment with the published sequence of human breast RHAMM.37 Alignment of RHAMM cDNA fragments produced three distinct RHAMM gene products from B cells and from BM plasma cells designated RHAMMFL, RHAMM−48, and RHAMM−147 (Fig3). B-cell and BM plasma cell RHAMM was 99% homologous to breast RHAMM. The two start codons, stop codon, and the two HA-binding domains, previously described for RHAMM,32 37 were all present in the aligned cDNA fragments cloned from MM B and plasma cells. RHAMM−48 and RHAMM−147 align with RHAMM, but have sequence deletions of 48 bp and 147 bp, respectively. The 48-bp deletion that characterizes RHAMM−48 lies between the two start codons. Comparison of the cDNA nucleotide sequences of B-cell RHAMMFL, RHAMM−48, and RHAMM−147 with plasma cell RHAMMFL, RHAMM−48, and RHAMM−147 showed that the RHAMM gene products from these two cell types were 99% homologous. Thus, three distinct RHAMM gene products were identified in MM B and plasma cells by sequence analysis.
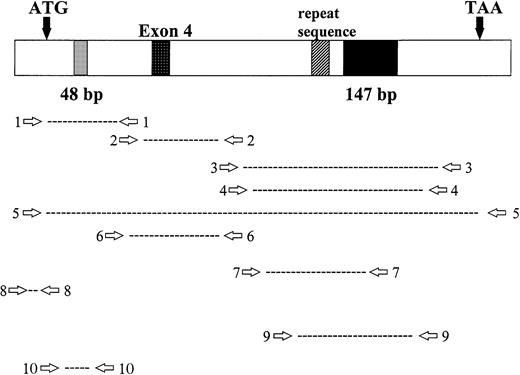
Location of RHAMM primer sets within RHAMM cDNA. Locations of the primers used to amplify RHAMM are identified with reference to the start (ATG) and stop (TAA) codons; the 48-bp and 147-bp deletion; exon 4; and the repeat sequence.
Location of RHAMM primer sets within RHAMM cDNA. Locations of the primers used to amplify RHAMM are identified with reference to the start (ATG) and stop (TAA) codons; the 48-bp and 147-bp deletion; exon 4; and the repeat sequence.
Comparison of B-cell and plasma cell RHAMM. RHAMM cDNA fragments were amplified from MM B cells and BM plasma cells by RT-PCR with RHAMM-specific primers and subsequently cloned and sequenced. The nucleotide sequences of the RHAMM fragments were aligned to generate the sequence of B-cell RHAMMFL and BM plasma cell RHAMMFL. The 48-bp deletion and the 147-bp deletion that characterize RHAMM−48 and RHAMM−147,respectively, are marked in bold lowercase. The 2 ATG start codons (base 18-20 and base 363-365) and TAA stop codon (base 2197-2199), previously described for breast RHAMM, plus the 2 HA-binding domains are underlined.
Comparison of B-cell and plasma cell RHAMM. RHAMM cDNA fragments were amplified from MM B cells and BM plasma cells by RT-PCR with RHAMM-specific primers and subsequently cloned and sequenced. The nucleotide sequences of the RHAMM fragments were aligned to generate the sequence of B-cell RHAMMFL and BM plasma cell RHAMMFL. The 48-bp deletion and the 147-bp deletion that characterize RHAMM−48 and RHAMM−147,respectively, are marked in bold lowercase. The 2 ATG start codons (base 18-20 and base 363-365) and TAA stop codon (base 2197-2199), previously described for breast RHAMM, plus the 2 HA-binding domains are underlined.
Cloning of RHAMM variants.
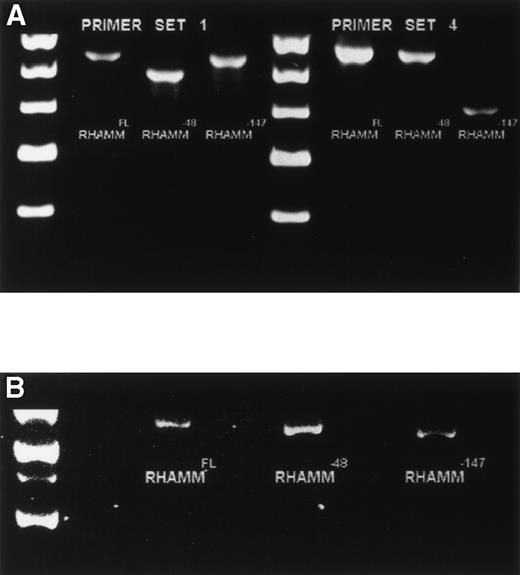
Because the identification of RHAMMFL, RHAMM−48, and RHAMM−147 was performed by sequence alignment of RHAMM cDNA fragments, we confirmed that each variant identified was transcribed into a complete RHAMM message in vivo. The complete ORF of RHAMM was amplified from BM plasma cell cDNA and screened for full-length RHAMMFL, RHAMM−48, or RHAMM−147 clones (Table 2). Full-length RHAMMFL, RHAMM−147, and RHAMM−48 clones were initially identified by PCR screening of colonies with primer set 1 (described earlier), which amplifies two products of 613 bp, corresponding to RHAMMFL or RHAMM−147, and of 565 bp corresponding to RHAMM−48. Next, clones scoring positive for RHAMMFL/RHAMM−147were rescreened by PCR with primer set 4, which amplifies two products: one of 666 bp, corresponding to RHAMMFL, and one of 519 bp, corresponding to RHAMM−147. The 519-bp PCR product that characterizes the 147-bp deletion of RHAMM−147 clones was not amplified from RHAMM−48 clones rescreened with primer set 4, but the 666-bp product was, showing that the 48-bp and 147-bp deletions are not found in the same RHAMM transcript and that RHAMM−48and RHAMM−147 are distinct RHAMM variants. Figure4A shows the pattern of PCR products amplified by primer sets 1 and 4 for each RHAMM isoform. A representative clone of each RHAMM isoform was sized by gel electrophoresis to confirm that RHAMMFL, RHAMM−48, and RHAMM−147 were indeed full-length RHAMM gene products (Fig 4B). The frequency of transcripts for each isoform expressed in MM was estimated by determining the frequency of clones derived from the ORF amplification (Table 2). RHAMMFL was the most frequent transcript, appearing in 49% of the randomly selected clones. RHAMM−48 was present in 47% of the clones, and RHAMM−147, detected in 4% of the clones, was relatively infrequent.
Frequency of Transcripts Encoding RHAMM Isoforms in Myeloma Plasma Cells
| Isoform . | Frequency of Clones . | |
|---|---|---|
| No. . | % . | |
| RHAMMFL | 56/115 | 49 |
| RHAMM−48 | 54/115 | 47 |
| RHAMM−147 | 5/115 | 4 |
| Isoform . | Frequency of Clones . | |
|---|---|---|
| No. . | % . | |
| RHAMMFL | 56/115 | 49 |
| RHAMM−48 | 54/115 | 47 |
| RHAMM−147 | 5/115 | 4 |
cDNA from MM plasma cells was amplified across the ORF and the products cloned; 115 clones were randomly picked and amplified with primer sets 1 and 4 to identify their insert.
Identification of ORF RHAMM isoforms from MM plasma cells. (A) Plasma cell ORF RHAMM was amplified, cloned, and colonies screened by PCR with primer set 1 and primer set 4. The amplification products from cloned RHAMMFL were 613 bp (lane 2) and 666 bp (lane 6), from RHAMM−48 were 565 bp (lane 3) and 666 bp (lane 7), and from RHAMM−147 were 613 bp (lane 4) and 519 bp (lane 8). Markers (lane 1 and 5) are 700, 600, 500, 400, and 300 bp. (B) A representative clone of each ORF RHAMM isoform was restriction-digested from the PCR TM II vector with EcoRV plusKpn I and sized on a 1% agarose gel. RHAMMFLis 2.319 kb; RHAMM−48 is 2.268 kb; and RHAMM−147 is 2.172 kb. Markers are 2.5, 2, 1.5, and 1 kb.
Identification of ORF RHAMM isoforms from MM plasma cells. (A) Plasma cell ORF RHAMM was amplified, cloned, and colonies screened by PCR with primer set 1 and primer set 4. The amplification products from cloned RHAMMFL were 613 bp (lane 2) and 666 bp (lane 6), from RHAMM−48 were 565 bp (lane 3) and 666 bp (lane 7), and from RHAMM−147 were 613 bp (lane 4) and 519 bp (lane 8). Markers (lane 1 and 5) are 700, 600, 500, 400, and 300 bp. (B) A representative clone of each ORF RHAMM isoform was restriction-digested from the PCR TM II vector with EcoRV plusKpn I and sized on a 1% agarose gel. RHAMMFLis 2.319 kb; RHAMM−48 is 2.268 kb; and RHAMM−147 is 2.172 kb. Markers are 2.5, 2, 1.5, and 1 kb.
RHAMM transcripts from MM cells include exon 4 and have a truncated “repeat” region in exon 8.
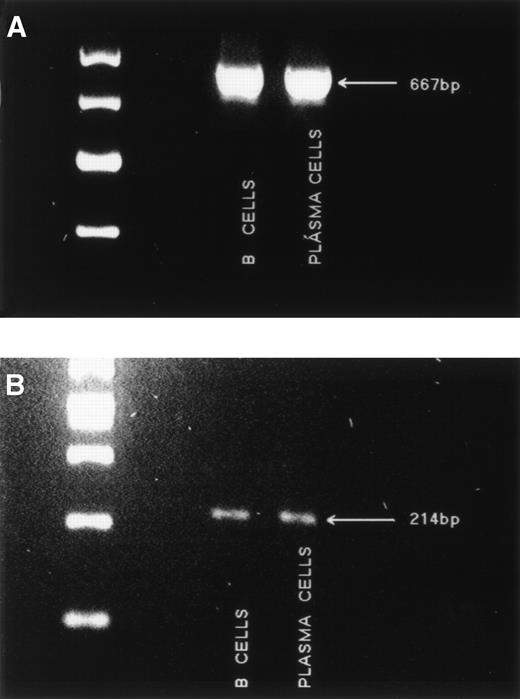
Murine RHAMM has been shown to include an isoform termed RHAMMv4 to designate the presence of exon 4,26 which is transforming for normal fibroblasts (Fb) and is associated with tumorigenicity.27 Transformed murine Fb also include a multiple repeat sequence within RHAMM exon 8.26 In the human, the RHAMM transcript detected in breast epithelium appears to include exon 4 and has only one copy of the “repeat” sequence found in mouse Fb.37 To characterize usage of exon 4 in RHAMM expressed by MM cells, we designed primers to amplify a region spanning exon 4 (primer set 6), such that any transcripts lacking exon 4 would appear as a smaller size than those incorporating exon 4. A single amplification product of 667 bp, incorporating exon 4, was detected in MM B cells and plasma cells (Fig5A). No small transcripts were detectable, suggesting that in MM, all RHAMM incorporates exon 4. Using primers spanning exon 8 (primer set 7), a 214-bp product (Fig 5B) indicates transcripts with one repeat sequence, and larger products would indicate transcripts having more than one of the repeated sequence. A single amplification product, of 214 bp, corresponding to a single sequence in the repeat region, was detected in MM B cells and plasma cells (Fig 5B).
Amplification of exon 4 and the “repeat” region from MM B cells and plasma cells. (A) The region spanning exon 4 was amplified from MM B cells and plasma cells with primer set 6. A single band of 667 bp was detected. Markers are 700, 600, 500, and 400 bp. (B) The “repeat” region of RHAMM was amplified from MM B cells and plasma cells with primer set 7. A single band of 214 bp was detected. Markers are 500, 400, 300, 200, and 100 bp.
Amplification of exon 4 and the “repeat” region from MM B cells and plasma cells. (A) The region spanning exon 4 was amplified from MM B cells and plasma cells with primer set 6. A single band of 667 bp was detected. Markers are 700, 600, 500, and 400 bp. (B) The “repeat” region of RHAMM was amplified from MM B cells and plasma cells with primer set 7. A single band of 214 bp was detected. Markers are 500, 400, 300, 200, and 100 bp.
Expression of RHAMM isoforms is detectable in B and plasma cells from all MM patients.
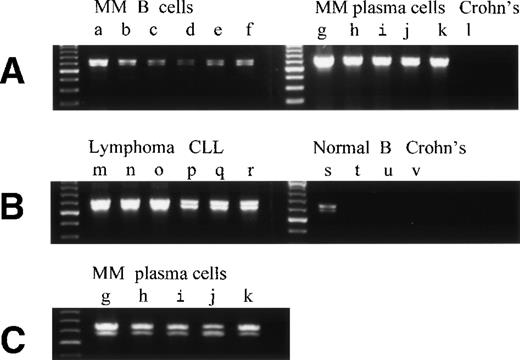
To confirm that RHAMMFL, RHAMM−48, and RHAMM−147 were expressed in B and plasma cells from all myeloma patients, we analyzed the pattern of RHAMM isoforms expressed by the sorted B cells from 11 individual MM patients and BM plasma cells from 11 individual MM patients. Figures6 and 7 show the pattern of RHAMM isoforms amplified by primer set 1 and primer set 3, respectively. RHAMMFL and RHAMM−48were amplified from the B cells of all 11 patients analyzed (six representative patients are shown in Fig 6A, lanes a through f) and from the BM plasma cells of all 11 patients analyzed (five representative patients are shown in Fig 6A and C, lanes g through k). RHAMM−147 was amplified from the BM plasma cells of six of six MM patients (Fig 7A, lanes 1 to 6, top panel) and from the B cells of four of five MM patients (Fig 7A, lanes 1 to 5, middle panel). Amplification and detection of MM B-cell genomic DNA, under identical conditions, did not generate products corresponding to RHAMMFL, RHAMM−48, or RHAMM−147, but a genomic interleukin-2 (IL-2) product was amplified with IL-2–specific primers, showing that the RHAMM sequences detected were not amplified genomic products. As an additional control, BM RNA was incubated with or without superscript in the RT step and then amplified; RHAMM amplification products were only detected “with Superscript” showing that the RHAMM sequences were derived from RNA and not DNA. Thus, the RHAMM transcripts identified here are expressed by all MM patients.
Overexpression of RHAMMFL and RHAMM−48 from MM B and plasma cells, B-lymphoma, and B-CLL, as compared with normal B cells and chronically activated B cells from patients with Crohn’s disease. (A) RHAMMFL and RHAMM−48 were amplified by RT-PCR from 500 ng of RNA from the B cells of 6 myeloma patients (a, b, c, d, e, f), the BM plasma cells of 5 myeloma patients (g, h, I, j, k), and the pooled B cells from 2 patients with Crohn’s disease (lane l). The same quantity of RNA (500 ng) and cDNA (5 μL) was used in the RT-PCR step for all cell populations analyzed so that the level of RHAMM expression could be compared between the different cell populations in the figure. The quality of RNA isolated from all B-cell populations was comparable as determined by amplification of CD19 transcripts from a separate aliquot of the same cDNA (5 μL) from which RHAMM was amplified (data not shown). A 613-bp fragment was amplified for RHAMMFL and a 565-bp fragment for RHAMM−48. (B) RHAMMFL and RHAMM−48 were amplified by RT-PCR from 500 ng of RNA from the malignant lymph nodes of 3 patients (m, n, o), the PBMC of 3 CLL patients (p, q, r), the B cells of 3 normal individuals (s, t, u), and the B cells from a patient with Crohn’s disease (v). The same quantity of RNA (500 ng) and cDNA (5 μL) was used in the RT-PCR step for all cell populations analyzed so that the level of RHAMM expression could be compared between the different cell populations in the figure. The quality of RNA isolated from all B-cell populations was comparable as determined by amplification of CD19 transcripts from a separate aliquot of the same cDNA (5 μL) from which RHAMM was amplified (data not shown). (C) Shows that the amplification product in A, from the BM plasma cells of patients g, h, I, j, and k, resolves into RHAMMFL and RHAMM−48. The amplification products were analyzed by gel electrophoresis and visualized by ethidium bromide staining.
Overexpression of RHAMMFL and RHAMM−48 from MM B and plasma cells, B-lymphoma, and B-CLL, as compared with normal B cells and chronically activated B cells from patients with Crohn’s disease. (A) RHAMMFL and RHAMM−48 were amplified by RT-PCR from 500 ng of RNA from the B cells of 6 myeloma patients (a, b, c, d, e, f), the BM plasma cells of 5 myeloma patients (g, h, I, j, k), and the pooled B cells from 2 patients with Crohn’s disease (lane l). The same quantity of RNA (500 ng) and cDNA (5 μL) was used in the RT-PCR step for all cell populations analyzed so that the level of RHAMM expression could be compared between the different cell populations in the figure. The quality of RNA isolated from all B-cell populations was comparable as determined by amplification of CD19 transcripts from a separate aliquot of the same cDNA (5 μL) from which RHAMM was amplified (data not shown). A 613-bp fragment was amplified for RHAMMFL and a 565-bp fragment for RHAMM−48. (B) RHAMMFL and RHAMM−48 were amplified by RT-PCR from 500 ng of RNA from the malignant lymph nodes of 3 patients (m, n, o), the PBMC of 3 CLL patients (p, q, r), the B cells of 3 normal individuals (s, t, u), and the B cells from a patient with Crohn’s disease (v). The same quantity of RNA (500 ng) and cDNA (5 μL) was used in the RT-PCR step for all cell populations analyzed so that the level of RHAMM expression could be compared between the different cell populations in the figure. The quality of RNA isolated from all B-cell populations was comparable as determined by amplification of CD19 transcripts from a separate aliquot of the same cDNA (5 μL) from which RHAMM was amplified (data not shown). (C) Shows that the amplification product in A, from the BM plasma cells of patients g, h, I, j, and k, resolves into RHAMMFL and RHAMM−48. The amplification products were analyzed by gel electrophoresis and visualized by ethidium bromide staining.
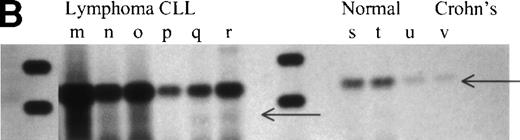
Overexpression of RHAMMFL and RHAMM−147 in MM, B-lymphoma, and B-CLL, as compared with normal B cells and chronically activated B cells from patients with Crohn’s disease. (A) RHAMMFL and RHAMM−147 were amplified by RT-PCR of 1μg RNA from the BM plasma cells of 6 myeloma patients, from the B cells of 5 myeloma patients, and from the B cells of 5 normal individuals. The same quantity of RNA (1 μg) and cDNA (5 μL) was used in the RT-PCR step for all cell populations analyzed so that the level of RHAMM expression could be compared between the different cell populations in the figure. The quality of RNA isolated from all cell populations was comparable as determined by amplification of histone transcripts from a separate aliquot of the same cDNA (1 μL) from which RHAMM was amplified (data not shown). Markers are 1,114, 900, and 692 bp. A 989-bp fragment was amplified for RHAMMFL and a 842-bp fragment for RHAMM−147. The amplification products were transferred to a nylon membrane and hybridized to a DIG-labeled RHAMM probe. BM plasma cell RHAMM was visualized by colorimetric detection; color development was for 1 hour. MM B-cell RHAMM and normal B cell RHAMM was detected by chemiluminescence; exposure was for 18 hours. (B) RHAMMFLand RHAMM−147 were amplified by RT-PCR of 500 ng RNA from the malignant lymph nodes of 3 patients (m, n, o), the PBMC of 3 CLL patients (p, q, r), the B cells of 3 normal individuals (s, t, u), and the B cells from a patient with Crohn’s disease (v). The same quantity of RNA (500 ng) and cDNA (5 μL) was used in the RT-PCR step for all cell populations analyzed so that the level of RHAMM expression could be compared between the different cell populations in the figure. The quality of RNA isolated from all B-cell populations was comparable as determined by amplification of CD19 transcripts from a separate aliquot of the same cDNA (5 μL) from which RHAMM was amplified (data not shown). A 989-bp fragment was amplified for RHAMMFL and a 842-bp fragment for RHAMM−147; arrows indicate their position. The amplification products were transferred to a nylon membrane and hybridized to a DIG-labeled RHAMM probe. RHAMM was visualized by colorimetric detection; color development was for 1 hour. Markers are 1,114 and 900 bp.
Overexpression of RHAMMFL and RHAMM−147 in MM, B-lymphoma, and B-CLL, as compared with normal B cells and chronically activated B cells from patients with Crohn’s disease. (A) RHAMMFL and RHAMM−147 were amplified by RT-PCR of 1μg RNA from the BM plasma cells of 6 myeloma patients, from the B cells of 5 myeloma patients, and from the B cells of 5 normal individuals. The same quantity of RNA (1 μg) and cDNA (5 μL) was used in the RT-PCR step for all cell populations analyzed so that the level of RHAMM expression could be compared between the different cell populations in the figure. The quality of RNA isolated from all cell populations was comparable as determined by amplification of histone transcripts from a separate aliquot of the same cDNA (1 μL) from which RHAMM was amplified (data not shown). Markers are 1,114, 900, and 692 bp. A 989-bp fragment was amplified for RHAMMFL and a 842-bp fragment for RHAMM−147. The amplification products were transferred to a nylon membrane and hybridized to a DIG-labeled RHAMM probe. BM plasma cell RHAMM was visualized by colorimetric detection; color development was for 1 hour. MM B-cell RHAMM and normal B cell RHAMM was detected by chemiluminescence; exposure was for 18 hours. (B) RHAMMFLand RHAMM−147 were amplified by RT-PCR of 500 ng RNA from the malignant lymph nodes of 3 patients (m, n, o), the PBMC of 3 CLL patients (p, q, r), the B cells of 3 normal individuals (s, t, u), and the B cells from a patient with Crohn’s disease (v). The same quantity of RNA (500 ng) and cDNA (5 μL) was used in the RT-PCR step for all cell populations analyzed so that the level of RHAMM expression could be compared between the different cell populations in the figure. The quality of RNA isolated from all B-cell populations was comparable as determined by amplification of CD19 transcripts from a separate aliquot of the same cDNA (5 μL) from which RHAMM was amplified (data not shown). A 989-bp fragment was amplified for RHAMMFL and a 842-bp fragment for RHAMM−147; arrows indicate their position. The amplification products were transferred to a nylon membrane and hybridized to a DIG-labeled RHAMM probe. RHAMM was visualized by colorimetric detection; color development was for 1 hour. Markers are 1,114 and 900 bp.
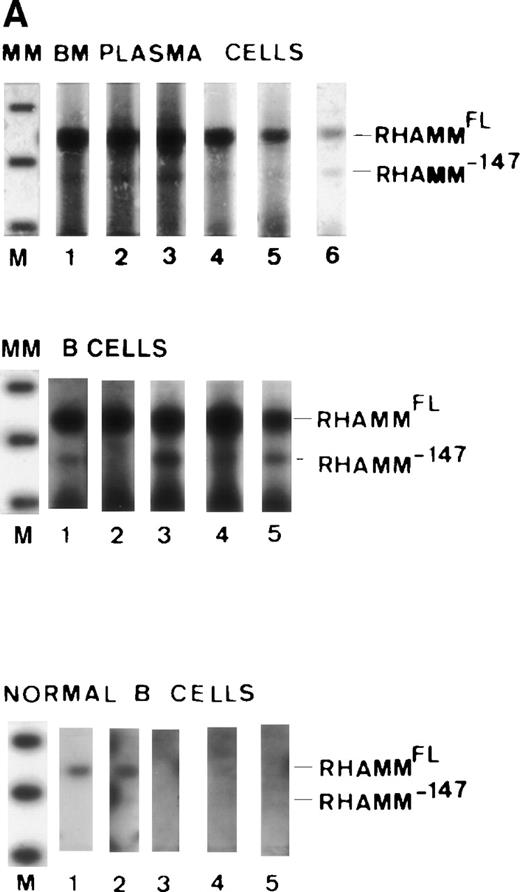
RHAMMFL and RHAMM−48 were easily visualized by ethidium bromide staining, while detection of RHAMM−147 necessitated the increased sensitivity of hybridization to a DIG-labeled RHAMM probe, which suggests that RHAMM−147 is less abundant in MM B cells and plasma cells than RHAMMFL and RHAMM−48, although for one MM patient, RHAMM−147 was easily detectable by ethidium bromide staining for both B and plasma cells (not shown). Because RHAMMFL and RHAMM−147 were amplified simultaneously, this suggests that RHAMM−147 is less abundant in MM B cells and plasma cells than RHAMMFL. However, the conclusion that RHAMM−147 is generally less abundant than RHAMMFL assumes that RHAMMFL/RHAMM−147 are amplified equally by primer set 3. To determine whether there was any difference in the efficiency of amplification of RHAMMFL/RHAMM−147 by primer set 3 or in the efficiency of amplification of RHAMMFL/RHAMM−48 by primer set 1, we amplified sequential dilutions of cloned RHAMM. Figure8A shows that equivalent amounts of DNA were amplified by primer set 1 from 100, 10, and 1 pg of RHAMMFL and RHAMM−48. Therefore, primer set 1 should equivalently amplify RHAMMFL and RHAMM−48 transcripts. Figure 8B shows that equivalent amounts of DNA were amplified by primer set 3 from 100 and 10 pg of RHAMMFL and RHAMM−147. A longer exposure (Fig 8C) of the hybridized membrane in Fig 8B shows that equivalent amounts of DNA were amplified from 1 pg of RHAMMFL and RHAMM−147. Therefore, primer set 3 should equivalently amplify RHAMMFL and RHAMM−147 transcripts present in MM B cells and BM plasma cells. In comparison to RHAMMFL expression, RHAMM−147 could either be expressed at a low level by all MM B cells/plasma cells or, alternatively, may be expressed by only a few cells within these populations.
Comparison of primer efficiency. (A) 100 pg, 10 pg, and 1 pg of RHAMMFL and RHAMM−48 were amplified with primer set 1. The amplification products for RHAMMFL(666 bp) and RHAMM−48 (613 bp) were analyzed by gel electrophoresis and visualized by ethidium bromide staining. (B and C) 100 pg,10 pg, and 1 pg of RHAMMFL and RHAMM−147 were amplified with primer set 3. The amplification products for RHAMMFL (989 bp) and RHAMM−147 (842 bp) were transferred to a nylon membrane and hybridized to a DIG-labeled RHAMM probe. RHAMM was detected by chemiluminescence; exposure was for 1 hour in (B) and for 10 hours in (C).
Comparison of primer efficiency. (A) 100 pg, 10 pg, and 1 pg of RHAMMFL and RHAMM−48 were amplified with primer set 1. The amplification products for RHAMMFL(666 bp) and RHAMM−48 (613 bp) were analyzed by gel electrophoresis and visualized by ethidium bromide staining. (B and C) 100 pg,10 pg, and 1 pg of RHAMMFL and RHAMM−147 were amplified with primer set 3. The amplification products for RHAMMFL (989 bp) and RHAMM−147 (842 bp) were transferred to a nylon membrane and hybridized to a DIG-labeled RHAMM probe. RHAMM was detected by chemiluminescence; exposure was for 1 hour in (B) and for 10 hours in (C).
To evaluate the expression of RHAMM and its splice variants in other B-cell malignancies, transcripts from three malignant B lymphomas and from three B-CLL were analyzed by RT-PCR (Fig 6B and Fig 7B). B-lymphoma cells have been previously shown to express surface RHAMM as measured by flow cytometry and immunohistochemistry, but B-CLL cells do not express surface RHAMM.22 RHAMMFL and RHAMM−48 were detectable for all patients (Fig6B, lanes m through r). RHAMM−147 was detectable for two of three lymphoma patients (Fig 7B, lanes m and o) and for two of three CLL patients (Fig 7B, lanes q and r). For lymphoma (m), RHAMM−147 was visible by ethidium bromide staining (not shown). The expression of RHAMM transcripts by B-CLL cells, which lack surface RHAMM,22 suggests that they may encode intracellular RHAMM.
RHAMMFL, RHAMM−48 or RHAMM−147 are overexpressed in malignant B cells (MM, lymphoma, and CLL) relative to resting or chronically activated normal B cells.
To determine which, if any, RHAMM species were overexpressed by malignant B-lineage cells, we compared the pattern of RHAMM transcripts in malignant B cells with those of resting B cells from normal donors, and of chronically activated B cells from patients with Crohn’s disease. The same quantity of RNA and cDNA was used in the RT-PCR step for all cell populations analyzed so that the level of RHAMM expression could be compared between nonmalignant and malignant B cells. Unlike B cells from normal donors or patients with nonmalignant disease, circulating B cells from patients with Crohn’s disease have a phenotype similar to that of MM B cells. This includes expression of CD45R0 by MM B and plasma cells.16,19 CD45R0 is absent from normal B cells, but is expressed in vivo on PBMC B cells from patients with Crohn’s disease.38 39 RHAMM−48 (Fig6A, lane l and Fig 6B, lane v) and RHAMM−147 (Fig 7B, lane v) were not detectable in these polyclonal B cells in Crohn’s disease, and RHAMMFL was weakly detectable by hybridization to a DIG-labeled probe (Fig 7B, lane v). RHAMMFLand RHAMM−48 were weakly detected by ethidium bromide staining in B cells from one of eight normal donors (three representative samples are shown in Fig 6B, lanes s through u). However, use of hybridization to a DIG-labeled RHAMM probe (Fig 7) showed that RHAMMFL transcripts were weakly detectable in populations of B cells from five of eight normal donors (Fig 7A, bottom panel, lanes 1 to 5, and Fig 7B, lanes s through u). In contrast, amplified RHAMMFL was easily detected in myeloma B cells from 11 of 11 patients by ethidium bromide staining (six representative samples are shown in Fig 6A, lanes a through f), although at lower levels than those for MM plasma cells, B lymphoma, or B-CLL. The quality of RNA isolated from normal B-cell populations was comparable to that isolated from malignant B-cell populations, because equivalent levels of histone or CD19 transcripts were amplified in all B-cell populations from a separate aliquot of the same cDNA from which RHAMM was amplified (data not shown). This work, together with the phenotypic expression of RHAMM by B-lineage cells in MM and lymphoma, suggests that all three RHAMM variants are overexpressed in malignant B or plasma cells, relative to B cells from normal donors or donors with Crohn’s disease.
In situ RT-PCR indicates expression of RHAMM transcripts by MM B or plasma cells, but few normal B cells.
To quantitate the number of cells expressing RHAMM, in situ RT-PCR was performed on MM plasma cells, MM B cells, and normal B cells (Table3) with primers (primer set 11) that would amplify all three RHAMM isoforms. For six patients, 22% to 36% of BM plasma cells expressed RHAMM (mean, 29%); for six patients, 8% to 13% of B cells expressed RHAMM (mean, 12%); and for four normal volunteers, 0% to 1.8% of B cells expressed RHAMM (mean, 1%). Comparison of the in situ RT-PCR values with the number of B cells expressing surface RHAMM protein, as detected phenotypically (Fig1),17 suggests that most RHAMM+ B and plasma cells have downregulated RHAMM mRNA, and/or that these cells have a low RHAMM mRNA copy number. Overall, this analysis indicates that in MM, cells with sufficient RHAMM transcripts to be detected in situ were more abundant in the BM plasma cell population than in the B-cell population, and confirms that detectable RHAMM transcripts are absent from nearly all normal B cells, consistent with their lack of surface RHAMM protein.
Individual B and Plasma Cells in MM But Not in Normal Donors Express RHAMM Transcripts as Measured by In Situ RT-PCR With Primers Annealing to RHAMM
| . | % Indicated Subset Expressing RHAMM Transcripts3-150 . | ||
|---|---|---|---|
| MM Plasma Cells . | MM B Cells . | Normal B Cells . | |
| 35.3 | 12.6 | 1.8 | |
| 22.7 | 13.4 | 1.1 | |
| 29.1 | 10.9 | <1 | |
| 26.5 | 15.1 | 1.7 | |
| 22.3 | 8.0 | ||
| 35.9 | 11.7 | ||
| Mean ± SE | 28.6 ± 2.43-151 | 11.8 ± 13-152 | 1.1 ± 0.4 |
| . | % Indicated Subset Expressing RHAMM Transcripts3-150 . | ||
|---|---|---|---|
| MM Plasma Cells . | MM B Cells . | Normal B Cells . | |
| 35.3 | 12.6 | 1.8 | |
| 22.7 | 13.4 | 1.1 | |
| 29.1 | 10.9 | <1 | |
| 26.5 | 15.1 | 1.7 | |
| 22.3 | 8.0 | ||
| 35.9 | 11.7 | ||
| Mean ± SE | 28.6 ± 2.43-151 | 11.8 ± 13-152 | 1.1 ± 0.4 |
Each data point represents the indicated type of sorted cells from an individual MM or normal donor, as described in Materials and Methods. MM plasma cells were from BM, and MM B cells and normal B cells were from blood.
P < .001 as compared with MM B cells.
P < .001 as compared with B cells from normal donors.
DISCUSSION
This study identifies three distinct variants transcribed from the RHAMM gene in MM B and plasma cells. Our results suggest that RHAMMFL, RHAMM−48, and RHAMM−147 are overexpressed in malignant B and plasma cells from myeloma, in B lymphomas, and in B-CLL cells, as compared with resting or chronically activated normal B cells. RHAMM has been linked to oncogenesis, enables the transforming properties of rasmutations in murine systems,27 and mediates motility of malignant human B cells.17,22 24 The overexpression of RHAMM variants in MM, lymphoma, and CLL implicates them in progression and spread of malignant B cells, and perhaps in the oncogenic events giving rise to myeloma and other B-cell malignancies.
In myeloma, RHAMMFL is virtually identical to the published sequence of human breast RHAMM.37 In addition, myeloma cells express two novel isoforms, RHAMM−48, which has a deletion of 48 bp, and RHAMM−147, which has a deletion of 147 bp. These correspond to coding deletions of 16 amino acids (aa) and 49 aa, respectively, in the RHAMM protein. The 48-bp deletion lies between two possible start codons and could alternatively represent a noncoding deletion, in the 5′ untranslated region of a RHAMM transcript, translated from the second start codon.
The significance of the 16-aa and 49-aa deletions to the interaction of RHAMM with HA, a prerequisite for transducing a motile signal,31 is not readily apparent because these deletions lie outside of the HA-binding domains. The deletion variants may encode conformationally altered RHAMM and/or have a cellular or intracellular localization different from that of RHAMMFL, which is known to be present on the cell surface as a GPI-linked receptor.26,27 In MM, surface and intracellular forms of RHAMM are detected,17 but their correlation with the splice variants identified here is as yet unknown. The nucleotide sequences of the HA-binding domains cloned from MM B and plasma cells translate into a (B[X7]B) HA-binding motif32,40 and thus encode two functional HA-binding domains. Because RHAMM appears to be part of an HA receptor complex,41 and has been shown to interact with src and ras family signaling molecules,31,42,43 deletion of key regulatory regions would have profound functional consequences that may facilitate malignant processes. Alternatively, deletion of the regions described may modify the affinity of the RHAMM receptor for its ligand, HA. This suggestion is not without precedent, because it has been documented that variant isoforms of the CD44 HA receptor have intact HA-binding domains yet differ in their ability to bind HA.44
The observation that overexpressed RHAMM is oncogenic in the absence of other known transforming events27 suggests that RHAMM is unique among adhesion receptors. Although CD44 isoforms are associated with increased metastatic spread,45-47 overexpression of CD44 itself has not been associated with oncogenesis. In this context, our observation that all of the RHAMM variants detected here are overexpressed in myeloma cells is provocative. Consistent with previous work showing strong surface expression of RHAMM, RHAMM is also overexpressed in B lymphoma.22 B-CLL cells, which lack surface RHAMM,22 have strong expression of RHAMM transcripts for all variants, suggesting that in B-CLL, RHAMM is intracellular. In contrast, resting B cells from normal donors and chronically activated B cells from patients with Crohn’s disease, which also lack surface RHAMM (not shown), have undetectable or weakly detectable RHAMM transcripts. In myeloma, the unusual expression of novel RHAMM variants, as well as of RHAMMFL, is consistent with the expression of surface RHAMM by the majority of B and plasma cells in 90 of 90 MM patients, and its relative absence from nearly all normal B cells. Expression of RHAMM appears to be a distinguishing feature of MM, lymphoma, and CLL, as compared with resting or chronically activated normal B cells.14 RHAMM may be involved in progression and spread of malignant B cells, based on our observations that motility by MM B and leukemic plasma cells is HA-dependent and mediated by RHAMM.17 However, the RHAMM isoforms involved in the motile process have yet to be identified. RHAMM has been shown to enable transforming mutations of ras; if RHAMM is inactivated by treatment with antisense or by a dominant negative RHAMM, ras-transformed cell lines acquire a normal phenotype and lose their tumorigeneic properties in vivo.27Thus, the overexpression of RHAMM in MM and its known role in migratory behavior suggest that deregulation of RHAMM isoforms may play a central role in the oncogenic events underlying myeloma.
Our study suggests that at some point during malignant transformation, B cells upregulate, or lose the ability to downregulate RHAMMFL, RHAMM−48, and RHAMM−147. In so doing, they may acquire the ability to move and metastasize to distant bone marrow sites. The trigger for such a switch is not yet apparent, but ras and RHAMM colocalize on the ruffles in migrating cells, both are involved in signaling via HA, and both are required to convert myeloma cell lines from sessile to motile behavior.48Ras is elevated in MM, and, aside from overexpression of RHAMM, ras mutations are the most common oncogenic abnormality in MM.49-53Rasinduces metastatic behavior in tumor cells by inducing CD44 variant isoform expression,54 suggesting it may also play a role in RHAMM expression and alternative splicing. Alternatively, because even wild-type ras is transforming when constitutively targeted to the membrane,55 deregulated RHAMM may directly or indirectly alter ras localization to give a transformed phenotype.
The molecular characterization of RHAMM isoforms in MM represents a primary step in defining the role RHAMM plays in mediating the metastatic spread of malignant B cells to distant BM sites. In this report, we have shown that overexpression of RHAMM and its splice variants characterizes malignant B and plasma cells. Overexpression and modulation of the balance between soluble, surface, and intracellular RHAMM isoforms may control the regulation of key signaling molecules and the behavioral characteristics of premalignant cells, leading to constitutive stimulation and malignancy.
ACKNOWLEDGMENT
We are grateful to Dr Bruce Yacyshyn for providing blood samples from patients with Crohn’s disease. Juanita Wizniak provided skilled expertise in flow cytometry and cell sorting. Darlene Paine and Karen Seeberger provided expert technical assistance.
Supported by a grant from The Alberta Cancer Board Research Initiatives Program.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Linda M. Pilarski, PhD, Department of Oncology, University of Alberta, Edmonton, AB, T6G1Z2, Canada.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal