The monocyte and granulocyte azurophilic granule proteinases elastase, proteinase 3, and cathepsin G are implicated in acute and chronic diseases thought to result from an imbalance between the secreted proteinase(s) and circulating serpins such as 1-proteinase inhibitor and 1-antichymotrypsin. We show here that the intracellular serpin, proteinase inhibitor 6 (PI-6), is present in monocytes, granulocytes, and myelomonocytic cell lines. In extracts from these cells, PI-6 bound an endogenous membrane-associated serine proteinase to form an sodium dodecyl sulfate (SDS)-stable complex. Using antibodies to urokinase, elastase, proteinase 3, or cathepsin G, we demonstrated that the complex contains cathepsin G. Native cathepsin G and recombinant PI-6 formed an SDS-stable complex in vitro similar in size to that observed in the extracts. Further kinetic analysis demonstrated that cathepsin G and PI-6 rapidly form a tight 1:1 complex (ka = 6.8 ± 0.2 × 106mol/L−1s−1 at 17°C;Ki = 9.2 ± 0.04 × 10−10 mol/L). We propose that PI-6 complements 1-proteinase inhibitor and 1-antichymotrypsin (which control extracellular proteolysis) by neutralizing cathepsin G that leaks into the cytoplasm of monocytes or granulocytes during biosynthesis or phagocytosis. Control of intracellular cathepsin G may be particularly important, because it has recently been shown to activate the proapoptotic proteinase, caspase-7.
IMPORTANT PHYSIOLOGICAL processes such as blood coagulation, fibrinolysis, complement activation, embryo implantation, extracellular matrix remodeling, and cell differentiation involve proteolysis mediated by serine proteinases.1 The activity of many of these proteinases can be controlled by one or more members of the serine proteinaseinhibitor (serpin) protein superfamily.1 The physiological significance of serpins is illustrated by the consequences of defects in their production or activity. Loss of the thrombin inhibitor, antithrombin III, results in a greatly increased risk of thrombosis; loss of the serpin C1 inhibitor, which is a regulator of the classical complement activation pathway, leads to angioedema; and loss of α1-proteinase inhibitor activity underlies emphysema and liver cirrhosis.2
Inhibitory serpins typically form 1:1 complexes with their target proteinases by acting as pseudosubstrates.1 The serpin undergoes a conformational change leading to stabilization of the serpin-proteinase complex, which becomes resistant to a variety of denaturants, including sodium dodecyl sulfate (SDS). The serpin pseudosubstrate region is known as the reactive center and consists of approximately 20 amino acids near the carboxy terminus. Cleavage of the serpin by the proteinase may occur in this region between two residues designated P1 and P1′. The identity of the P1 residue largely dictates the specificity of the serpin-proteinase interaction, whereas those surrounding the cleavage site contribute to the affinity of the interaction.
Although the vast majority of serpins are secreted glycoproteins involved in regulating extracellular proteinases, it has recently become clear that some function within cells. One example is the cowpox virus serpin CrmA, which inhibits both the cytotoxic lymphocyte granule proteinase, granzyme B, and the intracellular caspases involved in cytokine maturation and apoptosis.3,4 Apart from CrmA, several proteins belonging to the mammalian ovalbumin (ov) serpin group also appear to function intracellularly. At present, this group includes plasminogen activator inhibitor 2,5 the squamous cell carcinoma antigens-1 and -2,6,7 monocyte/neutrophil elastase inhibitor,8 maspin,9 proteinase inhibitor 6,10 proteinase inhibitor 8,11proteinase inhibitor 9,11,12 bomapin,13 and megakaryocyte maturation factor.14 The ov-serpins are unusual because they lack an amino terminal signal peptide normally required to initiate extracellular secretion of the protein, yet several appear to exist in both intracellular and extracellular forms.15 The physiological roles of these inhibitors are not fully understood, but some may function in processes associated with tumorigenesis and inflammation. For example, loss of maspin correlates with increased malignancy and metastasis of breast carcinomas9; increases in the levels and release of squamous cell carcinoma antigen occur in squamous cell carcinomas6; and increased expression and release of plasminogen activator inhibitor 2 occurs from monocytes during inflammation.16 Recent evidence suggests that proteinase inhibitor 9 is a regulator of granzyme B that may protect immune cells against the effects of this cytotoxin.12
We originally described the ov-serpin proteinase inhibitor 6 (PI-6) after a screen for novel thrombin inhibitors.17 Unlike the majority of ov-serpins, PI-6 is apparently restricted to the cytoplasm of cells and cannot be released via the conventional secretory pathway.18 It is present in most tissues in capilliary endothelial cells, platelets, and epithelial cells.10,18Sequence comparisons suggest that the P1 residue of PI-6 is Arg,10 but although this explains the interaction of PI-6 with thrombin, subsequent analyses have shown that PI-6 is only a moderately effective thrombin inhibitor that inhibits trypsin far more efficiently.19,20 Of additional interest is the finding that PI-6 is also a very good chymotrypsin inhibitor,21 due to the presence of Met at the P2 position in the reactive center.
The broad distribution and dual inhibitory capacity of PI-6 suggests that it fulfils an important regulatory or protective role associated with proteolysis within cells. Yet, despite extensive study in vitro, the identity of the proteinase(s) targeted by PI-6 in vivo remains obscure. We show here that PI-6 is also produced in monocytes and granulocytes and that it interacts with a membrane-associated proteinase expressed in these cells. We have identified this proteinase as the azurophilic granule component cathepsin G and have shown that PI-6 inhibits it very efficiently. We propose that PI-6 functions to protect the interior of cells against ectopic or inadvertant exposure to cathepsin G.
MATERIALS AND METHODS
Reagents and antibodies.
Preparation of recombinant PI-6 has previously been described.19 Recombinant cathepsin G was purchased from Calbiochem (La Jolla, CA), and the cathepsin G substrate N-succinyl-Ala-Ala-Pro-Phe-pNA was from Sigma (St Louis, MO). Anti-urokinase plasminogen activator antiserum was purchased from America Diagnostica (Sydney, Australia). Polyclonal anti–PI-6 antisera is described elsewhere.18Monoclonal anti–PI-6 antibody (3A) was produced to recombinant PI-6 by standard procedures.22 Hybridomas producing immunoreactive antibody were identified using enzyme-linked immunosorbent assay (ELISA) on immobilized recombinant PI-6 and indirect immunofluorescence on PI-6 transfected COS cells.23 Rabbit polyclonal antibodies24 recognizing cathepsin G, proteinase 3, and human leukocyte elastase were a kind gift from Dr J. Hoidal (University of Utah, Salt Lake City, UT). Monoclonal 3C10 anti-CD14 antibody was a kind gift from Dr R. Steinman (Rockefeller Institute, New York, NY).
Biotinylation of monoclonal anti–PI-6 antibody (3A).
One milligram of purified antibody was biotinylated by exposure to sulfo-NHS-Biotin according to the manufacturer’s instructions (EZ-Link; Pierce, Rockford, IL). Unbound biotin was removed by extensive dialysis against phosphate-buffered saline (PBS).
Cell culture.
HL60, THP-1, and U937 cells were cultured in RPMI-1640 media supplemented with 10% heat-inactivated fetal calf serum, 2 mmol/L L-glutamine, 100 U/mL penicillin, and 50 μg/mL streptomycin. Cultures were maintained in a 5% CO2, 95% air mixture at 37°C and passaged every 3 days.
Isolation of peripheral blood monocytes, neutrophils, and lymphocytes.
Human monocytes were purified from whole blood using Nycoprep 1.068 according to the manufacturer’s instructions. To isolate lymphocytes and neutrophils, erythrocytes were removed from whole blood containing 0.15% (wt/vol) EDTA by precipitation using 1.2% Dextran 500 (wt/vol) for 1 hour at room temperature. Neutrophils were pelleted from the leukocyte-rich plasma by centrifuging over Ficoll Hypaque (Pharmacia, Sydney, Australia) at 2,500g. The supernatant containing the lymphocytes was also retained. All cell preparations were washed repeatedly with PBS and lysed with 10 mmol/L Tris-HCl, 0.15 mmol/L NaCl, pH 7.6, containing 1% Triton X-100 and 1 μg/mL aprotinin, 150 μg/mL phenylmethyl sulfonyl fluoride (PMSF), 0.5 μmol/L leupeptin, and 1 μmol/L pepstatin. Total protein concentration was determined (Bio-Rad, Hercules, CA), and equal amounts of total protein were immunoblotted with rabbit polyclonal anti–PI-6 as described below.
Fluorescence-activated cell sorting (FACS) analysis.
All steps were performed at room temperature and during staining samples were kept in the dark. One hundred microliters of whole blood from a healthy volunteer was stained with a monoclonal antibody recognizing CD14 (3C10). After 30 minutes, cells were washed twice in PBS containing 1% neonatal calf serum and 0.02% sodium azide (FACS buffer; used in all subsequent washes). Cells were incubated with sheep antimouse Ig-PE (Silenus, Melbourne, Australia) for 30 minutes, washed twice, and then incubated in FACS Lysing Solution (Becton Dickinson, Mountain View, CA) for 10 minutes. Cells were collected and resuspended in FACS Permeabilizing Solution (Becton Dickinson) for 10 minutes, washed, and then blocked in 10% normal mouse serum for 30 minutes. Biotinylated monoclonal 3A anti–PI-6 antibody was then added for 30 minutes. Cells were washed twice and incubated with streptavidin-fluorescein isothiocyanate (FITC) (Amersham, Sydney, Australia) for 30 minutes. After two washes, cells were analyzed using a Becton Dickinson FACScan and CellQuest software.
Immunoblotting.
Cells were washed with ice-cold PBS and subsequently lysed with 10 mmol/L Tris-HCl, 0.15 mmol/L NaCl, pH 7.6, containing 1% Triton X-100. In some experiments, 1 μg/mL aprotinin, 150 μg/mL PMSF, 0.5 μmol/L leupeptin, 1 μmol/L pepstatin, 100 μmol/L EDTA, or a cocktail of all the proteinase inhibitors were included in the lysis buffer. To allow the PI-6 and cathepsin G interaction to occur, samples were incubated at 37°C for 10 minutes after the addition of lysis buffer. Cell lysates were centrifuged to remove cell debris, boiled in Laemmli sample buffer containing 0.1 mol/L dithiothreitol (DTT) for 5 minutes, and resolved on 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions.25 After electrophoresis, proteins were transferred to nitrocellulose membranes and incubated for 1 hour in blocking buffer (5% skim milk powder in 20 mmol/L Tris-HCl, 150 mmol/L NaCl, pH 7.6). Membranes were incubated for 1 hour with rabbit polyclonal anti–PI-6 antiserum or monoclonal 3A anti–PI-6 and washed in 20 mmol/L Tris-HCl, 150 mmol/L NaCl, pH 7.6. Horseradish peroxidase (HRP)-conjugated sheep antirabbit Ig (Silenus) or HRP-conjugated sheep antimouse Ig (Bio-Rad) were used as secondary antibodies and were detected using enhanced chemiluminescence (DuPont, Sydney, Australia).
Cell fractionation.
U937 cells were washed in PBS and resuspended in hypotonic buffer (50 mmol/L PIPES, 50 mmol/L KCl, 5 mmol/L EGTA, 2 mmol/L MgCl2, 5 mmol/L DTT, pH 7.6). Cells were sonicated on ice at 100 W five times for 30 seconds. Samples were centrifuged at 1,000g to remove nuclei and whole cells and then at 100,000g for 1 hour. The supernatant was kept (cytosol) and the pellet was extracted with 1% Triton X-100 in 10 mmol/L Tris-HCl, 150 mmol/L NaCl, pH 7.6, for 1 hour at 4°C (membrane). Protein concentrations were determined and equal amounts of cytosol and membrane were mixed and incubated at 37°C for 10 minutes before SDS-PAGE and immunoblotting with polyclonal anti–PI-6 antisera as described above.
35S-Methionine labeling and immunoprecipitation.
U937 cells (2 × 106) were [35S]methionine labeled for 3 hours, as previously described.18 Labeled cells were lysed and immunoprecipitated overnight at 4°C with 100 μL of 10% (wt/vol) protein A-sepharose (Pharmacia Biotech Inc, Sydney, Australia) and 1 μL of the indicated rabbit polyclonal antiserum, and immune complexes were analyzed by 10% SDS-PAGE according to Laemmli.25 Gels were enhanced in Amplify (Amersham Corp) and visualized by fluorography.
Immunoprecipitation of the PI-6/cathepsin G complex from U937 cells.
U937 cells (2 × 106) were washed with ice-cold PBS, lysed in 1 mL 50 mmol/L Tris, pH 7.4, 150 mmol/L NaCl, 5 mmol/L EDTA, 0.25% (wt/vol) gelatin (NETGEL) containing 1% Nonidet P-40, and incubated at 37°C for 10 minutes. A further 1 mL of NETGEL was added and extracts were immunoprecipitated with 1 μL of the appropriate antibody. Immune complexes were analyzed by 10% SDS-PAGE, and separated proteins were transferred to a nitrocellulose membrane, blocked for 1 hour as described previously, and immunoblotted with monoclonal 3A anti–PI-6 antibody. HRP-conjugated sheep antimouse Ig (Bio-Rad) was used as the secondary antibody and was detected using enhanced chemiluminescence.
Analysis of the PI-6/cathepsin G complex in vitro.
Recombinant PI-6 (2 ng) was mixed with 1 to 200 ng of cathepsin G in 20 mmol/L Tris, 0.15 mol/L NaCl and incubated at 37°C for 10 minutes. Samples were separated by SDS-PAGE under reducing conditions and immunoblotted with polyclonal anti–PI-6 antisera as described above.
Determination of kinetic parameters.
The activity of rPI-6 was measured by inhibition of active site-titrated thrombin using procedures described in Duranton et al.26 The active site concentration of cathepsin G was determined using α1-proteinase inhibitor as previously described26 using the substrate suc-Ala-Ala-Pro-Phe-pNA in 50 mmol/L Tris, 150 mmol/L NaCl, 0.1% (wt/vol) polyethylene glycol (PEG), pH 7.8, at 20°C. Determination of the stoichiometry of inhibition was performed as previously described27 using the substrate suc-Ala-Ala-Pro-Phe-pNA in 50 mmol/L Tris, pH 7.8, 150 mmol/L NaCl, 0.1% (wt/vol) PEG at 20°C. The inhibition (Ki) and second order association rate (ka) constants were determined as previously described.28 Briefly, the second order rate constant was measured by mixing cathepsin G and PI-6 at equimolar concentrations (10 nmol/L) for variable periods of time before the addition of substrate (0.2 mmol/L); the residual enzyme activity was then measured (Molecular Dynamics plate reader; Molecular Dynamics, Sunnyvale, CA) and the temperature of the incubation was maintained at 17°C. All data presented represent the mean of five determinations.
RESULTS
Expression of PI-6 in peripheral blood leukocytes.
PI-6 was originally detected in cytosolic extracts from K562 cells, which are human myeloleukemia cells that maintain the potential to differentiate along the monocytic, granulocytic, or erythroid lineages.17 It is also present in the megakaryocytic cell line MEG-01 and in platelets.10 These observations suggested that PI-6 is normally expressed in myeloid cell lineages. To examine the distribution of PI-6 in human leukocytes, we used FACS to analyze permeabilized peripheral blood leukocytes stained with a monoclonal antibody specific for PI-6. This antibody recognizes PI-6 by indirect immunofluorescence and immunoblotting23 but does not recognize the ov-serpins most closely related to PI-6, namely proteinase inhibitor 8, proteinase inhibitor 9, monocyte/neutrophil elastase inhibitor, and plasminogen activator inhibitor 2 (data not shown).
As shown in Fig 1, the monoclonal antibody marked the granulocytic (R2) population of leukocytes, but not the small lymphocyte (R1) population (T and B cells). Double-staining with an anti-CD14 monoclonal antibody confirmed that PI-6 is in the monocyte/granulocyte population. PI-6 was present in both CD14-high and CD14-low R2 cells, which correspond to monocytes and granulocytes, repectively.
PI-6 is expressed in peripheral blood monocytes and granulocytes. Leukocytes from fresh, normal peripheral blood were analyzed by FACS after staining for PI-6 and CD14. The left-hand panel shows a scatter plot of the distribution of leukocytes into the small lymphocyte (R1) and monocyte/granulocyte (R2) populations. The central and right hand panels show the results of two-color FACS analysis of the R1 and R2 populations simultaneously stained with the anti–PI-6 monoclonal antibody 3A (x-axis) and an antibody recognizing the monocyte/granulocyte differentiation antigen CD14 (y-axis). The R1 population is not significantly marked by either antibody, whereas the R2 population is stained by both.
PI-6 is expressed in peripheral blood monocytes and granulocytes. Leukocytes from fresh, normal peripheral blood were analyzed by FACS after staining for PI-6 and CD14. The left-hand panel shows a scatter plot of the distribution of leukocytes into the small lymphocyte (R1) and monocyte/granulocyte (R2) populations. The central and right hand panels show the results of two-color FACS analysis of the R1 and R2 populations simultaneously stained with the anti–PI-6 monoclonal antibody 3A (x-axis) and an antibody recognizing the monocyte/granulocyte differentiation antigen CD14 (y-axis). The R1 population is not significantly marked by either antibody, whereas the R2 population is stained by both.
Detection of a PI-6/proteinase complex in monocyte/macrophage cell lines.
To analyze the expression of PI-6 in cells of monocyte/macrophage lineage in more detail, we used the well-established and characterized human lines HL60, THP-1, and U937. These resemble myeloid cells arrested at different stages of differentiation. For example, HL60 cells resemble less mature precursor cells and can be induced to differentiate either along the monocyte/macrophage pathways by exposure to phorbol esters or tumor necrosis factor or along the granulocyte pathway by exposure to retinoic acid.29 By contrast, THP-1 and U937 cells resemble more mature cells that have acquired monocytic characteristics.30
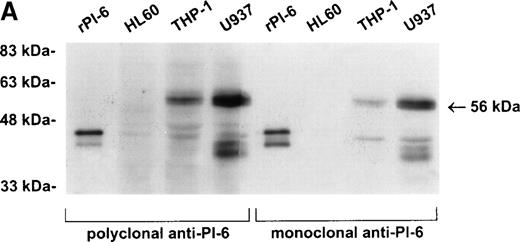
As shown in Fig 2A, PI-6 was detected in extracts of U937 and THP-1 cells, but surprisingly most of the material detected by both polyclonal and monoclonal antibodies to PI-6 was in the form of a higher molecular weight species. The size of this species (56 kD) is too small to represent a PI-6 dimer, but it is reminiscent of an SDS-resistant complex that may form when a serpin interacts with a serine proteinase.1 To determine whether the 56-kD species is present in normal monocytes or granulocytes, PBLs were fractionated into monocyte, neutrophil, and lymphocyte populations, and cytosolic extracts were prepared. Overall, the distribution of PI-6 immunoreactive material in the purified leukocytes was consistent with the FACS analysis (Fig 1). The 56-kD species was present in monocytes but not in neutrophils or lymphocytes (Fig 2B).
PI-6 and a 56-kD PI-6/proteinase complex are found in myelomonocytic cells. (A) HL60, THP-1, and U937 cells were lysed and equal amounts of protein were separated by 10% SDS-PAGE under reducing conditions and immunoblotted using polyclonal anti–PI-6 antisera or monoclonal anti–PI-6 antibody. (B) Monocytes, neutrophils, and lymphocytes were isolated from peripheral human blood as described in Materials and Methods. Cells were lysed in the presence of 1 μg/mL aprotinin, 150 μg/mL PMSF, 0.5 μmol/L leupeptin, and 1 μmol/L pepstatin to minimize degradation by plasma proteinases. Equal amounts of protein were separated by 12.5% SDS-PAGE under reducing conditions and immunoblotted using polyclonal anti–PI-6 antisera. Both gels contain 2 ng of recombinant PI-6 (rPI-6) loaded as a positive control. The lower band in the rPI-6 sample represents serpin cleaved in the reactive loop by nonspecific proteolysis during purification and storage.
PI-6 and a 56-kD PI-6/proteinase complex are found in myelomonocytic cells. (A) HL60, THP-1, and U937 cells were lysed and equal amounts of protein were separated by 10% SDS-PAGE under reducing conditions and immunoblotted using polyclonal anti–PI-6 antisera or monoclonal anti–PI-6 antibody. (B) Monocytes, neutrophils, and lymphocytes were isolated from peripheral human blood as described in Materials and Methods. Cells were lysed in the presence of 1 μg/mL aprotinin, 150 μg/mL PMSF, 0.5 μmol/L leupeptin, and 1 μmol/L pepstatin to minimize degradation by plasma proteinases. Equal amounts of protein were separated by 12.5% SDS-PAGE under reducing conditions and immunoblotted using polyclonal anti–PI-6 antisera. Both gels contain 2 ng of recombinant PI-6 (rPI-6) loaded as a positive control. The lower band in the rPI-6 sample represents serpin cleaved in the reactive loop by nonspecific proteolysis during purification and storage.
Establishment of a stable serpin-proteinase complex requires that the proteinase is active, and complex formation may be perturbed or abolished in the presence of compounds that bind the active site of the proteinase.1 To test whether a proteinase is present in the 56-kD species, extracts of U937 cells were prepared in the presence of a variety of protease inhibitors including EDTA, PMSF, aprotinin, leupeptin, and pepstatin. As shown in Fig3, the 56-kD species disappeared in the presence of the serine proteinase inhibitors PMSF and aprotinin, but not in the presence of inhibitors of other classes of proteases such as EDTA, leupeptin, and pepstatin. From these results we concluded that the 56-kD species represents PI-6 complexed with a serine proteinase produced by myeloid cells.
The 56-kD PI-6/proteinase complex contains a serine proteinase and is formed after lysis. U937 cells were lysed in the presence of 1 μg/mL aprotinin, 150 μg/mL PMSF, 0.5 μmol/L leupeptin, 1 μmol/L pepstatin, 100 μmol/L EDTA, or a mixture of all these inhibitors. Samples were incubated at 37°C for 10 minutes, separated by 10% SDS-PAGE, and immunoblotted using polyclonal anti–PI-6 antisera.
The 56-kD PI-6/proteinase complex contains a serine proteinase and is formed after lysis. U937 cells were lysed in the presence of 1 μg/mL aprotinin, 150 μg/mL PMSF, 0.5 μmol/L leupeptin, 1 μmol/L pepstatin, 100 μmol/L EDTA, or a mixture of all these inhibitors. Samples were incubated at 37°C for 10 minutes, separated by 10% SDS-PAGE, and immunoblotted using polyclonal anti–PI-6 antisera.
PI-6 was essentially undetectable by immunoblotting of cytosolic extracts from undifferentiated HL60 cells (Fig 2A), suggesting that PI-6 expression is induced as monocytes mature. Addition of recombinant PI-6 to HL60 cell extracts did not result in the appearance of the 56-kD PI-6–proteinase complex, indicating that the proteinase is also not present in these precursor type cells (data not shown). Treatment of HL60 cells with phorbol myristate acetate (PMA)-induced monocyte/macrophage differentiation as assessed by morphological alterations and acquisition of adhesiveness. Under these conditions, high levels of PI-6 were produced (data not shown). By contrast, exposure of HL60 cells to retinoic acid induced granulocytic differentiation as assessed by NBT assay,29and lower levels of PI-6 were detected under these conditions (data not shown). These results are consistent with the FACS analysis showing the presence of PI-6 in peripheral blood monocytes and granulocytes and suggest that PI-6 expression is induced at specific stages of differentiation of these cells.
Compartmentalization of PI-6 and the proteinase in U937 cells.
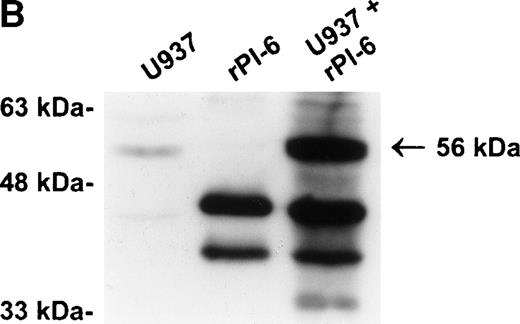
Our previous studies have demonstrated that PI-6 is a cytosolic serpin,17 18 which raised the possibility that the interacting proteinase is also cytosolic. An alternative scenario is that the proteinase is in a specific compartment within the cell, such as a membrane-bound granule, and that the interaction observed with PI-6 occurs primarily after lysis of the cells. To distinguish these possibilities, we fractionated U937 cells into membrane (100,000g pellet) and cytosolic components (after 100,000g supernatant) and analyzed the distribution of PI-6 and the complex. As shown in Fig 4A, PI-6 was present in the cytosolic fraction, but not in the membrane fraction. The complex was not detected in either the cytosolic or membrane fraction, but became apparent after the two fractions were mixed. This indicated that the proteinase is in a separate, membranous compartment that is normally inaccessible to PI-6. When recombinant PI-6 was added to the whole-cell extracts, the amount of complex formed was much larger than normally observed, indicating that the amount of endogenous PI-6 is limiting in the extracts (Fig 4B). Taken together, these results suggest that the U937 proteinase interacting with PI-6 is in a separate, membranous compartment, that it is in excess to the amount of PI-6 contained within the cytoplasm, and that the complex observed in U937 cell extracts is probably formed after lysis.
The protease is in a membrane bound subcellular compartment and is in excess to PI-6. (A) U937 cells were fractionated into cytosol and Triton X-100 soluble membrane fractions as described in Materials and Methods. Equal amounts of cytosolic protein, membrane protein, and a 1:1 mixture of both were incubated at 37°C for 10 minutes. Samples were separated by 10% SDS-PAGE under reducing conditions and immunoblotted using polyclonal anti–PI-6 antisera to visualize the 56-kD complex. The 63-kD band present in the first two lanes is nonspecific and was recognized by preimmune sera (not shown). (B) U937 cells were lysed and incubated at 37°C for 10 minutes with or without 2 ng recombinant PI-6 (rPI-6). Samples were separated by 10% SDS-PAGE under reducing conditions and immunoblotted using polyclonal anti–PI-6 antisera. Two nanograms of recombinant PI-6 was included in a separate lane as a positive control.
The protease is in a membrane bound subcellular compartment and is in excess to PI-6. (A) U937 cells were fractionated into cytosol and Triton X-100 soluble membrane fractions as described in Materials and Methods. Equal amounts of cytosolic protein, membrane protein, and a 1:1 mixture of both were incubated at 37°C for 10 minutes. Samples were separated by 10% SDS-PAGE under reducing conditions and immunoblotted using polyclonal anti–PI-6 antisera to visualize the 56-kD complex. The 63-kD band present in the first two lanes is nonspecific and was recognized by preimmune sera (not shown). (B) U937 cells were lysed and incubated at 37°C for 10 minutes with or without 2 ng recombinant PI-6 (rPI-6). Samples were separated by 10% SDS-PAGE under reducing conditions and immunoblotted using polyclonal anti–PI-6 antisera. Two nanograms of recombinant PI-6 was included in a separate lane as a positive control.
The PI-6/proteinase complex contains cathepsin G.
It is well established that myelomonocytic cells produce the serine proteinases urokinase plasminogen activator, elastase, proteinase 3, and cathepsin G (reviewed in Borregaard and Cowland31). Numerous studies have shown that these proteinases are contained within membrane-bound granules in resting cells and are released into phagocytic vesicles and secreted during inflammation. It is also known that expression of the elastase, cathepsin G, and proteinase 3 is differentiation stage-specific and is downregulated on terminal differentiation into macrophages.32
Because PI-6 can inhibit urokinase in vitro,19 we used urokinase-specific antiserum to determine by immunoblotting whether this proteinase is present in the 56-kD complex. Although urokinase was present in the U937 cells, we were unable to demonstrate any PI-6–urokinase interaction in the cell extracts, and experiments using purified proteinase and inhibitor indicated that a PI-6–urokinase SDS-stable complex would be much larger than 56 kD (data not shown). We therefore concluded that urokinase and PI-6 do not interact detectably in U937 cell extracts.
To determine whether the 56-kD complex contains elastase, proteinase 3, or cathepsin G, we obtained rabbit antisera specific for each of these proteinases24 (a kind gift of Dr J. Hoidal, Department of Internal Medicine, University of Utah Health Sciences Center, Salt Lake City, UT). As shown in Fig 5A, when U937 cells were metabolically labeled using 35S-methionine, elastase, proteinase 3, or cathepsin G, which are all approximately 30 kD in size, could be immuoprecipitated from cell extracts using the appropriate antiserum. The higher molecular (70 kD) species observed in the elastase immunoprecipitate probably represents an elastase-monocyte/neutrophil inhibitor complex (see Discussion). The 45-kD species seen in all the samples probably represents actin, which commonly contaminates immunoprecipitates.
PI-6 coimmunoprecipitates with cathepsin G from U937 cells as a 56-kD complex. (A) U937 cells were [35S]methionine labeled, lysed, and immunoprecipitated with rabbit polyclonal antibodies recognizing cathepsin G, proteinase 3, or human leukocyte elastase. Immune complexes were separated by 12.5% SDS-PAGE under reducing conditions. (B) U937 cells were lysed and incubated at 37°C for 10 minutes. Extracts were immunoprecipitated with rabbit polyclonal antibodies recognizing cathepsin G, proteinase 3, or human leukocyte elastase. Immune complexes were separated by 12.5% SDS-PAGE and separated proteins were immunoblotted using monoclonal 3A anti–PI-6 antibody.
PI-6 coimmunoprecipitates with cathepsin G from U937 cells as a 56-kD complex. (A) U937 cells were [35S]methionine labeled, lysed, and immunoprecipitated with rabbit polyclonal antibodies recognizing cathepsin G, proteinase 3, or human leukocyte elastase. Immune complexes were separated by 12.5% SDS-PAGE under reducing conditions. (B) U937 cells were lysed and incubated at 37°C for 10 minutes. Extracts were immunoprecipitated with rabbit polyclonal antibodies recognizing cathepsin G, proteinase 3, or human leukocyte elastase. Immune complexes were separated by 12.5% SDS-PAGE and separated proteins were immunoblotted using monoclonal 3A anti–PI-6 antibody.
Because these antisera do not recognize denatured antigen, they could not be used directly in immunoblotting experiments to detect the PI-6 complex in U937 cell extracts. Instead, we used the antisera to immunoprecipitate the target proteinases from cell extracts and then used the anti–PI-6 monoclonal antibody to determine by immunoblotting whether any PI-6 or complex had been coprecipitated. As a control, we also used rabbit anti–PI-6 serum to immunoprecipitate the complex from U937 extracts. Figure 5B shows that, of the four rabbit antisera used in the experiment, only the cathepsin G and PI-6 sera immunoprecipitated proteins that were detected using the PI-6 monoclonal antibody. The major species in each case was 56 kD in size, indicating that the PI-6 proteinase complex in U937 cells almost certainly contains cathepsin G.
Kinetics of the interaction of PI-6 and cathepsin G.
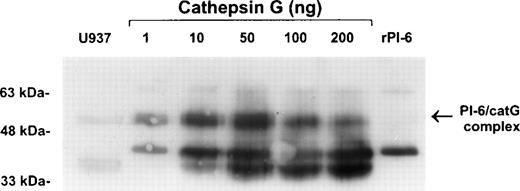
To confirm the interaction of PI-6 and cathepsin G, native human cathepsin G was incubated with purified recombinant PI-6 in varying ratios. This resulted in the formation of a complex identical in size to the complex formed in U937 cell extracts (Fig 6). As expected for a typical serpin-serine proteinase interaction, the amount of complex formed varied with the ratio of proteinase to inhibitor. As the amount of proteinase in the reaction increased, the amount of complex decreased and was accompanied by the appearance of lower molecular weight forms that probably represent the serpin cleaved in the reactive center by cathepsin G.
Recombinant PI-6 and native cathepsin G form an SDS-stable complex in vitro. Recombinant PI-6 (2 ng) was mixed with 1 to 200 ng of native cathepsin G and incubated at 37°C for 10 minutes. Samples were resolved by 10% SDS-PAGE under reducing conditions and then immunoblotted with polyclonal anti–PI-6 antisera.
Recombinant PI-6 and native cathepsin G form an SDS-stable complex in vitro. Recombinant PI-6 (2 ng) was mixed with 1 to 200 ng of native cathepsin G and incubated at 37°C for 10 minutes. Samples were resolved by 10% SDS-PAGE under reducing conditions and then immunoblotted with polyclonal anti–PI-6 antisera.
We then used standard procedures to analyze the kinetics of the PI-6 and cathepsin G interaction.26-28 The stoichiometry of inhibition of cathepsin G by PI-6 was found to be 1.0, and the inhibitory constant (Ki) was 9.2 ± 0.04 × 10−10 mol/L, indicating very tight 1:1 binding. The second order rate constant (ka) for the inhibition of cathepsin G by PI-6 was measured at 17°C as 6.8 ± 0.2 × 106 mol/L−1s−1. All attempts to accurately measure the ka at higher temperatures failed because the reaction was too fast. Under physiological conditions we estimate the lower limit for theka is 107mol/L−1s−1.
DISCUSSION
Mononuclear phagocytes and neutrophils play key roles in the elimination of bacterial and fungal pathogens via the generation of reactive oxygen species and secretion of azurophilic granule cytotoxins into phagocytic vesicles.31,33,34 Among the azurophilic granule cytotoxins are the neutral serine proteinases cathepsin G, proteinase 3, and elastase. Apart from contributing to the destruction of phagocytosed targets, these proteinases are sometimes secreted and have pronounced effects on the extracellular matrix, cytokine processing,35 chemotaxis,36 and platelet activation.37 In addition, it has been shown recently that cathepsin G also has the ability to proteolytically activate the intracellular cysteine proteinase, caspase-7, suggesting that in some circumstances cathepsin G may participate in apoptosis.38
Examples of extracellular targets of the granule proteinases include connective tissue components such as fibronectin, elastin, proteoglycans, and collagen types III and IV,39-42 and it has long been recognized that one or more of these proteinases are implicated in cell and tissue injury in several acute and chronic diseases, including emphysema, cystic fibrosis, asthma, arthritis, and psoriasis.43-46 The pathological effects of the proteinases are thought to arise through a localized imbalance between their levels and the levels of their major serum inhibitors, α1-proteinase inhibitor (α1-antitrypsin) and α1-antichymotrypsin. This hypothesis is supported by the observation that α1-proteinase inhibitor deficiency is associated with the development of emphysema.47
Clearly, control of these powerful proteinases is crucial for normal tissue homeostasis. In this study we have shown that monocytes and granulocytes produce PI-6 and that PI-6 is a potent inhibitor of cathepsin G. We believe that the PI-6–cathepsin G interaction is physiologically relevant because (1) the kinetics of the PI-6–cathepsin G interaction show that complex formation is rapid and tight, with the deduced association rate constant at the upper end of the scale of known serpin-proteinase interactions1; and (2) PI-6 and cathepsin G are produced in the same physiological compartment, suggesting that an interaction occurs in vivo. In contrast, the ov-serpin, squamous cell carcinoma antigen-2 (SCCA-2), has also recently been demonstrated to have cathepsin G inhibitory capacity.48 However, the interaction between SCCA-2 and cathepsin G is 100-fold less efficient than the PI-6–cathepsin G interaction, and there is no evidence to suggest that SCCA-2 and cathepsin G colocalize in a physiologically relevant compartment. Indeed, it is more likely that SCCA-2 is an inhibitor of mast cell chymase, because it is expressed in the epithelium where chymase-producing mast cells reside.48
The identity of the PI-6 P1 residue in the context of an interaction with cathepsin G is presently unresolved. PI-6 was initially described as an inhibitor of trypsin-like serine proteinases that by sequence comparisons with other serpins has a P1Arg.10 However, a subsequent study showed that it also inhibits chymotrypsin.21 Peptide analysis in this study demonstrated that PI-6 uses a Met upstream and adjacent to the Arg as the P1 residue for interactions with chymotrypsin and confirmed that it uses the P1 Arg for interactions with trypsin. Thus, PI-6 is a dual function inhibitor with overlapping reactive sites.
Because cathepsin G is considered to resemble chymotrypsin in overall structure and substrate preference,49 these results might imply that Met rather than Arg in the reactive center of PI-6 is crucial for cathepsin G binding. However, cathepsin G possesses an unusual active site pocket that may accomodate a variety of bulky P1 residues,50 and a very recent study has shown that it can efficiently cleave substrates after basic amino acids.51 According to this study, cathepsin G exhibits dual and equal trypsin and chymotrypsin specificities, hence it is conceivable that the PI-6–cathepsin G interaction can accomodate Arg or Met as the P1 residue. Distinguishing these possibilities will require peptide analysis of PI-6 cleaved by cathepsin G; however, the absence of Met in the corresponding position in mouse PI-652 suggests that Arg is the likely P1 residue.
Studies by Remold-O’Donnell et al8,53 54 have elegantly demonstrated that monocytes and neutrophils produce another ov-serpin, monocyte/neutrophil elastase inhibitor (M/NEI), that rapidly inactivates leukocyte elastase and, to a lesser extent, proteinase 3 and cathepsin G. M/NEI is present in the cytoplasm of monocytes and neutrophils and is thought to be released from cells during inflammation primarily to control elastase. Although cathepsin G has a different substrate preference to both elastase and proteinase 3 (which prefer small hydrophobic P1 residues), the hydrophobic nature of the PI-6 reactive center raises the possibility the latter proteinases also interact with PI-6. At present, we cannot formally exclude this, but the failure to coimmunoprecipitate elastase or proteinase 3 with PI-6 from U937 extracts suggests either that productive interactions do not occur or that cathepsin G competes very efficiently with elastase and proteinase 3 for a limited amount of PI-6.
Interestingly, M/NEI is highly homologous to PI-6 and the genes encoding these inhibitors are part of a cluster on human chromosome 6,55 suggesting a close evolutionary relationship and similarities or redundancy in function. Besides PI-6 and M/NEI, monocytes and neutrophils produce a third ov-serpin, plasminogen activator inhibitor 2 (PAI-2).5 This serpin is thought to be the major regulator of urokinase, which is present in the specific granules and is secreted during inflammation.31 PAI-2 exists predominantly within cells but can also be secreted on cell stimulation.16 The secreted form of PAI-2 is thought to regulate the extracellular activity of urokinase, but the function of the intracellular form is unknown.
Between them, PI-6 and M/NEI have the potential to efficiently regulate three major monocyte/granulocyte granule proteinases and to complement the serum serpins α1-proteinase inhibitor and α1-antichymotrypsin in vivo. It is interesting to consider why both extracellular and intracellular inhibitors of granule proteinases exist. We believe that the answer may lie in the dual functions of these proteinases—as enzymes acting within phagocytic vesicles to destroy microbes or other ingested material and as extracellular modulators of the inflammatory response. We postulate that, whereas the serum serpins act to contain extracellular collateral damage mediated by the granule proteinases, PI-6 and M/NEI act to contain intracellular damage occurring as microbes or tissue debris are ingested and destroyed. If the vacuole integrity is breached during phagocytosis, any proteinases released into the interior of the cell would be rapidly inactivated by PI-6 and M/NEI. This may be particularly important in light of the recent finding that cathepsin G can activate a pro-apoptotic proteinase (caspase-7)38 and thus has the potential to induce apoptosis if released into the cytoplasm of the host cell.
In support of this model, it is known that oxidative stress such as exposure to hydrogen peroxide can induce lysosomal rupturing in cultured fibroblasts, leading to the release of lysosomal contents, including proteases, into the cell (discussed in Brunk et al56). Although cells can apparently tolerate and repair lysosomal ruptures to a certain degree, as the amount of damage increases cells may undergo apoptosis (moderate damage) or necrosis (substantial damage). Given that myeloperoxidase is a key component of azurophilic granules and that phagocytic vesicles and lysosomes have many structural similarities, we argue that some leakage of phagocytic vesicle contents due to oxidative stress is likely to occur in monocytes, macrophages, and neutrophils. To avoid death and remain functional, these cells would require vesicle repair mechanisms similar to those described in fibroblasts56 as well as the means of countering the potentially cytotoxic components of leaking vesicles, such as cathepsin G and elastase.
In addition to countering proteinases leaking from phagocytic vesicles, PI-6 and M/NEI could also serve to inactivate any proteinase molecules that are misdirected into the cytoplasm during biosynthesis or degranulation. This model can also be extended to explain the function of intracellular PAI-2 by postulating it is present to control any urokinase (which is not directed into the phagocytic vesicle) that escapes into the cell during biosynthesis or degranulation.
Alternatively, it could be argued that PI-6 and M/NEI (perhaps like PAI-2) form an intracellular pool of inhibitors that can be rapidly released under specific conditions to operate at high local concentrations in the extracellular milieu. However, release of M/NEI from monocytes or neutrophils has not been convincingly demonstrated, whereas we have shown that PI-6 cannot be detected in media conditioned by U937 and other cells and that it cannot be released from cells through the classical secretory pathway.18 In addition, both M/NEI and PI-6 are sensitive to oxidation that indicates that they would be short-lived once outside the cell.8 10
Finally, although PI-6 is a very efficient cathepsin G inhibitor and our evidence points to a physiological role in monocytes and granulocytes, the fact remains that it is expressed in endothelial cells and many types of epithelial cells that do not produce cathepsin G and it is also a good inhibitor of trypsin-like serine proteinases.18 Thus, it is possible that the role of PI-6 extends beyond the control of cathepsin G and that it regulates one or more proteinases that have yet to be identified.
ACKNOWLEDGMENT
The authors are grateful to Dr J. Hoidal for the generous gift of antisera to cathepsin G, proteinase 3, and elastase and to Dr R. Steinman for the anti-CD14 antibody. We also thank C. O’Malley for advice and assistance with hematological procedures and Dr E. Randle-Barrett and Dr R. Pike for discussions.
Supported by a grant from the National Health and Medical Research Council, Australia.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Phillip I. Bird, PhD, Department of Medicine, Clive Ward Centre, Box Hill Hospital, Box Hill 3128, Australia; e-mail: Phil.Bird@med.monash.edu.au.





![Fig. 5. PI-6 coimmunoprecipitates with cathepsin G from U937 cells as a 56-kD complex. (A) U937 cells were [35S]methionine labeled, lysed, and immunoprecipitated with rabbit polyclonal antibodies recognizing cathepsin G, proteinase 3, or human leukocyte elastase. Immune complexes were separated by 12.5% SDS-PAGE under reducing conditions. (B) U937 cells were lysed and incubated at 37°C for 10 minutes. Extracts were immunoprecipitated with rabbit polyclonal antibodies recognizing cathepsin G, proteinase 3, or human leukocyte elastase. Immune complexes were separated by 12.5% SDS-PAGE and separated proteins were immunoblotted using monoclonal 3A anti–PI-6 antibody.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/6/10.1182_blood.v93.6.2089.406k10_2089_2097/5/m_blod40610005aw.jpeg?Expires=1767795775&Signature=qZQ5Ly1Uc2xD70DCsqw892QGqW58RpXNPNrnXO5HdN7Ia7KDo99s4rCB5oqSo0sjy6y~yjDQrmHt3M-L1vbs5AfTLv9nyOQ-lbM1yPSWd4DAJnWfbB~fM1dECxJ2ENf29pq-m1J7piOxVLptToiRkqfmd1iGkk~TS64b~V1fUziSflhHo4TDVtA1f9VAgilIGksFH6Fiz-LFFvS2fZoewW6S50C7NHd1bfnyNy6s~XTxI9drqO9M2BT8Sqs3IyPpa3K1SI7b03Sdois9fLy64icySy~TA8wnrXM64aNoEXog87OwNQWAd4u0vGSMhlERZcCe9XnubZf~JOpLeEiX9w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. PI-6 coimmunoprecipitates with cathepsin G from U937 cells as a 56-kD complex. (A) U937 cells were [35S]methionine labeled, lysed, and immunoprecipitated with rabbit polyclonal antibodies recognizing cathepsin G, proteinase 3, or human leukocyte elastase. Immune complexes were separated by 12.5% SDS-PAGE under reducing conditions. (B) U937 cells were lysed and incubated at 37°C for 10 minutes. Extracts were immunoprecipitated with rabbit polyclonal antibodies recognizing cathepsin G, proteinase 3, or human leukocyte elastase. Immune complexes were separated by 12.5% SDS-PAGE and separated proteins were immunoblotted using monoclonal 3A anti–PI-6 antibody.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/6/10.1182_blood.v93.6.2089.406k10_2089_2097/5/m_blod40610005bw.jpeg?Expires=1767795775&Signature=Gy7~ELVIEZSQzMefq3aCLk6qxIK2ISfPbb81vYV5gdxlSF8HSITaaYLCicgnGDfaQyVgl7FdvfyN3sjF0fNmuDzR6Zpb5h9Es-H~09TEoh~COV-6R3uk8nwBAEIp2lDNEu5QyI75urarl87scRla5NjcEER4EbhM-lRlkWKhkCV~YyigJqSWqyc52OnxG8I67NEOolBLBzhWCkVdT6JV8obNE8425LLjvCu9im5x-ExijBv2vopPf7nTmb9SjFvCUHGlVRvw0t3xSNNhatqIT7e2TEVYfytBqx~lrPqeQGCFFfYDgM984DXzCvd0nI1whLB4OVVGPBDpTlyO7RnXog__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal