Abstract
Bernard-Soulier syndrome is an uncommon bleeding disorder caused by a quantitative or qualitative defect in the platelet glycoprotein (GP)Ib/IX complex. The complex is composed of four subunits, GPIb, GPIbβ, GPIX, and GPV. Here we describe the molecular basis of a novel Bernard-Soulier syndrome variant in a patient in whom GPIb and GPIX were undetectable on the platelet surface. DNA sequence analysis showed normal sequence for GPIb, GPIX, and GPV. The GPIbβ gene has been mapped to the 22q11.2 region of chromosome 22 which was deleted from one chromosome of this patient. There was a single nucleotide deletion within the codon for Ala 80 in GPIbβ within the other allele. This mutation causes a translational frame shift that encodes for 86 altered amino acids and predicts a premature stop 15 amino acids short of the length of the wild-type protein. Transient coexpression of the mutant GPIbβ in 293T cells with wild-type GPIb and GPIX resulted in the surface expression of GPIb, but the absence of GPIX. Moreover, when a plasmid encoding the wild-type GPIbβ was transiently transfected into Chinese hamster ovary cells stably expressing GP, which retain the capacity to reexpress GPIX, there was a significant increase in the surface expression of GPIX. In contrast, when the mutant GPIbβ was transiently transfected into these cells, GPIX was not reexpressed on the plasma surface. Thus, a deletion of one copy of GPIbβ and a single nucleotide deletion in the codon for Ala 80 within the remaining GPIbβ allele causes the Bernard-Soulier phenotype through an interaction of GPIbβ with GPIX resulting in the absence of GPIb on the plasma membrane. The interaction of GPIbβ with GPIX is essential for the functional expression of GPIb.
THE PLATELET MEMBRANE glycoprotein (GP) GPIb/IX complex plays a major role in primary hemostasis. The complex is composed of four type 1 membrane-spanning proteins which belong to the leucine-rich motif (LRM) family of proteins.1 Each of these glycoproteins are encoded by a single copy gene located on different chromosomes.2-5 GPIb consists of two disulfide linked subunits, α (145 kD) and β (22 kD).6,7 GPIX (22 kD) binds to GPIb noncovalently, but it is not clear whether it binds to the α or β subunit on the platelet surface.8,9 GPV is also noncovalently associated with the GPIb/IX complex.10
Defects in the GPIb/IX complex which result in either a qualitative or quantitative abnormality cause the congenital bleeding disorder Bernard-Soulier syndrome (BSS)11 or platelet-type von Willebrand disease.12 BSS is usually inherited in an autosomal recessive manner and is characterized by a prolonged bleeding time, thrombocytopenia, and giant platelets.13 While platelets aggregate normally in response to agonists such as adenosine diphosphate (ADP), they do not aggregate or agglutinate in response to the agonist ristocetin, a process that depends on the interaction between von Willebrand factor (vWF) and the GPIb/IX complex.14
The α subunit of the GPIb/IX complex contains the binding site for vWF.15 Intuitively, it seems obvious that mutations which disrupt the binding site for vWF would cause BSS. Indeed the molecular defects resulting in BSS have been characterized in a number of cases16,17 to date. For example, mutations in the LRM of the α subunit of GPIb appear to cause BSS by disrupting the structure of the α subunit and hence the binding of vWF.16,18-20Other mutations in the α subunit which result in the clinical syndrome are due to premature terminations which result in a truncated protein21 or frameshifts which result in the protein not being inserted in the platelet membrane.22,23 Although a number of mutations have also been characterized in GPIX which result in BSS, it is not immediately apparent why they cause the clinical phenotype. The majority of these have been characterized in the LRM or the region flanking the LRM of GPIX.24-27 More detailed molecular analysis of these variants suggests that it is the interaction of the conserved LRM of GPIX in association with GPIbβ that is essential for the stability of the complex on the platelet surface.28
Although there is a considerable body of knowledge defining the interaction between GPIb and vWF, very little is known about the synthetic pathway of the GPIb complex. The precise mechanism of assembly and expression of the complex on the platelet surface has not been determined. Experimental data in vitro has suggested that the β-subunit is the critical unit linking GPIbα and GPIX and that the β-subunit is essential for surface expression of the complex.9 Moreover, experimental data derived from transfections in heterologous cells suggest that GPIbβ and GPIX appear to stabilize GPIbα by preventing its intracellular degradation.9 There has been a single case of a mutation within the promoter region of GPIbβ together with a deletion on the homologous chromosome in a patient with DiGeorge syndrome29that resulted in BSS. In this report we describe, to the best of our knowledge, the first mutation within the coding region for GPIbβ resulting in BSS.
MATERIALS AND METHODS
Case history.
The patient is a 13-year-old black male who has a long-standing diagnosis of BSS. The patient presented with thrombocytopenia at birth. Since then his platelet counts have ranged from 50,000 to 85,000 μL with giant “lymphocytoid-like” platelets present on peripheral blood smears. He has a life-long history of chronic easy bruisability, frequent hematomas, and severe epistaxis requiring transfusion on several occasions. It is unclear if the patient is the product of a consanguineous relationship.
Investigation of the patient’s thrombocytopenia included a normal bone marrow aspirate with the exception of increased numbers of megakaryocytes. Bone marrow chromosomes were normal and there were normal numbers of chromosomal breaks in the presence of mitomycin C. Serum immunoglobulins and coagulation screening were normal. Initial studies to investigate the diagnosis of BSS included a bleeding time of greater than 20 minutes when the platelet count was 80,000 μL. During an episode of severe epistaxis, both the bleeding time and platelet count were corrected and bleeding ceased following transfusion with platelet concentrate. Platelet function studies showed normal platelet aggregation in the presence of ADP and collagen, but platelets failed to agglutinate in the presence of 1.2 mg/mL ristocetin. The clinical diagnosis was confirmed by absent binding of the anti-GPIbα monoclonal antibody AP130 and normal binding of the anti-GPIIbIIIa monoclonal antibody AP230 to platelets.
Because BSS has previously been reported with a deletion in the DiGeorge/Velo-cardio-facial chromosomal region in 22q11.2,31 the patient was referred to Genetics for clinical evaluation and fluorescence in situ hybridization studies were performed. Apart from small stature, hypernasality, and learning disabilities, no other clinical abnormalities were noted.
Monoclonal antibodies (MoAbs) and reagents.
The anti-GPIbα antibody AP-1 blocks vWF binding to GPIbα.14 MBC 142.2, 142.6, and 142.11 are MoAbs raised against purified GPIbα that do not inhibit the binding of vWF to GPIbα.22 Anti-GPIX MoAbs FMC 25 and GRP were purchased from Harlan Bioproducts (Indianapolis, IN). AP2 is an MoAb against the GPIIb-IIIa complex.30 AK1, an MoAb which recognizes an epitope that requires the intact GPIb-IX complex32 was a generous gift of Dr Michael C. Berndt (Baker Medical Research Institute, Victoria, Australia). An affinity-purified platelet GPIbβ-specific rabbit polyclonal antibody8 was a generous gift of Dr Sandor S. Shapiro (Cardeza Foundation for Hematologic Research, Philadelphia, PA).
Blood.
Blood samples from the patient and control were collected into acid-citrate-dextrose (National Institutes of Health formula A). Platelets were isolated and washed three times by differential centrifugation, and resuspended in a buffer containing 96.5 mmol/L NaCl, 85.7 mmol/L glucose, 1.1 mmol/L EDTA, 8.5 mmol/L Tris with 50 ng/mL of prostaglandin E1 (PgE1; Sigma, St Louis, MO). Because Bernard-Soulier platelets are typically large and morphologically abnormal, a previously described sedimentation technique was used to isolate platelets from the patients.33
Platelet lysates were prepared by resuspending the platelet pellet in 500 μL of lysis buffer (96.5 mmol/L NaCl, 85.7 mmol/L glucose, 1.1 mmol/L EDTA, 8.5 mmol/L Tris, 5 mmol/L N-ethylmaleimide, 100 μg/mL leupeptin, 1 mmol/L phenylmethylsulfonyl fluoride, and 1% Triton X-100 [Pierce, Rockford, IL]). The lysate was vortexed for 3 minutes and then centrifuged at 4°C for 10 minutes at 16,000g. Aliquots of platelet lysate and platelet-poor plasma were frozen at −80°C until analyzed.
Immunoblotting.
Platelet lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on an 8% or 4% to 20% gradient gel according to Laemmli.34 The separated proteins were electroblotted onto a polyvinylidine difluoride membrane (Novex, San Diego, CA) as described by Towbin et al,35 blocked in phosphate-buffered saline (PBS) containing 5% powdered milk, then incubated overnight with the anti-GPIbα MoAb MBC 142.11 or the affinity-purified anti-GPIbβ polyclonal antibody. The membrane was washed three times with PBS containing 5% powdered milk, incubated with a goat anti-mouse or donkey anti-rabbit IgG, conjugated with horseradish peroxidase, washed again three times, and treated with SuperSignal Substrate (Pierce).
Flow cytometry of whole blood.
Platelets were analyzed by flow cytometry using anti-GPIbα MoAbs AP1 and 142.2, an anti-GPIIbIIIa MoAb AP2 and anti-GPIX MoAbs FMC 25 and GRP. Whole blood was diluted 1:10 with PBS and divided into 50-μL aliquots. Five microliters of 20 μg/mL of each primary antibody was added to the blood and incubated for 20 minutes at room temperature. A phycoerythrin (PE)-conjugated donkey anti-mouse was then added to the samples and incubated for 10 minutes at room temperature in the dark. An additional 1.5 mL of PBS was added to the samples which were then analyzed in a Becton Dickinson FACScan flow cytometer (San José, CA).
PCR amplification of genomic DNA.
Genomic DNA was isolated from peripheral blood lymphocytes as described.36 DNA was amplified by the polymerase chain reaction (PCR) using primer pairs based on the published genomic sequence of GPIbα, GPIbβ, GPIX, and GPV. For DNA sequence analysis the full-length coding region for mature GPIbα was amplified with primers 162-181 and 2634-2653.37 For GPIbβ, primers 8-30 and 767-7916 for GPIX primers 792-816 and 1547-156038 and for GPV primers 51-74 and 1877-189939 were used for amplification. The target sequences were amplified in a 50-μL reaction volume containing 500 to 1,000 ng of genomic DNA, 30 pmol of each primer, and 0.2 mmol/L of each dNTP in a reaction buffer consisting of 60 mmol/L Tris-HCl pH 9.0, 15 mmol/L (NH4)2SO4, 2 mmol/L MgCl2, 1 U of Taq polymerase (Perkin Elmer, Foster City, CA), and 4% (vol/vol) dimethyl sulfoxide (DMSO). PCR amplification was performed in a programmable thermal cycler (model 9600; Perkin Elmer) for 35 cycles of 45 seconds of denaturation at 96°C, annealing for 1 minute at 60°C, and extension for 1 minute at 72°C. PCR products containing the entire coding regions of GPIbα, β, and IX were cloned into the pCRII cloning vector using the TA cloning kit (Invitrogen, San Diego, CA).
Fluorescent in situ hybridization (FISH).
FISH was performed using a DNA sequence (TUPLE1) known to be deleted in many DiGeorge/velo-cardio-facial syndrome patients.40Hybridization and wash procedures suggested by the manufacturer (Vysis, Downers Grove, IL) were followed. Following recommended washing, 20 metaphases were scored for the presence of the TUPLE1 fluorescent signal. Examination and photography were performed on a Zeiss Axioplan with 100 W mercury epi-illumination (Gr/Zeiss, Thornwood, NY).
DNA sequencing.
Direct sequence analysis of the entire coding region of PCR-amplified GPIbα, GPIbβ, GPIX, and GPV from the subject was performed using the Prism Ready Reaction DyeDeoxy terminator cycle sequencing kit (Perkin Elmer, Foster City, CA) and an Applied Biosystems (Foster City, CA) Model 373A DNA Sequencer. Sequencing primers were synthesized on a Model 394/DNA synthesizer (Applied Biosystems).
Transient expression.
For transient expression studies, 293T cells were used. The parent 293T-cell line is a human renal epithelial cell, transformed with SV40 large T antigen.41 293T cells were maintained at 37°C in a 5% CO2 humidified chamber in modified Eagle media (Sigma) supplemented with 10% fetal calf serum.
An XhoI/MluI restriction fragment, containing the entire coding region for GPIbα was inserted into the mammalian expression vector pCI-NEO (Promega, Madison, WI). EcoRI restriction fragments containing the entire coding region of both GPIbβ and GPIX amplified from genomic DNA were inserted into pCI-Neo. Constructs containing the wild-type GPIbα, GPIbβ, GPIX, and mutant GPIbβ were sequenced to ensure that no additional mutations had been introduced and that they were inserted in the expression vector in the correct orientation. Expression plasmids were introduced into 293T cells in the presence of lipofectamine (GIBCO-BRL, Gaithersburg, MD).
Transient expression studies were also performed in Chinese hamster ovary (CHO) αIX cells (kindly provided by Dr José A. López, Baylor College of Medicine, Houston, TX). CHO αIX cells are CHO cells that stably surface-express human GPIbα.42When these cells are additionally transfected with GPIbβ, the surface expression of GPIX then becomes readily detectable.9 Cells from this stable cell line were additionally transiently transfected with either the plasmid pCI-Neo alone, the wild-type GPIbβ, or the construct containing the mutation within GPIbβ. Expression plasmids were introduced into CHO αIX cells in the presence of lipofectamine and lipofectamine plus (GIBCO-BRL), following the protocol of Felgner et al.43 In brief, either 1.5 × 106 of CHOαIX cells or 4 × 106 293T cells were plated in 100-mm dishes and grown overnight. Eight milliliters of OPTI-MEM–reduced serum media (GIBCO-BRL) containing 36 μg of lipofectamine and 6 μg of the appropriate plasmid DNA was added to the CHOαIX cells or 120 μg of lipofectamine and 8 μg DNA to the 293T cells. Following transfection after 5 hours incubation the transfection media was removed, 8 mL of culture media was added, and incubation was reinitiated at 37°C for 60 hours.
Flow cytometry studies.
Transfected cells were detached from tissue culture plates with 3 mmol/L EDTA, centrifuged at 250g, and resuspended in Hanks’ balanced salt solution with 1% bovine serum albumin and 1% normal donkey serum. Cells, 3 × 105, were transferred to each well of a 96-well V bottom plate (Dynatech, Chantilly, VA) and incubated with either a rabbit anti-glycocalicin polyclonal antibody (5 μg/mL), the anti-IX MoAbs FMC-25 or GRP (5 μg/mL), or the complex-specific MoAb AK1 (ascites, 1:1,200). The cells were then washed twice and incubated for an additional 30 minutes in a darkened room with a 1:100 dilution of PE-conjugated affinity-purified F(ab′)2 donkey anti-mouse or anti-rabbit IgG (Jackson Immunoresearch Laboratories, West Grove, PA). The cells were then washed twice, resuspended in 2% paraformaldehyde, allowed to incubate at least 1 hour at 4°C, and analyzed in a Becton Dickinson FACScan flow cytometer.
RESULTS
GPIbα is not detectable on the platelet surface.
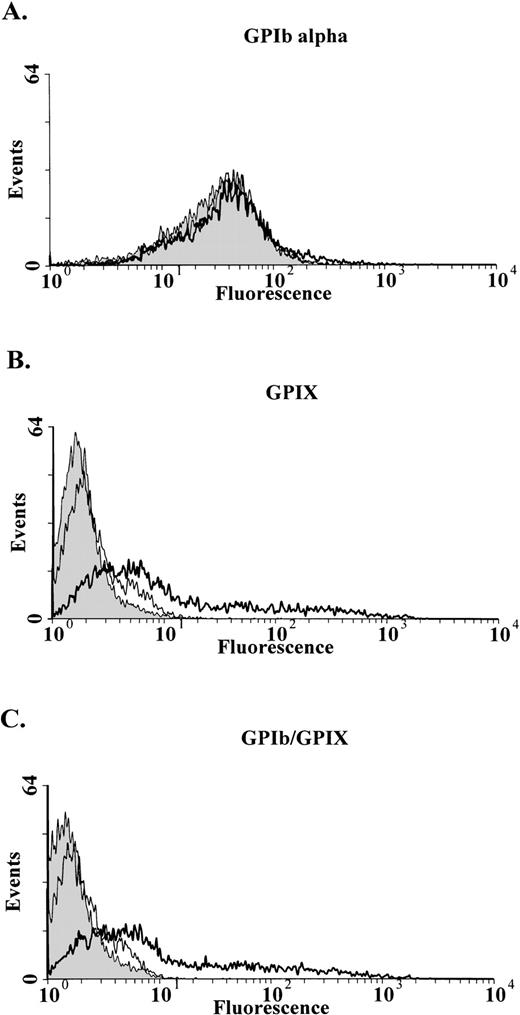
Fluorescence-activated cell sorter analysis showed that the binding of the anti-GPIIbIIIa antibody was increased compared with normal platelets (Fig 1A). This result is typically seen in large platelets.44 In contrast, the anti-GPIbα MoAb AP1 failed to bind to the patient’s platelets (Fig1B). Similarly the MoAb 142.2, which does not inhibit binding of vWF to GPIb, also did not bind the patient’s platelets (data not shown). GPIX was also undetectable on the platelet surface with either of the MoAbs FMC 25 (Fig 1C) or GRP (data not shown).
Flow cytometric analysis of patient’s platelets. Analysis was performed on whole blood with MoAbs against GPIIbIIIa (A) GPIb (B), and GPIX (C). As expected with platelets larger than normal there is an increase in surface fluorescence when the patient’s platelets (shaded area) are reacted with the anti-GPIIbIIIa MoAb AP2 compared with a normal control (clear area). There is no detectable GPIb (B) or GPIX (C) on the platelets of the patient (shaded area) compared with a normal control (clear area).
Flow cytometric analysis of patient’s platelets. Analysis was performed on whole blood with MoAbs against GPIIbIIIa (A) GPIb (B), and GPIX (C). As expected with platelets larger than normal there is an increase in surface fluorescence when the patient’s platelets (shaded area) are reacted with the anti-GPIIbIIIa MoAb AP2 compared with a normal control (clear area). There is no detectable GPIb (B) or GPIX (C) on the platelets of the patient (shaded area) compared with a normal control (clear area).
Immunoblot analysis of platelet lysate with the MoAb 142.6 confirmed the absence of GPIbα (data not shown). Similarly, GPIbβ was not detectable by immunoblot analysis of platelet lysate (Fig2).
Western blot analysis of GPIbβ in platelet lysate. Platelet lysate from a normal individual and the patient were analyzed by immunoblotting with an anti-GPIbβ polyclonal antibody. GPIbβ is readily detectable in the normal control and is absent in the patient.
Western blot analysis of GPIbβ in platelet lysate. Platelet lysate from a normal individual and the patient were analyzed by immunoblotting with an anti-GPIbβ polyclonal antibody. GPIbβ is readily detectable in the normal control and is absent in the patient.
The patient has a deletion of one chromosome containing the GPIbβ gene and a mutation in the other.
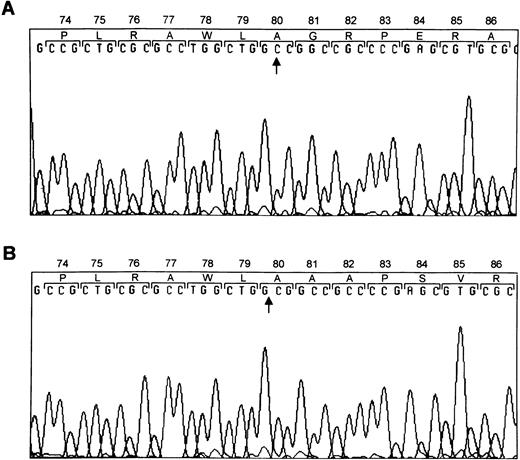
To determine the molecular basis for the absence of GPIbα on the platelet surface of this patient we initially amplified the entire coding region of GPIbα from the patient’s genomic DNA. The PCR-amplified DNA of the patient was of the predicted size according to the published sequence37 and direct sequence analysis confirmed that the sequence was normal. We then sequenced the PCR-amplified DNA for the coding regions of GPIbβ, GPIX, and GPV. The sequence of GPV and GPIX were also normal. Sequence analysis of PCR-amplified DNA from GPIbβ showed that either nucleotide 336 or 337 of the cDNA6 was deleted (Fig3). This deletion causes a shift in the reading frame beginning at amino acid residue 81, encodes 86 new amino acids, and predicts a premature stop codon 15 residues short of the wild-type protein. Twenty of 20 metaphases scored for the presence of TUPLE1 showed signal on only one chromosome 22, identified by the control sequence ARSA at 22q13. This indicates that the sequences detected by probe TUPLE1 are deleted in this patient.
Mutation within GPIbβ. DNA sequence analysis of GPIbβ from a normal individual (A) and the patient (B). Sequence analysis of PCR-amplified DNA showed that either nucleotide 336 or 337 was deleted.
Mutation within GPIbβ. DNA sequence analysis of GPIbβ from a normal individual (A) and the patient (B). Sequence analysis of PCR-amplified DNA showed that either nucleotide 336 or 337 was deleted.
Expression of the GPIb/IX complex.
Transfected alone, GPIbα is not expressed on the cell surface in appreciable quantities.45 Studies examining the role of GPIbβ and GPIX have shown that all three subunits are required for the efficient expression of the ligand-binding subunit on the surface of transfected cells.45 Therefore, to investigate the effect of this mutation on surface expression, plasmids encoding GPIbα, GPIbβ, and GPIX were transiently transfected into 293T cells. These cells were then incubated with either the anti-GPIbα polyclonal antibody, the complex specific MoAb AK1, or the anti-GPIX MoAb FMC25 followed by a PE-conjugated donkey anti-mouse antibody or donkey anti-rabbit antibody, and then analyzed by flow cytometry. As shown in Fig 4A, a significant fraction of the cells transfected with the wild-type constructs exhibited an appreciable increase in surface fluorescence when reacted with the anti-GPIbα polyclonal antibody compared with a mock control. In contrast, in 293T cells transfected with the wild-type GPIbα, wild-type GPIX, and mutant GPIbβ, there was significantly less GPIbα detectable on the cell surface compared to the transfection with the three wild-type constructs. In cells transfected with the three wild-type subunits, GPIX was readily detectable on the cell surface (Fig 4B), whereas GPIX was not detected on the cells transfected with the wild-type GPIbα, GPIX, and mutant GPIbβ. Thus, in the presence of the mutant GPIbβ, GPIX is not detectable on the cell surface. Although GPIbα was detectable on the surface of cells transfected with the wild-type GPIbα, GPIX, and mutant GPIbβ, it did not react with the complex specific antibody AK1 (Fig 4C). These results suggested an important interaction between GPIbβ and GPIX since GPIX was not detected on the cell surface in the presence of the mutant GPIbβ. Furthermore, the complex specific antibody AK1 failed to recognize the residual amount of GPIbα that was expressed in the cells transfected with the wild-type GPIbα, GPIX, and mutant GPIbβ.
Analysis of GPIb and GPIX in 293T cells transfected with GPIb, GPIbβ, and GPIX. 293T cells were transiently transfected with the wild-type GPIb, GPIbβ, and GPIX or with the wild-type GPIb GPIX and mutant GPIbβ. The cells were then analyzed with an anti-GPIb polyclonal antibody, the anti-GPIX antibody FMC25 or the complex specific antibody AK1 (each figure is representative of four different experiments). (A) In the cells transfected with the wild-type GPIb, GPIbβ, and GPIX, there is a significant increase in fluorescence when the cells are reacted with the anti-GPIb polyclonal antibody (bold lines) compared with mock transfected cells (shaded area). When cells are transfected with the mutant GPIbβ and wild-type GPIb and GPIX, GPIb is detectable on the cell surface (thin line), but significantly reduced compared with the triple wild-type transfections. (B) When cells are transfected with the wild-type GPIb, GPIbβ, and GPIX, GPIX is readily detectable on the cell surface (bold line). However, there is no significant difference between the mock transfected cells (shaded area) and in cells transfected with the wild-type GPIb, GPIX, and mutant GPIbβ (thin line). Thus, GPIX is not detectable when the mutant GPIbβ is transfected with the wild-type GPIb and GPIX (thin line). (C) GPIb that is expressed on the cell surface of the wild-type triple transfection is recognized by the complex specific antibody AK1 (bold lines). In contrast there is no difference between the mock transfected cells (shaded area) and in cells transfected with the wild-type GPIb, GPIX, and mutant GPIbβ (thin line). Thus, GPIb that is expressed on the cell surface (A) in the transfections involving the mutant GPIbβ is not recognized by AK1, confirming the lack of surface expression of GPIX.
Analysis of GPIb and GPIX in 293T cells transfected with GPIb, GPIbβ, and GPIX. 293T cells were transiently transfected with the wild-type GPIb, GPIbβ, and GPIX or with the wild-type GPIb GPIX and mutant GPIbβ. The cells were then analyzed with an anti-GPIb polyclonal antibody, the anti-GPIX antibody FMC25 or the complex specific antibody AK1 (each figure is representative of four different experiments). (A) In the cells transfected with the wild-type GPIb, GPIbβ, and GPIX, there is a significant increase in fluorescence when the cells are reacted with the anti-GPIb polyclonal antibody (bold lines) compared with mock transfected cells (shaded area). When cells are transfected with the mutant GPIbβ and wild-type GPIb and GPIX, GPIb is detectable on the cell surface (thin line), but significantly reduced compared with the triple wild-type transfections. (B) When cells are transfected with the wild-type GPIb, GPIbβ, and GPIX, GPIX is readily detectable on the cell surface (bold line). However, there is no significant difference between the mock transfected cells (shaded area) and in cells transfected with the wild-type GPIb, GPIX, and mutant GPIbβ (thin line). Thus, GPIX is not detectable when the mutant GPIbβ is transfected with the wild-type GPIb and GPIX (thin line). (C) GPIb that is expressed on the cell surface of the wild-type triple transfection is recognized by the complex specific antibody AK1 (bold lines). In contrast there is no difference between the mock transfected cells (shaded area) and in cells transfected with the wild-type GPIb, GPIX, and mutant GPIbβ (thin line). Thus, GPIb that is expressed on the cell surface (A) in the transfections involving the mutant GPIbβ is not recognized by AK1, confirming the lack of surface expression of GPIX.
To further evaluate the interaction between the mutant GPIbβ, GPIbα, and GPIX, wild-type and mutant GPIbβ were transfected into CHOαIX cells and the expression of GPIbα and GPIX were compared. In this stable cell line GPIbα was readily detectable on the cell surface in mock transfected cells and in cells transfected with the wild-type and mutant GPIbβ (Fig 5A). Furthermore, there was no difference in the expression of GPIbα when the CHOαIX cells were additionally transfected with plasmid alone, the wild-type or mutant GPIbβ (Fig 5A). When the wild-type GPIbβ was transfected into these cells there was a marked increase in the surface expression of GPIX compared with the mock transfected CHOαIX cells or CHOαIX transfected with the mutant GPIbβ (Fig 5B). When the CHOαIX cells transfected with the wild-type GPIbβ were reacted with the complex specific antibody AK1, there was a marked increase in surface fluorescence compared to the cells transfected with either the mock control or the mutant GPIbβ (Fig 5C).
Analysis of GPIb and GPIX in CHOIX cells transiently transfected with GPIbβ. (A) CHOIX cells were additionally transfected with the wild-type GPIbβ, the mutant GPIbβ, or mock transfected with the expression plasmid alone. GPIb is readily detectable in cells transfected with plasmid alone (shaded area), the mutant (thin lines), or wild-type GPIbβ (bold lines). (B) There was a significant increase in the surface expression of GPIX when the wild-type GPIbβ (bold lines) is transfected into CHOIX cells, compared with the mock control (shaded area) or cells transfected with the wild-type GPIb, GPIX, and mutant GPIbβ (thin line). (C) CHOIX cells transfected with the wild-type GPIbβ (bold lines) were reacted with the complex specific antibody AK1. Again, there is a marked increase in surface fluorescence compared with either the mock transfected cells (shaded area) or the cells transfected with the mutant GPIbβ (thin lines).
Analysis of GPIb and GPIX in CHOIX cells transiently transfected with GPIbβ. (A) CHOIX cells were additionally transfected with the wild-type GPIbβ, the mutant GPIbβ, or mock transfected with the expression plasmid alone. GPIb is readily detectable in cells transfected with plasmid alone (shaded area), the mutant (thin lines), or wild-type GPIbβ (bold lines). (B) There was a significant increase in the surface expression of GPIX when the wild-type GPIbβ (bold lines) is transfected into CHOIX cells, compared with the mock control (shaded area) or cells transfected with the wild-type GPIb, GPIX, and mutant GPIbβ (thin line). (C) CHOIX cells transfected with the wild-type GPIbβ (bold lines) were reacted with the complex specific antibody AK1. Again, there is a marked increase in surface fluorescence compared with either the mock transfected cells (shaded area) or the cells transfected with the mutant GPIbβ (thin lines).
DISCUSSION
The results of this investigation show that a single nucleotide deletion within the coding region of GPIbβ results in BSS. The wild-type GPIbβ protein contains 181 amino acids. The deletion described in this investigation causes a shift in the reading frame beginning at amino acid residue 81 within GPIbβ, encodes for 86 altered amino acids, and predicts a premature stop codon 15 residues short of the wild-type protein. We were unable to detect GPIbα, GPIbβ, or GPIX on the patient’s platelets. The absence of detectable GPIbα on the patient’s platelets appears to be caused by a defective interaction of the mutant β-subunit with GPIX.
The functional role of GPIbα within the GPIb/IX complex has been well studied. Investigations using both recombinant peptides,46site-directed mutagenesis,47 and naturally occurring mutations48 have shown the importance of GPIbα in binding vWF. In contrast, the role of each of the subunits in the assembly and coordinate surface expression of the complex is less clear. Although GPV has a role in the binding of thrombin49 to the platelet surface, to date no mutations have been described in GPV which have resulted in BSS. Furthermore, GPV does not appear to be essential for the surface expression of a functional complex.45
The role of GPIX in the surface expression of the complex is less clear. Mutations have been described in GPIX which result in the Bernard-Soulier phenotype.24-27,50 More detailed characterization of these mutations have suggested that it is the interaction of the LRM of GPIX with GPIbβ that is essential for the surface expression of GPIbα.28 In each of the cases with mutations in GPIX, there have been barely detectable amounts of GPIbα in platelets.24-27,50 GPIX mutations appear to disrupt the GPIb/IX structure as evidenced by exposure of a cryptic site on GPIbβ25 and decreased expression of GPIX through its failure to interact with GPIbβ.26 Sae-Tung et al28 investigated the interaction between GPIbβ and GPIX. They showed that the expression of wild-type GPIX on the cell surface was considerably greater when it was cotransfected with GPIbβ than when it was transfected alone.28 In contrast, when mutant GPIX polypeptides were transfected alone or in combination with GPIbβ, GPIX was not detectable on the cell surface. The results of the present investigation also support a direct interaction between GPIbβ and GPIX in the surface expression of GPIX and confirm the corollary experiments of Sae-Tung et al.28 In the present investigation we have shown that residual amounts of GPIbα were expressed on the surface of 293T cells when cotransfected with the wild-type GPIX and mutant GPIbβ, whereas in the patient no GPIbα was detected on the platelet surface. The apparent difference between the phenotype observed in the patient and the results obtained in vitro probably result from using a eukaryotic expression vector in which transcription is greatly exaggerated.22 Because platelets lack a nucleus they do not have the ability to synthesize new proteins; consequently, any GPIbα that may be made in this patient is below the physiologically relevant range and results in the described phenotype.
GPIbβ contains a single LRM; the flanking regions of this motif are homologous to GPIbα and GPIX. On the carboxyl terminus of GPIbα ligand-binding region there are four cysteines that form two disulfide bonds that are essential for binding of vWF. The precise disulfide bonding pattern for GPIbβ has not been determined; however, it has been assumed to be similar to GPIbα based on homology with GPIbα and other members of the LRM family.7 There are nine cysteines in the extracellular region of the β chain, one of these is involved in a disulfide bond linking the α and β chain. The remaining eight cysteine residues probably form four intrachain disulfide bonds. The frameshift caused by this mutation occurs within a postulated disulfide loop on the carboxy terminus of the LRM, effectively disrupting this region. Hydropathy analysis of the mutant protein showed no potential transmembrane region. A somewhat similar mutation has been described by Kunishima et al.44 They described a patient with two independent single nucleotide substitutions in the GPIbβ gene, one converting Tyr to Cys at residue 88 and the other converting Ala to Pro at residue 108. Their patient had giant platelets with a low expression of GPIbα and GPIX on the platelet surface. Furthermore, GPIbβ did not appear to be covalently linked to GPIbα because of impaired disulfide bonding. However, the patient described by Kunishima et al44 did not have thrombocytopenia and the patient’s platelets were able to bind vWF normally in the presence of ristocetin and botrocetin. In the patient described by Kunishima et al,44 immunoprecipitation studies suggested a direct association between GPIbα and GPIX.
Wu et al,8 using purified platelet complex and platelets, have also shown a noncovalent association of GPIX with GPIbα in the platelet membrane complex. In contrast to the results of Wu et al,8 López et al9 showed in transfected cells that GPIbβ was essential for the efficient synthesis and the surface expression of both GPIX and GPIbα and that GPIbβ is the critical unit linking GPIbα and GPIX. The results of the present investigation with a naturally occurring mutation within GPIbβ directly support the conclusion of López et al9 that GPIbβ is the critical subunit linking GPIbα and GPIX.
The GPIbβ gene has been localized to chromosome 22q11.2.3This is within a region in 22q11 that is deleted in 90% of patients with DiGeorge/Velo-cardio-facial syndrome.29 While BSS is almost always inherited in an autosomal recessive manner, Ludlow et al29 reported a patient with a mutation within the promoter for GPIbβ, which was also unmasked by a chromosomal deletion resulting in the DiGeorge/Velo-cardio-facial syndrome. Because of the association between a microdeletion in the DiGeorge chromosomal region of one allele of chromosome 22 and BSS,29 our patient was referred to Genetics where the diagnosis of Velo-cardio-facial syndrome was made on the basis of small stature, learning disabilities, and hypernasality. As in our patient, the diagnosis of Velo-cardio-facial syndrome in patients with subclinical features is often overlooked.51
In summary, we have identified a novel mutation within the coding region for GPIbβ that results in BSS. This mutation appears to cause the observed phenotype by disrupting the critical interaction of GPIbβ with GPIX. The interaction of GPIbβ with GPIX is essential for the normal platelet surface expression of the ligand binding GPIbα.
Supported by US Public Health Service Grants No. HL56027 (D.K.), HL44612, and HL33721 (R.R.M.), and Grant No. 95007200 from the National American Heart Association (D.K.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Dermot Kenny, MD, Department of Clinical Pharmacology, Royal College of Surgeons in Ireland, 123 St Stephen’s Green, Dublin 2, Ireland; e-mail: dkenny@rcsi.ie.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal