Signal transducers and activators of transcription (STATs) belong to a family of transcription factors that were originally identified as mediators of cytokine-induced gene expression. Recent evidence, however, has shown that certain members of the STAT family, including STAT3, are also involved in cellular transformation. Here we show that STAT5 also plays a role in cellular transformation by the BCR-Abl oncogene. In BCR-Abl transformed K562 cells, STAT5A and 5B are constitutively phosphorylated on tyrosine and are transcriptionally active. Moreover, expression of a dominant negative form of STAT5 shows that active STAT5 is necessary for the growth in soft agar of these cells. These results show that besides STAT3, STAT5 can also be involved in cellular transformation.
JANUS KINASES (JAKs) are a family of cytoplasmic tyrosine kinases, which are associated with cytokine and growth factor receptors and play a major role in cytokine signaling.1-3 On ligand binding, the JAKs are activated and subsequently phosphorylate a number of substrates including cytokine receptors. The phosphorylated receptors provide docking sites for the SH-2 domain containing STAT transcription factor family (signal transducers and activators of transcription). Subsequently, STATs are phosphorylated on a single tyrosine residue by the JAKs, after which the STATs dimerize, migrate into the nucleus, and regulate gene transcription. Although the signaling pathway seems rather simple, the availability of four different JAKs (JAK1, JAK2, JAK3, and Tyk2) and at least eight different STATs (STAT1α, STAT1β, STAT2, STAT3, STAT3β, STAT4, STAT5A, STAT5B, and STAT6) all with different DNA binding and transactivation properties allows cellular specificity in this signaling pathway.1-3
Several studies have shown that STAT proteins are essential for cytokine-regulated processes such as cellular proliferation, differentiation, as well as survival.4,5 However, more recently, it has become evident that aberrant activation of STAT proteins is often associated with cellular transformation by various oncoproteins.6 Cells transformed by v-Abl or BCR-Abl contain constitutively activated STAT1, STAT3, and STAT5.7-14 Similarly, activation of STAT1, 3, and 5 was observed in several cell lines transformed by v-Src,15-22while the Eyk oncoprotein induces activation of STAT1 and STAT3.23 More importantly, activation of STATs 1, 3, and 5 was reported in several human malignancies, including lymphomas, leukemias, and breast carcinoma.19,24-29 Although these observations strongly suggest that STAT1, 3, and 5 can play a role in cellular transformation, an essential role for STAT activation was only demonstrated in v-Src transformed cells. Dominant negative variants of STAT3 clearly inhibited transformation of fibroblasts by v-Src.18 22 Evidence for an obligatory role of STAT1 and STAT5 in cellular transformation is currently lacking.
The BCR/ABL chimeric oncogene, a constitutively active tyrosine kinase, which is generated from the Philadelphia chromosome translocation (t(9;22)(q34;q11)), causes chronic myelogenous leukemia (CML).30 Although it is clear that the tyrosine kinase activity of BCR/ABL is essential for transformation,31,32the actual mechanism by which BCR/ABL transforms cells remains largely unknown. Interestingly, BCR/ABL constitutively activates several signaling pathways33 that are also used by several cytokines, including those of the interleukin-5 (IL-5)/IL-3/granulocyte-macrophage colony-stimulating factor (GM-CSF) cytokine family.34 These pathways include the RAS-Erk2, the PI3Kinase, and the JAK/STAT pathways.33 In addition, BCR/ABL activates the phosphatases SHP1 and SHP2, which also play a pivotal role in IL-5/IL-3/GM-CSF signaling. Among the STATs, which are found to be activated in various BCR/Abl-expressing cell lines, STAT5 seems to be the most prominent.7-14 Because STAT5 was shown to be necessary for cellular proliferation induced by the IL-5/IL-3/GM-CSF cytokine family35 and overexpression of constitutively active STAT5 stimulates cell proliferation,36 it seems conceivable that it also might contribute to cellular transformation by BCR/Abl.
In this report, we have investigated the contribution of STAT5 to cellular transformation by the BCR/Abl oncogene. In K562 cells, we show that STAT5 (A and B) is constitutively phosphorylated on tyrosine and bind to a STAT5 binding site. Similarly, reporter constructs containing STAT5 binding sites are active in these cells. Importantly, we show that blocking STAT5 activity with a dominant negative expression vector significantly decreases soft agar growth of K562 cells, as well as transformation of fibroblasts by v-Src. These results suggest that STAT5 plays an important role in cellular transformation.
MATERIALS AND METHODS
Cell culture.
U937 and K562 cells were maintained in RPMI 1640 supplemented with 8% fetal calf serum (FCS) (Hyclone, Greiner, Logan, UT). NIH 3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 8% FCS (Life Technologies, Breda, The Netherlands). Focus assays on NIH-3T3 cells were performed as described previously.
Reagents and antibodies.
The phosphotyrosine monoclonal antibody (MoAb) 4G10 was obtained from UBI (Lake Placid, NY). The MoAb anti-STAT1 (S21120, directed against amino acids [aa] 592-731) was obtained from Transduction laboratory (Lexington, KY). The polyclonal antibodies (pAb) directed against STAT3 (C-20, aa 750-769), STAT5A (L-20, aa 774-793), STAT5B (C-17, aa 711-727), and STAT5 (N-20, aa 5-24) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Synthetic oligonucleotides and plasmids.
The following oligonucleotide was used in this study (only the upper strands are shown); for bandshift assays: β-casein (5′-AGCTTAGATTTCTAGGAATTCAA ATCA-3′). The expression plasmids pMXmSTAT5A, pMXmSTAT5A, Δ75037 (a kind gift of Dr Fabrice Gouilleux, Tumor Biology Center, Freiburg, Germany), pSG5-STAT3, pSG5-STAT3β, and pMvSrc were described previously.22,38 Full-length cDNAs of the different STATs were excised from the plasmids described above and cloned into the LNCX expression vector (Clontech, Palo Alto, CA), which contains a neomycin resistance gene. The reporter constructs 4xIREtkCAT, 4xGAStkCAT, 4xSIEtkCAT, and 4xβCaseintkCAT were previously described.38 39
Transfection of K562 cells.
For transient assays, K562 cells were electroporated with 2 μg of reporter plasmid and 10 μg of bluescript SK− carrier DNA. For some experiments, dominant negative expression vectors for STAT3 or STAT5 were cotransfected instead of bluescript SK−. Cells were harvested for CAT assays 48 hours after transfection. Chloramphenicol acetyltransferase (CAT) assays were performed as described previously.38
For soft agar assays, 24 hours posttransfection K562 cells were treated with 1 mg/mL G418 for 10 days. Cells were then plated in soft agar and scored as was described previously,40 except that RPMI 1640 with 10% FCS was used as medium.
Gel retardation assay.
Nuclear extracts were prepared following a previously described procedure.41 Oligonucleotides were labeled by filling in the cohesive ends with [a-32P]deoxycytidine triphosphate (dCTP) using Klenow fragment of DNA polymerase I. Gel retardation assays were performed according to published procedures.38 Supershift analysis was performed by preincubating 10 μg of nuclear extract with 1 μg of anti-STAT antibody for 30 minutes on ice before addition of the binding buffer and 32P-labeled probe.
Immunoprecipitation/Western blotting.
K562 cells (10.106 per sample) were washed with ice-cold phosphate-buffered saline and subsequently lysed in 750 μL radioimmunoprecipitation assay (RIPA) buffer (150 mmol/L NaCl, 20 mmol/L Tris pH 7.4, 5 mmol/L EDTA, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, 1 mmol/L phenylmethylsulphonylfluoride [PMSF], 2 μg/mL leupeptin, 4 μg/mL aprotinin, 1.5 μg/mL pepstatin, 1 μg/mL trypsin inhibitor, and 50 mmol/L NaF) for 10 minutes on ice. Cell lysates were centrifuged to remove the insoluble material, and supernatants were precleared with protein A-Sepharose beads for 15 minutes at 4°C. Thereafter, supernatants were incubated with STAT antibodies for 1.5 hours at 4°C. Protein A-Sepharose was added to the reaction mixture and the incubation was continued for 45 minutes. Beads were collected by centrifugation and washed eight times with cold RIPA buffer, resuspended in Laemmli sample buffer, and boiled for 3 minutes. Protein samples were separated on 8% SDS-polyacrylamide gels, and electrotransferred to Immobilon-P membranes (Millipore, Bedford MA). Membranes were blocked in TBST-buffer (150 mmol/L NaCl, 10 mmol/L Tris pH 8.0, 0.3% Tween 20) containing 5% bovine serum albumin (BSA) for 30 minutes and probed with either an antiphosphotyrosine MoAb 4G10 or the anti-STAT antibodies described above for 1 hour. After three washes with TBST, the membranes were incubated for 1 hour with either peroxidase-conjugated rabbit antimouse antibodies (after MoAb) or peroxidase-conjugated swine antirabbit antibodies (DAKO, Glostrup, Denmark) (after pAb), followed by five washes with TBST. Proteins were visualized with enhanced chemiluminescence (ECL; Amersham, Buckinghamshire, UK).
RESULTS AND DISCUSSION
Previously, it was demonstrated that BCR/Abl activates STAT5, although activation of STAT1 was also reported in some cell types.7-14 To determine which STAT family members were activated in BCR/Abl-expressing K562 cells, we performed immunoprecipitation/Western blotting experiments using antiserum specific for STAT1, 3, 5A, and 5B. Figure1A shows that both STAT5A and 5B are phosphorylated on tyrosine residues in these cells. The second band observed in the STAT5B immunoprecipitation is likely to be caused by serine phosphorylation of STAT5B (data not shown). By contrast, activation of STAT1 or 3 could not be detected. These results were confirmed by gel-shift analysis. Figure 1B shows that STAT5A and 5B constitutively bind to the STAT binding site from the β-casein promoter and from the FcγRI promoter (not shown), while DNA binding of STAT1 and STAT3 could not be observed. These results clearly show that STAT5A and 5B are the major targets for BCR/Abl in K562 cells.
Constitutive activation of STAT5A and 5B in K562 cells. (A) K562 cells (3.106 per lane) and U937 cells (negative control) were lysed in RIPA buffer, after which the tyrosine phosphorylation state of different STAT molecules was assessed by immunoprecipitation and Western blotting. Probing the blot with an antiphosphotyrosine antibody clearly shows that only STAT5A and STAT5B are phosphorylated on tyrosine in K562 cells. (B) Nuclear extracts from K562 cells were analyzed in a gel shift assay using the STAT binding site from the β-casein promoter as a probe. Supershift analysis clearly shows that STAT5A and 5B constitutively bind to the β-casein site in K562 cells.
Constitutive activation of STAT5A and 5B in K562 cells. (A) K562 cells (3.106 per lane) and U937 cells (negative control) were lysed in RIPA buffer, after which the tyrosine phosphorylation state of different STAT molecules was assessed by immunoprecipitation and Western blotting. Probing the blot with an antiphosphotyrosine antibody clearly shows that only STAT5A and STAT5B are phosphorylated on tyrosine in K562 cells. (B) Nuclear extracts from K562 cells were analyzed in a gel shift assay using the STAT binding site from the β-casein promoter as a probe. Supershift analysis clearly shows that STAT5A and 5B constitutively bind to the β-casein site in K562 cells.
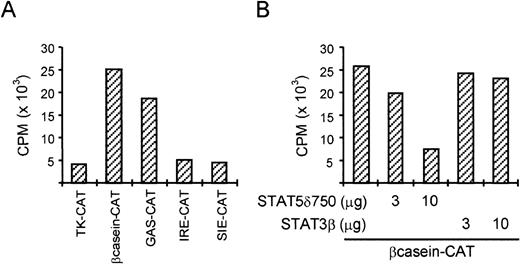
Although tyrosine phosphorylation and DNA binding of STAT5 was previously reported in BCR/Abl-expressing cells, transcriptional activation of STAT5-dependent promoters was not shown. We therefore transfected various STAT-dependent reporter constructs into K562 cells. Figure 2A shows that constructs containing STAT binding sites from the β-casein promoter (which can bind STAT1 and STAT5) and from the FcγRI promoter (GAS, which can bind STATs 1, 3, and 5) are more active than the TK-CAT control reporter. By contrast, the IRE-CAT and SIE-CAT reporters, which cannot bind STAT5, are comparable to the TK-CAT control reporter. These results suggest that STAT5 is indeed transcriptionally active in K562 cells. To further extend these results, we transfected K562 cells with the β-casein-CAT reporter together with either dominant-negative STAT5 (STAT5δ750) or STAT3β (Fig 2B). While STAT3β did not significantly alter the activity of the β-casein-CAT reporter, STAT5δ750 caused a strong repression of CAT activity, further suggesting that STAT5 is transcriptionally active in these cells.
STAT5 is transcriptionally active in K562 cells. (A) K562 cells were transfected with different reporter plasmids (2 μg) by electroporation. Two days posttransfection, cells were harvested and CAT activity was determined. The two reporters containing STAT5 binding sites (β-casein-CAT and GAS-CAT) were more active compared with the empty reporter and the reporters containing STAT1 and STAT3 binding sites (IRE-CAT and SIE-CAT). (B) K562 cells were transfected with as described in (A) with 2 μg β-casein CAT and increasing amounts of dominant negative (DN) STAT5 (δ750) or STAT3 (STAT3β). Only DN STAT5 is able to (partially) block the activity of the β-casein-CAT reporter in K562 cells.
STAT5 is transcriptionally active in K562 cells. (A) K562 cells were transfected with different reporter plasmids (2 μg) by electroporation. Two days posttransfection, cells were harvested and CAT activity was determined. The two reporters containing STAT5 binding sites (β-casein-CAT and GAS-CAT) were more active compared with the empty reporter and the reporters containing STAT1 and STAT3 binding sites (IRE-CAT and SIE-CAT). (B) K562 cells were transfected with as described in (A) with 2 μg β-casein CAT and increasing amounts of dominant negative (DN) STAT5 (δ750) or STAT3 (STAT3β). Only DN STAT5 is able to (partially) block the activity of the β-casein-CAT reporter in K562 cells.
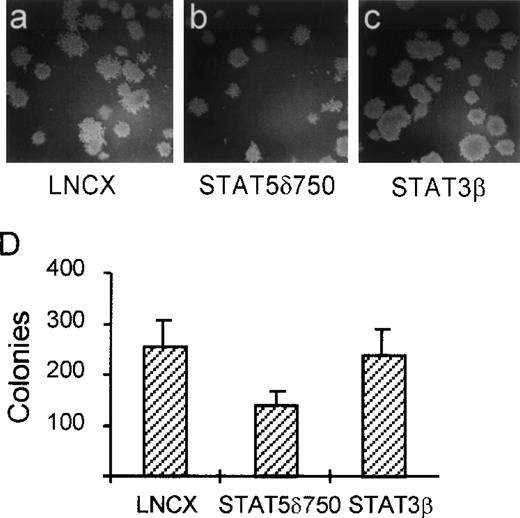
In contrast with most nontransformed hematopoietic cells, one of the transformed properties of K562 cells is their ability to grow in semisolid media (soft agar). To determine whether activation of STAT5 is involved in this property of K562 cells, we used STAT5δ750. K562 cells were transfected with STAT5δ750, STAT3β, or the empty expression vector (LNCX), after which the transfected cells were selected in G418-containing media for 10 days. Thereafter, cells were plated in dishes containing soft agar. Figure 3 shows that K562 cells transfected with an empty expression vector (LNCX) grow efficiently in soft agar (Fig 3A and D). By contrast, K562 cells transfected with STAT5δ750 form fewer and smaller colonies in soft agar (Fig 3B and D), suggesting that STAT5 is indeed involved in this aspect of cellular transformation by BCR/Abl. As a control, cells transfected with STAT3β grew as efficiently in soft agar as vector controls, further showing that STAT3 is not involved in transformation by BCR/Abl (Fig 3C and D).
STAT5 is involved in soft agar growth of K562 cells. K562 cells were electroporated with STAT5δ750 (B and C), the empty expression vector (LNCX, A and D), or STAT3β (C and D), after which the cells were selected on G418 (1 mg/mL) for 10 days. Cells were then plated in soft agar. Seven to 10 days later, colonies were counted as described in Materials and Methods. STAT5δ750 partially inhibits the capacity of K562 cells to grow in soft agar, while STAT3β did not have an effect.
STAT5 is involved in soft agar growth of K562 cells. K562 cells were electroporated with STAT5δ750 (B and C), the empty expression vector (LNCX, A and D), or STAT3β (C and D), after which the cells were selected on G418 (1 mg/mL) for 10 days. Cells were then plated in soft agar. Seven to 10 days later, colonies were counted as described in Materials and Methods. STAT5δ750 partially inhibits the capacity of K562 cells to grow in soft agar, while STAT3β did not have an effect.
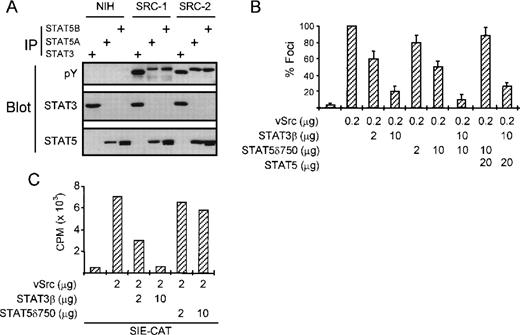
We next wanted to investigate whether STAT5 also contributes to transformation by other oncogenes. Recently, we and others have shown that STAT3 is involved in cellular transformation by v-Src.18 22 Figure4A shows that besides STAT3, STAT5A and 5B are also tyrosine-phosphorylated in v-Src–transformed NIH-3T3 cells, but not in the parental NIH-3T3 cells. However, in contrast to K562 cells, we failed to detect substantial STAT5 DNA binding activity in v-Src–transformed cells (data not shown). To investigate whether STAT5 is causally involved in v-Src–induced transformation, we performed focus assays in NIH-3T3 cells. Transfection of NIH-3T3 cells with v-Src efficiently induces focus formation in these cells (Fig 4B). As we have previously shown, this can be strongly repressed by cotransfecting STAT3β. Interestingly, cotransfection of STAT5δ750 also represses v-Src–dependent focus formation in NIH-3T3 cells, albeit much less efficiently than STAT3β. The combination of STAT3β and STAT5δ750 was somewhat more potent in repression focus formation than either plasmid alone. In addition, the repression observed with STAT5δ750 could be overcome by cotransfection of wild-type STAT5, further suggesting that STAT5 indeed plays a role in transformation by v-Src. To rule out potential a-specific effects of STAT5δ750 on STAT3-dependent signaling, we performed cotransfection of a STAT3-dependent reporter construct (SIE-CAT) together with v-Src, STAT3β, and STAT5δ750. Figure 4C shows that STAT3β efficiently blocks v-Src–induced SIE-CAT activity. By contrast, STAT5δ750 only slightly reduced v-Src–induced SIE-CAT activity, suggesting that STAT5δ750 does not repress STAT3 function in these cells. These results show that although STAT3 is the major STAT involved in transformation by the v-Src oncogene, STAT5 is also likely to play a minor role.
STAT5 is also involved in transformation by the v-Src oncogene. (A) The tyrosine phosphorylation status of STAT3 and STAT5 was analyzed in NIH 3T3 cells and two derivatives that were transformed by the v-Src oncogene. STAT3 and STAT5 are both constitutively phosphorylated on tyrosine in v-Src–transformed, but not parental NIH-3T3 cells. (B) NIH 3T3 cells were transfected with v-Src expression plasmid and increasing concentrations of DN-STAT3 and DN-STAT5 expression vectors or wild-type STAT5 as a control. Two weeks after transfection, foci were scored and represented as percent compared with v-Src alone. Both DN STAT3 and DN STAT5 partially block transformation of NIH 3T3 by v-Src, although DN STAT3 is much more potent. Wild-type STAT5 could overcome the effect of STAT5δ750, but not STAT3β. (C) NIH-3T3 cells were transfected with the STAT3-dependent SIE-CAT reporter plasmid and increasing amounts of STAT3β or STAT5δ750. Only STAT3β is able to block v-Src–induced SIE-CAT activity.
STAT5 is also involved in transformation by the v-Src oncogene. (A) The tyrosine phosphorylation status of STAT3 and STAT5 was analyzed in NIH 3T3 cells and two derivatives that were transformed by the v-Src oncogene. STAT3 and STAT5 are both constitutively phosphorylated on tyrosine in v-Src–transformed, but not parental NIH-3T3 cells. (B) NIH 3T3 cells were transfected with v-Src expression plasmid and increasing concentrations of DN-STAT3 and DN-STAT5 expression vectors or wild-type STAT5 as a control. Two weeks after transfection, foci were scored and represented as percent compared with v-Src alone. Both DN STAT3 and DN STAT5 partially block transformation of NIH 3T3 by v-Src, although DN STAT3 is much more potent. Wild-type STAT5 could overcome the effect of STAT5δ750, but not STAT3β. (C) NIH-3T3 cells were transfected with the STAT3-dependent SIE-CAT reporter plasmid and increasing amounts of STAT3β or STAT5δ750. Only STAT3β is able to block v-Src–induced SIE-CAT activity.
Taken together, we have shown that besides STAT3, STAT5 is also involved in transformation mediated by at least two oncoproteins. The mechanism by which active STAT5 contributes to cellular transformation remains to be determined, but is likely to involve constitutive activation of STAT5 target genes, which are somehow involved in the control of proliferation (eg, c-fos). In this respect, it is noteworthy that blocking STAT5 function in mouse BaF3 cells results in a significant decrease in IL-3–dependent proliferation,35while a constitutively active form of STAT5 renders IL-3–dependent cells partially IL-3–independent.36 On the other hand, because STAT5 is involved in the regulation of the antiapoptotic Bcl2 homologue A1,42 enhanced survival and escape from apoptosis of cells containing active STAT5 might also contribute to cellular transformation. The availability of constitutively active STAT5 variants36 43 will undoubtedly give new insights into the mechanism by which STAT5 contributes to cellular transformation.
Supported by a research grant from GlaxoWellcome b.v.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Rolf P. de Groot, PhD, Department of Pulmonary Diseases, Room G03.550, University Hospital Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands; e-mail:R.deGroot@hli.azu.nl.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal