In adult acute myeloid leukemia (AML), the weight of the contribution of the combined activity of Pgp and MRP1 to drug resistance is not known. To address this question, we compared the activity of these proteins to the in vitro resistance to daunorubicin (DNR), etoposide, and cytosine arabinoside (Ara-C), using the calcein-AM uptake and the 3-[4, 5-di-methyl-thiazol-2, 5-diphenyl] tetrazolium bromide (MTT) assay in 80 adult AML patients. We found no correlation or only a weak correlation between the in vitro drug resistance to DNR and etoposide and MRP1 or Pgp expression or function when tested separately. However, a strong correlation was observed between the simultaneous activity of MRP1 and Pgp (quantified as the modulation of calcein-AM uptake by cyclosporin A and probenecid) and the LC50 of DNR (r = .77, P < .0001). This emphasized the role of these two proteins, not separately, but together in the resistance to DNR. In contrast, Mvp/LRP expression did not correlate with the LC50 of DNR. A high level of simultaneous activity of Pgp and MRP1 was predictive of a poor treatment outcome (for achievement of CR [P = .008], duration of relapse-free survival [RFS; P = .01], and duration of overall survival [OS; P = .02]). In addition, high LC50 of DNR and high LC50 of etoposide together were also predictive of a poor treatment outcome (for duration of RFS [P= .02] and duration of OS [P = .02]). The unfavorable cytogenetic category was more closely associated with the combined activity of both MRP1 and Pgp (P = .002) than with the activity of Pgp or MRP1 separately. This could explain the poor prognosis and the in vitro resistance to daunorubicin in this group of patients. These data suggest that treatment outcome may be improved when cellular DNR and etoposide resistance can be circumvented or modulated. Modulation of not only Pgp but also MRP1 could be essential to attain this aim in adult AML.
MULTIDRUG RESISTANCE (MDR) of some cancers, particularly acute myeloid leukemia (AML), remains a major obstacle to successful chemotherapy. The best-characterized resistance mechanism in AML which has been shown to be associated with poor outcome is mediated by the MDR1 gene and expression of membrane P glycoprotein.1 But alternative proteins, such as the more recently recognized multidrug-associated protein (MRP1)2 or the lung-resistance protein (Mvp/LRP),3 may also contribute to the resistance to anthracyclines and etoposide in AML. However, the role of these two proteins are still under discussion.4-10In several publications, the expression of Pgp did not correlate with its function of drug efflux.1,11 For this reason, determining the functional role appears to be more informative than quantification of MDR proteins. In previous studies, we have shown in cell lines and in AML that the quantification of calcein-acetoxymethylester (calcein-AM) uptake (with or without specific modulator[s] of MRP1 and/or Pgp) can be used to assess the activity of both MRP1 and Pgp.10,12 Calcein-AM, which is a substrate of both Pgp and MRP1, becomes fluorescent after the cleavage of calcein-AM by cellular esterases, producing a measurable fluorescent derivate calcein in flow cytometry.10 12-16
Despite its association with clinical resistance, MDR1expression was not correlated with in vitro resistance to daunorubicin (DNR) and etoposide, using the quick and semi-automatized 3-[4, 5-di-methyl-thiazol-2, 5-diphenyl] tetrazolium bromide (MTT) assay, in several studies.17-19 However, both we and others have shown that MDR1 and MRP1 gene overexpression emerged in a sequential manner during selection of different leukemic cell lines by drugs.20-22 The overexpression of the MRP1 gene preceded that of the MDR1 gene; afterward, MRP1 andMDR1 were co-overexpressed. In light of these results,MRP1 gene overexpression is probably an early event in the development of drug resistance, and clinical trials that modulated only Pgp might have limited or no success. In addition, both we and van der Kolk et al have shown that MRP1 was functional in fresh leukemic blast cells.10,23 Therefore, it is important to study the combined activity of Pgp and MRP1. To date, no study has analyzed the correlations between the simultaneous activity of Pgp and MRP1 and in vitro and in vivo drug resistance to DNR or etoposide, even though coexpression of these two proteins is common in adult AML.4Understanding of these relationships can help unravel the mechanisms of resistance to anthracyclines and etoposide which are clinically relevant in adult AML. In addition, such studies can emphasize the contribution of the combined activity of Pgp and MRP1 in comparison to other mechanisms.
Therefore, we have studied the contribution of Pgp, MRP1, and Mvp/LRP expression and Pgp and MRP1 function (using calcein-AM) to the in vitro resistance to DNR and etoposide and to the in vivo treatment result in 80 adult AML patients.
MATERIALS AND METHODS
Patients.
Between July 1995 and December 1997, 80 samples from adult AML patients (60 de novo and 20 relapsed AML) were successfully tested. The diagnosis was based on French-American-British (FAB) criteria.24,25 Immunophenotyping was performed by using flow cytometry. Promyelocytic leukemia (AML3) patients were excluded from the study (because of retinoic acid treatment). For each patient, several clinical and biological characteristics were analyzed (age, white blood cell [WBC] count at diagnosis, CD34 expression, and karyotype). Unfavorable karyotypes were defined as t(9;22) or abnormalities of chromosomes 5 or 7, abnormalities of 11q2.3 band, or complex abnormalities. Inversion in chromosome 16 (inv 16) or t(8;21) indicated good prognosis, and the other karyotypes, including normal, indicated intermediate prognosis.26 Only untreated de novo AML patients (60 patients) were analyzed for treatment outcome. De novo AML patients, in our department, were included in the European Organization for the Research and Treatment of Cancer (EORTC) leukemia cooperative group protocols (AML-13 for patients ≥60 years old, and AML10 for patients <60 years old). In induction phase, all the patients received a standard dose of cytosine arabinoside (Ara-C) (100 mg/m2/d × 10 days), etoposide, and one anthracycline (DNR, idarubicin, or mitoxantrone × 3 days) at random.
Level of MDR1, MRP1, and Mvp/LRP mRNA expression.
The level of MDR1, MRP1, and Mvp/LRP mRNA expression measured by reverse transcriptase-polymerase chain reaction (RT-PCR) was described elsewhere.4,10,27 The variations between samples in the cDNA synthesis were normalized by their relative quantities of β2 microglobulin (β2m) amplified by 23 cycles of PCR. The normalized yield of MDR products relative to β2m were then compared with those of A549 cells for MRP1 and Mvp/LRP (a cell line that expressed a high level of MRP1 and Mvp/LRP) and to those of HL60/DNR for MDR1 (a cell line that expressed a high level of MDR1), which were defined as 1 arbitrary unit. All samples contained more than 80% of leukemic cells. Percentage of blast cells was determined by the May-Grünwald-Giemsa staining and by immunophenotyping performed by flow cytometry. We performed this test in 75 of the 80 patients. Correlations with clinical outcome were largely performed using results of RT-PCR as a continuous variable, in accordance with consensual recommendations.28-30
Levels of Pgp, MRP1, and Mvp/LRP protein expression.
Pgp, MRP1, and Mvp/LRP protein expression was measured by labeling fresh viable cells with the UIC2, MRPm6, and LRP56 monoclonal antibodies (MoAbs), respectively, and phycoerythrin (PE)-labeled second antibody as described before.10 The expression of MDR proteins was established with blast cells selected by CD34 antibody (HPCA2 clone; Becton Dickinson, Le Pont de Claix, France) (two-color assays) or other markers (for example CD33/CD7, CD33/CD2, CD33/CD19, or CD33/CD22 by three-color assays) whenever possible, or with physical characteristics only if blast cells did not express characteristic markers. Fluorescence was analyzed on a FACSORT flow cytometer (Becton Dickinson). Values were expressed as adjusted for control, ie, the ratio of MoAb fluorescence/control antibody fluorescence. We performed this test in 75 of the 80 patients. Correlations with clinical outcome were largely performed using the fluorescence ratio as a continuous variable, in accordance with consensual recommendations.28-30
Functional analysis of Pgp and MRP1 using calcein-AM.
Cells exposed to the nonfluorescent calcein-AM become fluorescent after the intracytoplasmic cleavage of calcein-AM by cellular esterases which produced the fluorescent derivate calcein. Both Pgp and MRP1 actively extruded calcein-AM.12,13 When we measured calcein-AM uptake by flow cytometry, we assessed the amount of fluorescent calcein that had been converted from nonfluorescent calcein-AM. When the Pgp and/or MRP1 proteins were active, less calcein-AM was retained and less was converted to fluorescent calcein. Therefore, calcein-AM uptake (with specific modulators of Pgp and/or MRP1) could be used to assess whether Pgp and/or MRP1 were functional.10,12-15,31,32 In our previous studies, calcein-AM uptake ± cyclosporin A (CsA) provided in AML cells a functional test as specific and sensitive as Rh123 ± CsA,10 the most specific and sensitive Pgp functional test.15,31 Calcein-AM uptake ± probenecid also provided a functional test for MRP1 in leukemic cells. Probenecid was used as specific modulator of MRP1 activity.10,12 33
We performed this functional test in 40 adult AML patients among the 60 de novo AML patients. Cells were incubated with 0.1 μmol/L of calcein-AM for 15 minutes at 37°C in RPMI medium without or with modulators (only CsA [2 μmol/L] for Pgp function, only probenecid [2 mmol/L] for MRP1 function, or both CsA and probenecid together to assess the simultaneous activity of Pgp and MRP1). Cells were washed twice in cold phosphate-buffered saline (PBS) and samples were analyzed with a FACSORT flow cytometer. One example is shown in Fig 1. All samples were analyzed without fixation. All the data were calculated as the ratio of drug fluorescence with modulator(s) divided by drug fluorescence without modulator after subtraction of the fluorescence of the control. Dead cells were gated out following scatter characteristics.34The function of MDR proteins was established with blast cells selected as above. Correlations with clinical outcome were largely performed using fluorescence ratio as a continuous variable, in accordance with several consensual recommendations.28-30
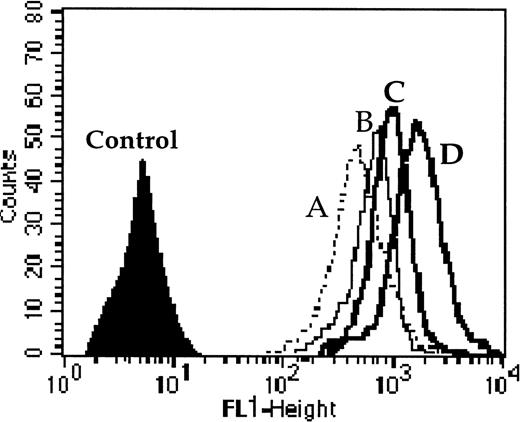
One example of Pgp and MRP1 activity quantified by the effect of probenecid ± CsA (modulators of MRP1 and Pgp, respectively) on the level of calcein-AM uptake. Cell fluorescence (A) without modulator, (B) with probenecid, (C) with CsA, and (D) with both probenecid and CsA together. The results were calculated as the ratio of drug fluorescence with modulator divided by drug fluorescence without modulator after subtraction of the fluorescence of the control. For this example the ratios were 1.51 with probenecid (which quantified MRP1 activity); 1.7 with CsA (which quantified Pgp activity); and 2.66 with both probenecid and CsA (which quantified the combined activity of MRP1 and Pgp).
One example of Pgp and MRP1 activity quantified by the effect of probenecid ± CsA (modulators of MRP1 and Pgp, respectively) on the level of calcein-AM uptake. Cell fluorescence (A) without modulator, (B) with probenecid, (C) with CsA, and (D) with both probenecid and CsA together. The results were calculated as the ratio of drug fluorescence with modulator divided by drug fluorescence without modulator after subtraction of the fluorescence of the control. For this example the ratios were 1.51 with probenecid (which quantified MRP1 activity); 1.7 with CsA (which quantified Pgp activity); and 2.66 with both probenecid and CsA (which quantified the combined activity of MRP1 and Pgp).
MTT cytotoxicity test.
In vitro sensitivity of cells to DNR, Ara-C, and etoposide was determined by planting 2 × 105 cells in a 200-μL growth medium, without any specific growth factor, containing several dilutions of the drug in 96-well microtiter plates. Each concentration of drugs was repeated in six wells. After incubation for 3 days at 37°C with 5% CO2, cell viability was determined using the MTT assay as described by Plumb et al.35 Briefly, 20 μL of MTT (5 mg/mL in PBS) was added to each well and incubated for 6 hours. The medium and MTT were then removed from the wells by centrifugation, and formazan crystals were dissolved in 200 μL of dimethyl sulfoxide (DMSO). The absorbance was recorded in a microplate reader (Model MR5000; Dynatech Laboratories, France) at the wavelength of 550 nm. The effect of drug on growth inhibition could be assessed as: % of Growth Inhibition = 1 − [(Absorbance of Drug-Treated Cells/Absorbance of Untreated Cells) × 100]. The lethal concentration 50% (LC50) was determined as the drug concentration that resulted in a 50% growth inhibition. Samples were considered evaluable if the drug-free control wells contained more than 80% of leukemic cells before and more than 70% of leukemic cells after 3 days of culture. The MTT assay gave reliable results under these conditions.36 Percentage of blast cells was determined by the May-Grünwald-Giemsa stain and by immunophenotyping that was performed by flow cytometry. In vitro drug resistance was defined as the LC50 more than the plasma peak concentration achieved in pharmacologic studies (etoposide 60 μmol/L, DNR 0.85 μmol/L, Ara-C 4 μmol/L).37-39
Statistical analysis.
Clinical and biological factors were investigated for their influence on remission rate by the χ2 or Fisher’s exact tests for binary variables and by the Mann Whitney U test for continuous variables. Correlations among levels of expression of continuous variables were estimated using the Spearman rank coefficient. The rate of (1) relapse-free survival (RFS) was measured from establishment of complete remission (CR) until relapse or death from any cause, with observation censored for patients last known alive without report of relapse; and (2) overall survival (OS) was measured from diagnosis until death from any cause, with observation censored for patients last known alive. RFS and OS were estimated by the method of Kaplan and Meier40 and compared by the log-rank test and the Breslow-Gehan-Wilcoxon test. However, data were largely reported and analyzed as continuous variables, as in the consensus recommendations.28-30 Analyses of prognostic factors for treatment outcomes were based on proportional hazards regression models for RFS and OS.41 Significance was defined as a two-tailedP value of ≤.05.
RESULTS
Expression and function of MDR variables.
We have performed both mRNA detection by RT-PCR and protein detection by flow cytometry in 70 of 80 patients. The correlation between RT-PCR and flow cytometry was good for MRP1 gene expression (r= .87, P < .0001) (Fig 2A). All the negative samples in RT-PCR (a sensitive technique) had a fluorescence ratio of MRP1 protein expression ≤1.4 and all samples with a fluorescence ratio >1.4 expressed MRP1 mRNA (Fig 2A). Therefore, we have used this threshold of positivity (1.4) for MRP1 protein expression. With this cut-off, 34% of patients expressed MRP1 protein. There was also a strong correlation between MRP1 expression by RT-PCR or flow cytometry and MRP1 activity (Fig 2B, r = .81,P < .0001; Fig 2C, r = .81, P < .0001, respectively). In some cases, the ratios of fluorescence in the MRP1 protein activity assay ranged from 0.72 to 1. This was probably caused by variations in cellular uptake of calcein-AM in the experiments. Conversely, fluorescence ratios of up to 1.28 can also represent some experimental variability. Therefore, we have used this threshold of positivity (1.28) for MRP1 protein activity (Fig 2B and C). With this cut-point, 27% of patients expressed a functional MRP1 protein.
(A) Correlation between MRP1 mRNA expression by RT-PCR and MRP1 protein expression by flow cytometry. All patients negative by RT-PCR (a very sensitive technique) expressed a fluorescence ratio from 0.81 to 1.4 in the MRP1 protein detection assay. Therefore, the threshold of positivity of 1.4 (horizontal dotted line) was used (with this cut-off of fluorescence ratio, 34% of patients expressed MRP1 protein). (B) Correlation between the effect of probenecid on calcein-AM uptake and MRP1 mRNA expression by RT-PCR in 40 patients. Three patients positive by RT-PCR assay were negative in the MRP1 activity assay. (C) Correlation between the effect of probenecid on calcein-AM uptake and MRP1 protein expression by flow cytometry in 40 patients. One patient positive by flow cytometry assay was negative in the MRP1 activity assay. In some cases, the ratios of fluorescence in MRP1 protein activity assay ranged from 0.72 to 1 (B and C). This was probably caused by variations in cellular uptake of calcein-AM in the experiments. Conversely, the fluorescence ratios of up to 1.28 can also represent some experimental variability (area between the two vertical dotted lines) (B and C). With these thresholds of positivity, there was 7.5% discordance between RT-PCR and functional assays (3 of 40 samples were MRP1+/activity−) and 2.5% between protein detection and functional assays (1 of 40 samples was MRP1+/activity−).
(A) Correlation between MRP1 mRNA expression by RT-PCR and MRP1 protein expression by flow cytometry. All patients negative by RT-PCR (a very sensitive technique) expressed a fluorescence ratio from 0.81 to 1.4 in the MRP1 protein detection assay. Therefore, the threshold of positivity of 1.4 (horizontal dotted line) was used (with this cut-off of fluorescence ratio, 34% of patients expressed MRP1 protein). (B) Correlation between the effect of probenecid on calcein-AM uptake and MRP1 mRNA expression by RT-PCR in 40 patients. Three patients positive by RT-PCR assay were negative in the MRP1 activity assay. (C) Correlation between the effect of probenecid on calcein-AM uptake and MRP1 protein expression by flow cytometry in 40 patients. One patient positive by flow cytometry assay was negative in the MRP1 activity assay. In some cases, the ratios of fluorescence in MRP1 protein activity assay ranged from 0.72 to 1 (B and C). This was probably caused by variations in cellular uptake of calcein-AM in the experiments. Conversely, the fluorescence ratios of up to 1.28 can also represent some experimental variability (area between the two vertical dotted lines) (B and C). With these thresholds of positivity, there was 7.5% discordance between RT-PCR and functional assays (3 of 40 samples were MRP1+/activity−) and 2.5% between protein detection and functional assays (1 of 40 samples was MRP1+/activity−).
The correlation between RT-PCR and flow cytometry was also strong forMDR1 gene expression (r = .95, P < .0001) (Fig 3A). All the negative samples in RT-PCR had a fluorescence ratio of Pgp expression ≤1.3 and all samples with a fluorescence ratio >1.3 expressed MDR1 mRNA (Fig 3A). Therefore, we have used this threshold of positivity (1.3) for Pgp expression. With this cut-point, 51% of patients expressed MDR1. There was also a correlation between MDR1 mRNA expression and Pgp function (Fig 3B, r = .63, P < .0001) and between Pgp expression and Pgp function (Fig 3C, r = .66, P < .0001). In some cases, the ratios of fluorescence in Pgp activity assay ranged from 0.61 to 1. This was probably because of variations in cellular uptake of calcein-AM in the experiments. Conversely, the fluorescence ratios of up to 1.39 can also represent some experimental variability. Therefore, we have used this threshold of positivity (1.39) for Pgp activity (Fig 3B and C). With this cut-point, 35% of patients expressed a functional Pgp.
(A) Correlation between MDR1 mRNA expression by RT-PCR and Pgp expression by flow cytometry (r = .95, P < .0001). All patients negative by RT-PCR expressed a fluorescence ratio from 0.7 to 1.3 in the Pgp detection assay. Therefore, the threshold of positivity of 1.3 (horizontal dotted line) was used (with this cut-off of fluorescence ratio, 51% of patients expressed Pgp). (B) Correlation between the effect of CsA on calcein-AM uptake and MDR1 mRNA expression by RT-PCR in 40 patients (r = .63, P < .0001). (C) Correlation between the effect of CsA on calcein-AM uptake and Pgp expression by flow cytometry in 40 patients (r = .66,P < .0001). In some cases, the ratios of fluorescence in Pgp activity assay ranged from 0.61 to 1. This was probably caused by variations in cellular uptake of calcein-AM in the experiments. Conversely, the fluorescence ratios of up to 1.39 can also represent some experimental variability (area between the two vertical bold lines) (B and C). With these thresholds of positivity, 14 patients (35%) were Pgp+/activity+, 12 patients (30%) were Pgp−/activity−. However, as previously described,1,6 11discrepant cases were identified, including 13 samples (32.5%) Pgp+/activity− and 1 sample (2.5%) Pgp−/activity+.
(A) Correlation between MDR1 mRNA expression by RT-PCR and Pgp expression by flow cytometry (r = .95, P < .0001). All patients negative by RT-PCR expressed a fluorescence ratio from 0.7 to 1.3 in the Pgp detection assay. Therefore, the threshold of positivity of 1.3 (horizontal dotted line) was used (with this cut-off of fluorescence ratio, 51% of patients expressed Pgp). (B) Correlation between the effect of CsA on calcein-AM uptake and MDR1 mRNA expression by RT-PCR in 40 patients (r = .63, P < .0001). (C) Correlation between the effect of CsA on calcein-AM uptake and Pgp expression by flow cytometry in 40 patients (r = .66,P < .0001). In some cases, the ratios of fluorescence in Pgp activity assay ranged from 0.61 to 1. This was probably caused by variations in cellular uptake of calcein-AM in the experiments. Conversely, the fluorescence ratios of up to 1.39 can also represent some experimental variability (area between the two vertical bold lines) (B and C). With these thresholds of positivity, 14 patients (35%) were Pgp+/activity+, 12 patients (30%) were Pgp−/activity−. However, as previously described,1,6 11discrepant cases were identified, including 13 samples (32.5%) Pgp+/activity− and 1 sample (2.5%) Pgp−/activity+.
As previously reported,10 there was a weak correlation between Mvp/LRP and MRP1 protein expression (r = .29, P= .04), but no correlation between Mvp/LRP and Pgp (r = .03,P = .84) and between MRP1 and Pgp expression (r = .25, P = .10) (data not shown). In contrast, there was a weak correlation between MRP1 and Pgp activity (r = .39,P = .008) (Fig 4).
Graphic representation of samples (A) with simultaneous activity of both Pgp and MRP1 (6 patients; 15%); (B) with MRP1 activity, without activity of Pgp (5 patients; 12.5%); (C) with Pgp activity, without activity of MRP1 (7 patients; 17.5%); and (D) without activity of both Pgp and MRP1 (22 patients; 55%). The area included between the two horizontal bold lines represents the samples with a negative activity of Pgp (fluorescence ratios of samples which ranged between 0.61 and 1.39 represent experimental variability). The area included between the two vertical dotted lines represented the samples without activity of MRP1 (fluorescence ratios of samples which ranged between 0.72 and 1.28 represent experimental variability). There was a weak correlation (r = .39, P = .008) between MRP1 and Pgp activity. *Two samples.
Graphic representation of samples (A) with simultaneous activity of both Pgp and MRP1 (6 patients; 15%); (B) with MRP1 activity, without activity of Pgp (5 patients; 12.5%); (C) with Pgp activity, without activity of MRP1 (7 patients; 17.5%); and (D) without activity of both Pgp and MRP1 (22 patients; 55%). The area included between the two horizontal bold lines represents the samples with a negative activity of Pgp (fluorescence ratios of samples which ranged between 0.61 and 1.39 represent experimental variability). The area included between the two vertical dotted lines represented the samples without activity of MRP1 (fluorescence ratios of samples which ranged between 0.72 and 1.28 represent experimental variability). There was a weak correlation (r = .39, P = .008) between MRP1 and Pgp activity. *Two samples.
We also analyzed 40 patients for combined Pgp and MRP1 activity (Fig4). In these patients, we found 6 (15%) samples with simultaneous activity of Pgp and MRP1, 22 (55%) samples without Pgp and MRP1 activity, 13 samples (32%) with Pgp activity, and 11 samples (27%) with MRP1 activity. Therefore, 18 (45%) samples had functional activity of one or both proteins (Fig 4).
The percentage of patients who expressed Pgp or MRP1 (protein or function) is shown in Table 1.
Summary of Expression and Activity of Pgp and/or MRP1 in Adult AML
| . | % of Positive Patients . |
|---|---|
| Pgp expression | 51 |
| MRP1 expression | 34 |
| Pgp function | 35 |
| MRP1 function | 27 |
| Functional activity of Pgp and MRP1 together | 15 |
| Functional activity of one or both proteins | 45-52* |
| . | % of Positive Patients . |
|---|---|
| Pgp expression | 51 |
| MRP1 expression | 34 |
| Pgp function | 35 |
| MRP1 function | 27 |
| Functional activity of Pgp and MRP1 together | 15 |
| Functional activity of one or both proteins | 45-52* |
MDR parameters and other in vitro resistance variables.
No statistically significant correlation was found between the level of Pgp expression and the LC50 of DNR (r = .29, P = .10), etoposide (r = .09, P = .62), and Ara-C (r = .12, P = .37) and between the level of MRP1 expression and the LC50 of DNR (r = .33, P = .07), etoposide (r = .23, P = .12) and Ara-C (r = .14, P = .50) (data not shown). There was also no correlation between the level of Mvp/LRP expression and the LC50 of DNR, etoposide, and Ara-C (data not shown).
Similarly, we found no correlation or only a weak correlation between the LC50 and both the effect of CsA on the level of calcein-AM uptake, which quantified only Pgp activity (r = .36, P = .02 for DNR; r = .03, P = .85 for etoposide; and r= .25, P = .16 for Ara-C) (Fig 5A and B) and the effect of probenecid on the level of calcein-AM uptake, which quantified only MRP1 activity (r = .47, P = .002 for DNR; r = .24, P= .14 for etoposide and r = −.08, P = .61 for Ara-C) (Fig 5C and D). In contrast, there was a strong correlation between the LC50 of DNR and the combined effect of probenecid and CsA (which quantified the simultaneous activity of Pgp and MRP1) on the level of calcein-AM uptake (r = .77, P < .0001) (Fig 5E), a weak correlation between the LC50 of etoposide and the combined effect of probenecid and CsA on the level of calcein-AM uptake (r = .54, P = .007) (Fig 5F) and no correlation between the LC50 of Ara-C and the combined effect of probenecid and CsA on the level of calcein-AM uptake (r = .27, P = .20) (data not shown).
Correlations between Pgp activity (measured by the effect of CsA on calcein-AM uptake) (y-axis, A and B) and the LC50 of DNR (x-axis, A) and the LC50 of etoposide (x-axis, B); between MRP1 activity (which was measured by the effect of probenecid on calcein-AM uptake) (y-axis, C and D) and the LC50 of DNR (x-axis, C) and the LC50 of etoposide (x-axis, D); and between the simultaneous activity of MRP1 and Pgp (which was quantified by the combined effect of probenecid and CsA on calcein-AM uptake) (y-axis, E and F) and the LC50 of DNR (x-axis, E) and the LC50 of etoposide (x-axis, F). When the combined effect of CsA/probenecid on calcein-AM uptake is analyzed, some cases had a fluorescence ratio which ranged from 0.59 to 1 (E and F). As for protein detection, this was probably caused by variations in cellular uptake of calcein-AM in the experiments. A similar variation in the opposite direction (1.41) can also represent some experimental variability. Therefore, we have used this threshold of positivity (1.41) for the simultaneous activity of Pgp and MRP1 (horizontal dotted line). With this cut-off, 52% of patients expressed combined activity of both MRP1 and Pgp.
Correlations between Pgp activity (measured by the effect of CsA on calcein-AM uptake) (y-axis, A and B) and the LC50 of DNR (x-axis, A) and the LC50 of etoposide (x-axis, B); between MRP1 activity (which was measured by the effect of probenecid on calcein-AM uptake) (y-axis, C and D) and the LC50 of DNR (x-axis, C) and the LC50 of etoposide (x-axis, D); and between the simultaneous activity of MRP1 and Pgp (which was quantified by the combined effect of probenecid and CsA on calcein-AM uptake) (y-axis, E and F) and the LC50 of DNR (x-axis, E) and the LC50 of etoposide (x-axis, F). When the combined effect of CsA/probenecid on calcein-AM uptake is analyzed, some cases had a fluorescence ratio which ranged from 0.59 to 1 (E and F). As for protein detection, this was probably caused by variations in cellular uptake of calcein-AM in the experiments. A similar variation in the opposite direction (1.41) can also represent some experimental variability. Therefore, we have used this threshold of positivity (1.41) for the simultaneous activity of Pgp and MRP1 (horizontal dotted line). With this cut-off, 52% of patients expressed combined activity of both MRP1 and Pgp.
MDR parameters and in vivo resistance.
Sixty untreated AML patients were evaluable for clinical response. Sixty-five percent of patients achieved CR. Variables influencing CR are shown Table 2. CR rate significantly decreased with increasing MDR1 gene expression (0.197 ± 0.022 v 0.092 ± 0.087,P = .04 by RT-PCR; 3.93 ± 2.27 v 2.10 ± 0.59,P = .04 by flow cytometry) and with increasing MRP1 gene expression (0.756 ± 0.312 v 0.212 ± 0.341,P = .05 by RT-PCR; 1.90 ± 0.43 v 1.38 ± 0.53, P = .05 by flow cytometry). However, CR rate was not associated with the level of Mvp/LRP expression by both assays (RT-PCR and flow cytometry) (Table 2). Patients who achieved CR also had a lower activity of Pgp (1.17 ± 0.27 v 1.55 ± 0.37,P = .05), a lower activity of MRP1 (1.12 ± 0.36 v1.39 ± 0.22, P = .05) and a lower simultaneous activity of MRP1 and Pgp (1.35 ± 0.47 v 2 ± 0.54, P = .008) than patients who did not (Table 2). While the CR rate significantly decreased with increasing LC50 of DNR (0.56 ± 1.32 v 0.29 ± 0.28, P = .05), it was not associated with the level of LC50 of etoposide and Ara-C. When the threshold of positivity was used for in vitro MDR variables, we obtained the same results (Table 3).
Parameters Influencing Achievement of CR
| Parameters . | Patients Who Achieved CR (patients) . | Patients Refractory to Treatment (patients) . | P Value . |
|---|---|---|---|
| In Vitro Resistance Parameters | |||
| MDR1 expression | |||
| RT-PCR* | 0.092 ± 0.087 | 0.197 ± 0.022 | .04# |
| Flow cytometry† | 2.10 ± 0.59 | 3.93 ± 2.27 | .04# |
| MRP1 expression | |||
| RT-PCR* | 0.212 ± 0.341 | 0.756 ± 0.312 | .05# |
| Flow cytometry† | 1.38 ± 0.53 | 1.90 ± 0.43 | .05# |
| Mvp/LRP expression | |||
| RT-PCR* | 0.57 ± 0.51 | 0.69 ± 0.66 | NS |
| Flow cytometry† | 3.7 ± 1.7 | 4.4 ± 3.1 | NS |
| Modulatory effect of CsA on calcein-AM uptake‡,2-153 | 1.17 ± 0.27 | 1.55 ± 0.37 | .05# |
| Modulatory effect of probenecid on calcein-AM uptake‡,2-155 | 1.12 ± 0.36 | 1.39 ± 0.22 | .04# |
| Modulatory effect of CsA + probenecid on calcein-AM uptake‡,2-154 | 1.35 ± 0.47 | 2 ± 0.54 | .008# |
| LC50 DNR (μmol/L) | 0.29 ± 0.28 | 0.56 ± 1.32 | .05# |
| LC50 AraC (μmol/L) | 5.0 ± 8.8 | 17.2 ± 28.5 | .06# |
| LC50 etoposide (μmol/L) | 14.9 ± 17.3 | 20.4 ± 28.5 | NS# |
| Clinical and Biological Parameters | |||
| Age (yr) | 48 ± 16 | 62 ± 22 | .03# |
| WBC at diagnosis (×109/L) | 77 ± 72 | 139 ± 111 | .03# |
| CD34 (% of positive patients) | 54 | 64 | NS2-160 |
| Cytogenetic (%) | .052-164 | ||
| Good | 88 | 12 | |
| Intermediate | 58 | 42 | |
| Poor | 40 | 60 | |
| Parameters . | Patients Who Achieved CR (patients) . | Patients Refractory to Treatment (patients) . | P Value . |
|---|---|---|---|
| In Vitro Resistance Parameters | |||
| MDR1 expression | |||
| RT-PCR* | 0.092 ± 0.087 | 0.197 ± 0.022 | .04# |
| Flow cytometry† | 2.10 ± 0.59 | 3.93 ± 2.27 | .04# |
| MRP1 expression | |||
| RT-PCR* | 0.212 ± 0.341 | 0.756 ± 0.312 | .05# |
| Flow cytometry† | 1.38 ± 0.53 | 1.90 ± 0.43 | .05# |
| Mvp/LRP expression | |||
| RT-PCR* | 0.57 ± 0.51 | 0.69 ± 0.66 | NS |
| Flow cytometry† | 3.7 ± 1.7 | 4.4 ± 3.1 | NS |
| Modulatory effect of CsA on calcein-AM uptake‡,2-153 | 1.17 ± 0.27 | 1.55 ± 0.37 | .05# |
| Modulatory effect of probenecid on calcein-AM uptake‡,2-155 | 1.12 ± 0.36 | 1.39 ± 0.22 | .04# |
| Modulatory effect of CsA + probenecid on calcein-AM uptake‡,2-154 | 1.35 ± 0.47 | 2 ± 0.54 | .008# |
| LC50 DNR (μmol/L) | 0.29 ± 0.28 | 0.56 ± 1.32 | .05# |
| LC50 AraC (μmol/L) | 5.0 ± 8.8 | 17.2 ± 28.5 | .06# |
| LC50 etoposide (μmol/L) | 14.9 ± 17.3 | 20.4 ± 28.5 | NS# |
| Clinical and Biological Parameters | |||
| Age (yr) | 48 ± 16 | 62 ± 22 | .03# |
| WBC at diagnosis (×109/L) | 77 ± 72 | 139 ± 111 | .03# |
| CD34 (% of positive patients) | 54 | 64 | NS2-160 |
| Cytogenetic (%) | .052-164 | ||
| Good | 88 | 12 | |
| Intermediate | 58 | 42 | |
| Poor | 40 | 60 | |
Abbreviation: NS, not significant.
Variations between samples in cDNA synthesis were normalized by their relative quantities of β2m amplified by 23-cycle PCR. The normalized yield of MDR products relative to β2m were then compared with those of A549 cells for MRP1 and LRP and with those of HL60 Pgp for MDR1, which were defined as 1 arbitrary unit.
Values were expressed as adjusted for control, ie, the ratio of MoAbs fluorescence divided by control antibody fluorescence.
Data were calculated as the ratio of drug fluorescence with modulator divided by drug fluorescence without modulator after subtraction of the fluorescence of the control.
Quantified Pgp activity.
Quantified MRP1 activity.
Quantified the simultaneous activity of Pgp and MRP1.
#Using U Mann Whitney test.
Using χ2 test.
Using Kruskal Wallis test.
In Vitro MDR Variables Influencing Achievement of CR (using the threshold of positivity)
| Variables3-150 (thresholds of positivity)3-151 . | Response to Treatment . | P Value3-152 . | |
|---|---|---|---|
| No. . | % CR . | ||
| Pgp expression (1.3) | |||
| Positive | 31 | 51 | .02 |
| Negative | 29 | 79 | |
| MRP1 expression (1.4) | |||
| Positive | 20 | 45 | .02 |
| Negative | 40 | 75 | |
| Modulatory effect of CsA on calcein-AM uptake (1.39) | |||
| Positive | 14 | 42 | .03 |
| Negative | 26 | 76 | |
| Modulatory effect of probenecid on calcein-AM uptake (1.28) | |||
| Positive | 11 | 36 | .02 |
| Negative | 29 | 75 | |
| Modulatory effect of CsA + probenecid on calcein-AM uptake (1.41) | |||
| Positive | 18 | 33 | .0001 |
| Negative | 22 | 90 | |
| Variables3-150 (thresholds of positivity)3-151 . | Response to Treatment . | P Value3-152 . | |
|---|---|---|---|
| No. . | % CR . | ||
| Pgp expression (1.3) | |||
| Positive | 31 | 51 | .02 |
| Negative | 29 | 79 | |
| MRP1 expression (1.4) | |||
| Positive | 20 | 45 | .02 |
| Negative | 40 | 75 | |
| Modulatory effect of CsA on calcein-AM uptake (1.39) | |||
| Positive | 14 | 42 | .03 |
| Negative | 26 | 76 | |
| Modulatory effect of probenecid on calcein-AM uptake (1.28) | |||
| Positive | 11 | 36 | .02 |
| Negative | 29 | 75 | |
| Modulatory effect of CsA + probenecid on calcein-AM uptake (1.41) | |||
| Positive | 18 | 33 | .0001 |
| Negative | 22 | 90 | |
None of the other in vitro resistance parameters, Mvp/LRP expression, LC50 DNR, LC50 etoposide, or LC50 Ara-C, had prognostic significance.
Using χ2 test.
RFS and OS decreased significantly with increasing LC50 of Ara-C (P = .02 and P = .001, respectively), and with increasing simultaneous activity of Pgp and MRP1 (P = .01 andP = .02, respectively) (Table 4). However, the expression (by both RT-PCR and flow cytometry) and the activity of Pgp and MRP1 separately did not influence RFS or OS (data not shown).
Significant Prognostic Variables for RFS and OS
| Variables4-150 . | RFS (P value) . | OS (Pvalue) . |
|---|---|---|
| In Vitro Resistance Parameters | ||
| Effect of both probenecid and CsA on calcein-AM uptake | .01 | .02 |
| LC50 Ara-C | .02 | .001 |
| Clinical and Biological Parameters | ||
| Age | .03 | .01 |
| Cytogenetics | .04 | .01 |
| WBC | .03 | .04 |
| Variables4-150 . | RFS (P value) . | OS (Pvalue) . |
|---|---|---|
| In Vitro Resistance Parameters | ||
| Effect of both probenecid and CsA on calcein-AM uptake | .01 | .02 |
| LC50 Ara-C | .02 | .001 |
| Clinical and Biological Parameters | ||
| Age | .03 | .01 |
| Cytogenetics | .04 | .01 |
| WBC | .03 | .04 |
Only variables with P values ≤.05 on log regression were recorded in the table. The other variables with a P value >.05 were MDR1, MRP1, and Mvp/LRP expression (by RT-PCR and flow cytometry); Pgp and MRP1 activity (the effect of either CsA or probenecid on calcein-AM uptake, respectively); LC50 DNR and LC50 etoposide; and CD34 expression.
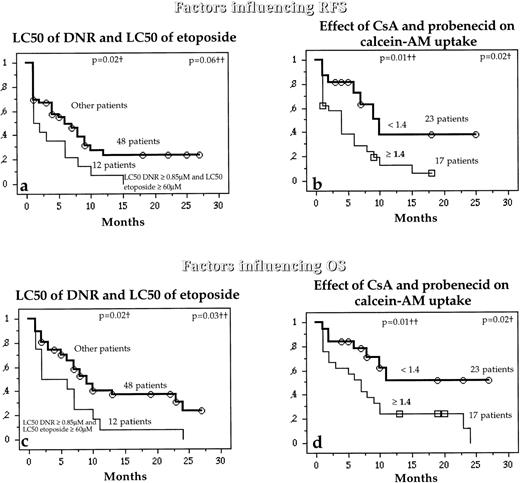
Interestingly, while the LC50 of DNR and LC50 of etoposide were not separately associated with RFS and OS, patients with both high LC50 DNR (≥0.85 μmol/L) and high LC50 etoposide (≥60 μmol/L) had higher risks of relapse or death than other patients (Fig 6a and c). Similarly, patients with high combined activities of Pgp and MRP1 also had a higher risk of relapse or death than other patients (Fig 6b and d).
Relation between the in vitro drug resistance to DNR and etoposide together (a and b) or the simultaneous effect of both CsA and probenecid on calcein-AM uptake (b and d) and the probability (which analyzed the combined activity of Pgp and MRP1) of RFS and OS in untreated de novo AML patients. †Compared by the log-rank test; ††compared by the BreslowGehan-Wilcoxon test.
Relation between the in vitro drug resistance to DNR and etoposide together (a and b) or the simultaneous effect of both CsA and probenecid on calcein-AM uptake (b and d) and the probability (which analyzed the combined activity of Pgp and MRP1) of RFS and OS in untreated de novo AML patients. †Compared by the log-rank test; ††compared by the BreslowGehan-Wilcoxon test.
Other prognostic factors and correlation with in vitro resistance variables.
The effect of other well-known variables such as age, cytogenetics, WBC count at diagnosis, and CD34 expression on clinical response were also analyzed. RFS and OS were significantly poorer for patients with unfavorable cytogenetics (P = .04 and P = .01, respectively) and decreased significantly with increasing age (P = .03 and P = .01, respectively) and increasing WBC (P = .03 and P = .04, respectively) (Table 4). CD34 expression was not a prognostic factor for RFS and OS.
Table 5 shows significant associations between older age and high mRNA MDR1 expression (P= .01), high Pgp expression (P = .01), high effect of CsA on calcein-AM uptake (P = .009), high level of LC50 of DNR (P = .05), and high level of LC50 of Ara-C (P = .04). Unfavorable cytogenetic effect was correlated with a high effect of both probenecid and CsA on calcein-AM uptake (P = .002), a high level of LC50 of DNR (P = .03), and a high level of LC50 of Ara-C (P = .02). High WBC count at diagnosis was associated with a high level of Mvp/LRP (P = .02).
Significant Associations Between Well-Known Clinical and Biological Variables on Clinical Response in AML and In Vitro Resistance Variables
| Characteristics (clinical and biological variables) . | Associated With5-150 . | P Value5-151 . |
|---|---|---|
| Older age | High MDR1 expression (RT-PCR) | .01 |
| High Pgp expression (flow cytometry) | .01 | |
| High effect of CsA on calcein-AM uptake | .009 | |
| High LC50 of Ara-C | .04 | |
| High LC50 of DNR | .05 | |
| High WBC at diagnosis | High Mvp/LRP expression (flow cytometry) | .02 |
| Unfavorable cytogenetics | Effect of both CsA and probenecid on calcein-AM uptake | .002 |
| High LC50 of Ara-C | .02 | |
| High LC50 of DNR | .03 |
| Characteristics (clinical and biological variables) . | Associated With5-150 . | P Value5-151 . |
|---|---|---|
| Older age | High MDR1 expression (RT-PCR) | .01 |
| High Pgp expression (flow cytometry) | .01 | |
| High effect of CsA on calcein-AM uptake | .009 | |
| High LC50 of Ara-C | .04 | |
| High LC50 of DNR | .05 | |
| High WBC at diagnosis | High Mvp/LRP expression (flow cytometry) | .02 |
| Unfavorable cytogenetics | Effect of both CsA and probenecid on calcein-AM uptake | .002 |
| High LC50 of Ara-C | .02 | |
| High LC50 of DNR | .03 |
The variables analyzed were: MDR1, MRP1, andMvp/LRP gene expression (measured by RT-PCR and flow cytometry), Pgp function (measured by the effect of CsA on calcein-AM uptake), MRP1 function (measured by the effect of probenecid on calcein-AM uptake), the simultaneous function of both Pgp and MRP1 (measured by the combined effect of CsA and probenecid on calcein-AM uptake), and the in vitro resistance to DNR, etoposide, and Ara-C (measured by the MTT assay).
Only associations with P value ≤.05 on log regression were recorded in the table.
DISCUSSION
While both MRP1 and Pgp may confer resistance to different families of drugs in AML, including anthracyclines and etoposide, the relative importance of these two genes is not known. Several studies reported the sequential expression of MRP1 and Pgp in drug-selected cell lines.20-22 At clinically relevant concentrations of doxorubicin or homoharringtonine, resistance to these drugs was related to MRP1 overexpression, but not to MDR1 expression in human myeloid leukemia cell lines.20,22 Only when cell lines were exposed to drugs for a prolonged time period or selected for relatively high-level drug resistance did Pgp/MDR1 overexpression become apparent. Similar findings occur in other cell lines (murine leukemia, small cell lung cancer cells).21,42,43 In light of these results,MRP1 gene overexpression is probably an early event in the development of drug resistance, and MRP1 and Pgp could be coexpressed in AML cells exposed to pharmacological doses of cytotoxic drugs. In clinical samples of AML, MRP1 overexpression ranged from 7% to 30%.4,7,8,44-49 MRP1 overexpression was more frequent in drug-refractory or relapsed patients than in drug-sensitive patients in one study,4 but not in others.46,47 Similarly, the coexpression and correlation between MRP1 and MDR1 are under debate. These contradictory results might be partially caused by differences in the composition of samples and experimental methods, as well as differences in the definition of overexpression. To date, few data on the coexpression of these two genes in AML cells have been reported, and the combined functionality of these two proteins in clinical samples has not been studied. We have shown, in previous studies, that calcein-AM uptake can be used to assess whether MRP1 and/or Pgp are functional and to assess the simultaneous activity of MRP1 and Pgp in fresh leukemic cells.10 12 In our present study, 45% of the AML samples studied exhibited a functional activity of one or both proteins. Taken together, these previous and present reports suggest that MRP1 and Pgp need to be considered together and that clinical trials that selectively modulate Pgp are likely to achieve limited success. Therefore, we analyzed the contribution of the combined activity of Pgp and MRP1 to in vitro and in vivo resistance to chemotherapy in AML patients.
In our study, the absence of or only weak correlations between MRP1 or Pgp expression or function (tested separately) and in vitro drug resistance to DNR and etoposide were in agreement with other data17-19 and could be partly explained by the separate analysis for the two resistance genes. In agreement with this, we have found a good correlation between the simultaneous activity of MRP1 and Pgp and in vitro resistance to DNR, which emphasizing the role of these two proteins together in the resistance to DNR in adult AML. In addition, probenecid, the modulator of MRP1 used in this study, has been associated with an increased accumulation of DNR and with the correction of the altered distribution of DNR in leukemic cell lines.33
Similarly, we have shown that the combined activity of MRP1 and Pgp was a prognostic factor for treatment outcome (achievement of CR, and duration of RFS and OS), but not MRP1 or Pgp separately (for RFS and OS). In several other studies, the prognostic value of MRP1 expression is discussed.4,7,8,44-49 However, in these studies only one technique was used and functionality was not assessed. As for MDR1,15 28-30 an elaboration of consensus recommendations would be required.
In contrast, preliminary studies using the MTT assay for the prediction of chemoresistance in adult AML suggest that it may be helpful for risk-group stratification in adult AML.50,51 In addition, this test has a strong value in the prediction of clinical response in childhood leukemias.52,53 It was also shown that in vitro drug resistance determined with the differential staining cytotoxicity (DiSC) assay, based on the same concept as the MTT assay, was related to survival in adult AML.54 Therefore, we used the MTT assay to assess the in vitro resistance to drugs. In our study, patients who exhibited both high LC50 of DNR and high LC50 of etoposide, but not etoposide alone, had a poorer prognosis than other patients, underlying the importance of these two drugs. In clinical trials, it was unclear whether the addition of etoposide improved treatment outcome in adult AML.55-57 But in Bishop’s randomized study, which included 264 patients, there was an additional benefit from the use of etoposide specifically confined to patients ages less than 55 years.58 There has not been another large randomized trial comparing anthracycline + Ara-C ± etoposide as induction chemotherapy. Nevertheless, our finding is in accordance with the fact that etoposide could be effective in the treament of AML. We also showed that the combined activity of MRP1 and Pgp in resistance to etoposide appears less important than their role in resistance to DNR.
We have shown that the well-known prognostic factors in AML,59 age and cytogenetics, were associated with both Pgp and MRP1 expression and function. Age has already been correlated with Pgp expression and function.6 7 In our study, the unfavorable cytogenetic category was better associated with the combined activity of both MRP1 and Pgp than with the activity of Pgp or MRP1 separately. This could explain the poor prognosis and the in vitro resistance to daunorubicin in this group of patients. Modulation of not only Pgp but also MRP1 could be essential, in this category of patients, to improve the results of treatment.
As in our previous report,10 we found a good correlation between mRNA expression detected by RT-PCR and protein expression detected by flow cytometry for both MDR1 and MRP1genes. As previously described, we have identified discrepant cases between Pgp expression and function.1,6,11 In contrast, for MRP1, discrepancy between protein expression and function were uncommon. 28-30 We recommend the detection of MRP1 expression by flow cytometry. In addition, a functional test using calcein-AM uptake assay could be used to assess the activity of MRP1 (we found 7% of cases with a discrepancy between MRP1 expression and function) and the simultaneous activity of both MRP1 and Pgp. Other functional tests can assess the activity of MRP1.60 A critical evaluation of these different assays would be useful.
Unlike the results obtained with MRP1 and Pgp, we found, as recently reported by both Leith et al7 and us,10 that the level of Mvp/LRP expression is not correlated with treatment outcome in adult AML, in contrast to other studies.5,9 In our study, the level of Mvp/LRP expression was not correlated with in vitro resistance to DNR or etoposide. Therefore, a causal mechanistic relationship of Mvp/LRP with drug resistance is still lacking. But, as we recently reported, the discrepancy in the clinical significance of Mvp/LRP expression may be related to the methodology used.61
In conclusion, all the data presented here support the hypothesis that the modulation of both Pgp and MRP1, by agents such as probenecid and PSC833, may simultaneously increase the percentage of CR, the percentage of RFS, and survival duration in adult AML patients by increasing blast cell DNR ± etoposide cytotoxicity. The concentration of probenecid that reverses MRP1 function is clinically achievable in vivo.33 However, because of the relatively small patient numbers in our study, a multicenter study will be required to evaluate all of these resistance parameters.
Supported in part by a grant from ARC (Grant No. 9637).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Ollivier Legrand, MD, PhD, Hôpital Hôtel Dieu, 1 place du parvis Notre Dame, Service d’hématologie, 181 Paris Cedex 04, France; e-mail:olivier.legrand@htd.ap-hop-paris.fr.