Abstract
Herein, we show that CD34, c-kit double-positive (CD34+c-kit+) cells from the aorta-gonad-mesonephros (AGM) region of the developing mouse are multipotent in vitro and can undergo both B-lymphoid and multimyeloid differentiation. Molecular analysis of individual CD34+c-kit+ cells by single-cell reverse transcriptase–polymerase chain reaction (RT-PCR) shows coactivation of erythroid (β-globin) and myeloid (myeloperoxidase [MPO]) but not lymphoid-affiliated (CD3, Thy-1, and λ5) genes. Additionally, most cells coexpress the stem cell–associated transcriptional regulators AML-1, PU.1, GATA-2 and Lmo2, as well as the granulocyte colony-stimulating factor receptor (G-CSF-R). These results show that the CD34+c-kit+ population from the AGM represents a highly enriched source of multipotent hematopoietic cells, and suggest that limited coactivation of distinct lineage-affiliated genes is an early event in the generation of hematopoietic stem and progenitor cells during ontogeny.
CONSIDERABLE INTEREST currently surrounds intraembryonic hematopoietic progenitor cells located in the aorta-gonad-mesonephros (AGM) region and its anlage, the para-aortic splanchnopleura. The interest focuses both on the origin of intraembryonic stem cells from mesodermal elements and on their own functional capacity in terms of hematopoietic output.1
With regard to origin, the precise relationship of these intraembryonic stem cells to extraembryonic or yolk sac–derived progenitors remains an area of considerable debate.2 However, recent evidence suggests that hematopoietic stem-cell activity, at least within the AGM, may derive from a classic induction of bipotent endothelial-hematopoietic precursors (hemangioblasts) mediated by oncostatin-M.3 A better molecular characterization of early hematopoietic stem cells may help in clarifying their origin.
With respect to output, AGM cells are the first cells within the developing mouse embryo capable of long-term reconstitution of lethally irradiated adult recipients.4 Recent studies using newborn animals as recipients have shown that paraaortic splanchnopleural cells and yolk-sac cells from day 9 embryos have long-term reconstitution potential.5 In all cases, the activity resides within the CD34+ c-kit+ population,6,7 which is consistent, to an extent, with descriptive studies of CD34 expression in developing mouse and human embryos.8-10 These investigations identified CD34+ cells both in the mesenchyme surrounding the dorsal aorta at the level of the AGM and budding from the internal wall of the aorta in the same region.
Our own interest in AGM-derived stem cells relates to their potential utility as a model system for the analysis of hematopoietic lineage specification. Although the CD34+c-kit+populations from both day 11 mouse AGM and fetal liver clearly contain hematopoietic stem cells, large numbers of cells from these populations are required to produce long-term reconstitution of adult recipients: injection of 4 × 103 CD34+c-kit+AGM cells per mouse reconstitutes about 30% of the recipients, whereas 10 times more CD34+c-kit+ fetal liver cells are required to obtain the same result.6 The reason for this low efficiency of long-term reconstitution is not immediately clear, although it may partly reflect a lack of compatibility between embryonic/fetal stem cells and the adult microenvironment.5,11 Alternatively, the CD34+c-kit+ population may be heterogeneous at the functional level, the molecular level, or both. Indeed, several recent studies have emphasized the importance of single-cell approaches in delineating mechanisms in lineage specification,12 and to this end, we have analyzed the functional potential of individual AGM-derived CD34+c-kit+ progenitors and some aspects of their “ground state” at the molecular level.
MATERIALS AND METHODS
Cell preparation.
The liver and the AGM region were dissected from F1 embryos of CBAxC57BL/10 crosses with the embryonic age determined as described previously,13 with the production of a vaginal plug denoting day 0. Cell suspensions were prepared by passing the organs through a 70-μm mesh nylon cell strainer (Becton Dickinson Labware, Franklin Lakes, NJ). Adult bone marrow was obtained by flushing the femurs of 3-week-old CBA female mice with Dulbecco’s modified Eagle’s medium (GIBCO-BRL, Paisley, UK).
Flow cytometry and sorting.
The method for the reaction of cell suspensions with antibodies for flow cytometry has been described previously.14 In our case, cell preparations were treated with combinations of antibodies to the CD34 molecule (clone RAM34, conjugated to fluorescein isothiocyanate [FITC]) and c-kit (CD117, clone 2B8, conjugated to phycoerythrin [PE]) or antibodies to IgM (clone R6-60.2, FITC) and B220 (CD45R, clone RA3-6B2, PE). All antibodies were obtained from Pharmingen (San Diego, CA).
Flow-cytometric analysis was performed on a FACScan flow cytometer using Lysis II software (Becton Dickinson, Mountain View, CA) with 10,000 events recorded for each sample. Dead cells were identified and excluded from the analysis by addition of propidium iodide to the sample. Single CD34+c-kit+ cells were sorted using a FACStar flow sorter fitted with an automated cell-deposition unit under the control of a Consort 30 computer system (Becton Dickinson).
Determination of differentiation potential.
The ability of single CD34+c-kit+ cells to undergo differentiation into different hematopoietic lineages was tested using a modification of a previously described method.15 Briefly, single CD34+c-kit+ cells were sorted into 96-well microtest tissue culture plates (Becton Dickinson) containing a layer of irradiated mouse S17 stromal cells. These were then cultured in Opti-MEM (GIBCO-BRL) with interleukin-7 (IL-7), c-kit ligand (description follows), and 2-mercaptoethanol and fed and checked every 7 days. After 14 to 34 days in culture, the clones were split and either grown in the presence of WEHI-conditioned medium (description follows) and c-kit ligand (myeloid differentiation conditions) or on S17 stroma in the presence of IL-7 (pre-B-cell conditions). After 14 days, cytospins were made for the cells grown in myeloid conditions, and these were analyzed by microscopy after staining with the May-Grünwald-Giemsa technique, whereas those grown in pre-B-cell conditions were analyzed by flow cytometry for the presence of mature B-cell markers.
Cytokines.
IL-7 was obtained from J558 myeloma cells transfected with c-DNA (a gift from Dr A. Rolink, Basel, Switzerland) and used at a final concentration of 50 to 100 U/mL. Chinese hamster ovary cells were stably transfected with cDNA that codes for the c-kit ligand (Genetics Institute, Boston, MA), and the supernatant was titrated against c-kit–dependent mast cells and thus used at a 1:250 dilution. WEHI-conditioned medium was produced as described previously16 and was titrated against FDCP-mix cells and used at a 1:10 dilution.
Analysis of single cells by reverse transcriptase–polymerase chain reaction.
As previously described,17 single cells were deposited into 96-well polymerase chain reaction (PCR) plates containing 4 μL lysis buffer (0.4% Nonidet P-40, 60 μmol/L dNTPs, 25 μmol/L dithiothreitol, and 0.5 U/μL RNAsin [Promega, Madison, WI]) and lysed for 15 minutes on ice. Cell lysates were reverse-transcribed using multiple (up to 8) pairs of gene-specific primers and 48 U MMLV-RT per reaction in the buffer provided by the supplier (GIBCO-BRL). The first-round PCR with 35 cycles was performed by addition of 40 μL PCR buffer and 1.25 U Taq polymerase (Cetus, Emeryville, CA). One-microliter aliquots of first-round PCRs were further amplified using fully nested gene-specific primers. Aliquots of second-round PCR products were subjected to gel electrophoresis and visualized by ethidium bromide staining. The primers used in first-round PCRs were as follows: CD34, TTGACTTCTGCAACCACGGA and TAGATGGCAGGCTGGACTTC18; c-kit, GGCTCATAAATGGCATGCTC and TATCTCCTCGAGAACCTTCC19; erythropoietin receptor (EPO-R), CGCTACACCTTCGCTGTTCG and CAAACTCGCTCTCTGGGCTT20; granulocyte colony-stimulating factor receptor (G-CSF-R), ACAGGAGTGTGAACTTCGCT and TTGCTTCTTCTGACACCACG21; Lmo2, TGGATGAGGTGCTGCAGATA and CCCATTGATCTTGGTCCACT; AML-1, ACTTCCTCTGCTCCGTGCTA and GTCCACTGTGATTTTGATGGC; PU-1, GATGGAGAAAGCCATAGCGA and TTGTGCTTGGACGAGAACTG; GATA-2, TTCTTCTGCAGGGGGTAGTGTAG and GGTGACTTCTCTTGCATGCACTT; myeloperoxidase (MPO), CGCTTCTCCTTCTTCACTGG and CTGCCATTGTCTTGGAATCG22; β-globin, AGACCTATCCTCTGCCTCTG and CCACTCCAGCCACCA(CG)CTTC23; Thy-1, AGAAGGTGACCAGCCTGACA and GTTCTGAACCAGCAGGCTTA; CD3δ, TTGTGACCTGCAATACCAGC and TTGGTTGATCACAGGGGAA; and λ5, AGTTCTCCTCCTGCTGCTGC and CCCACCACCAAAGACATACC.24 The primers used in second-round PCRs were as follows: CD34, ATCCCCATCAGTTCCTACCA and GTAGGCAGTATGCCAGTTGG; c-kit, ACAGGAGCAGAGCAAAGGTG and CGACCACAAAGCCAATGAGC; EPO-R, TCTGGAGTGCCTGGTCTGAG and GCTCTCTGGGCTTGGGATGC; G-CSF-R, TACCAGCCACAGCTCAAAGG and ACGTGTCCAGTCTGATGGTG; Lmo2, GTACTGGCATGAGGATTGCC and TGTCGGAGTTGATGAGAAGGT; AML-1, ACTCACTGGCGCTGCAACAA and AAGCTCTTGCCTCTACCGCT; PU-1, AACCACTTCACAGAGCTGCA and CAAGCCATCAGCTTCTCCAT; GATA-2, GACTATGGCAGCAGTCTCTTCC and GGTGGTTGTCGTCTGACAATT; MPO, ACTGGCCTCAACTGCGAGAC and GTGTATTGACAGCCAGCAGC; β-globin, A(AG)(AG)GT(GC)AAGGCCCATGGCAA and CA(CG)CTTCTG(GC)(AC)AGGCAGCCT; Thy-1, CACCAAGGATAACTCCATCC and GCCACACTTGACCAGCTTGT; CD3δ, GCATCTAGATGGAACGGTGG and CTTCACGATCTCGAAGAGGC; and λ5, GGGTCTAGTGGATGGTGTCC and CAAAACTGGGGCTTAGATGG.
Reverse transcriptase–PCR analysis of sorted populations.
Total RNA was extracted from 1.5 × 103CD34+c-kit+ sorted AGM cells and from 1 × 106 MEL, WEHI-3B, or S17 cells. cDNA synthesis was initiated using random primers, and reaction products were resuspended in water at a concentration corresponding to approximately 50 cells/μL. To ensure that the PCR was performed during the exponential phase, serial dilutions of the sample were then submitted to 40 cycles of PCR using HPRT, MPO, or β-globin first-round primers. The products were then loaded on agarose gels, and the gels were Southern-blotted and probed with the following radiolabeled probes: HPRT probe, CAGTCAACGGGGGACATAAAAG; MPO probe, GTGTATTGACAGCCAGCAGC; and β-globin probe, CA(CG)CTTCTG(GC)(AC)AGGCAGCCT. Degeneracy in the globin probe allows hybridization to products derived from embryonic and adult β-globin genes. Hybridization was performed at 42°C, and the filters were washed at the same temperature in 1× SSC (1× SSC is 0.15 mol/L NaCl plus 0.015 mol/L sodium citrate) and revealed using a phosphor screen and phosphoimager (Molecular Dynamics, Sunnyvale, CA).
MadgeBio gel apparatus.
Single-cell PCR products were analyzed by electrophoresis on agarose MadgeBio gels (MadgeBio Ltd, Nottingham, UK). These 96-well gels are structured with the exact same format as 96-well microtiter plates. Samples are transferred directly from the PCR plate into Madge gels, and electrophoresis is performed at an angle of 18.4° to the axis of the rows such that the tracks pass across 3 column gaps, thereby providing a 2.65-cm path length. Extra wells for size standards allow size determination of the PCR products.
RESULTS
Differentiation potential in vitro.
Single-cell suspensions of day 11 AGM and fetal liver were labeled with anti–mouse CD34 and anti–mouse c-kit antibodies (see Materials and Methods), and double-positive cells were obtained by FACSorting. Because CD34+c-kit+ cells represent a small percentage of AGM cells, the sorting gate was defined using fetal liver samples, in which the CD34+c-kit+ population is more readily identifiable (Fig 1A). In addition, a small gate was set to obtain the highest purity possible. The fidelity of cell sorting in both AGM and fetal liver samples was confirmed by subsequent validation of CD34 and c-kit gene expression by RT-PCR analysis of single sorted cells. Furthermore, these control experiments showed that in the case of the AGM samples, an average of 40% of the wells did not contain any cell. This inefficiency in sorting is not entirely surprising given the low percentage of CD34+c-kit+ cells in the AGM population.
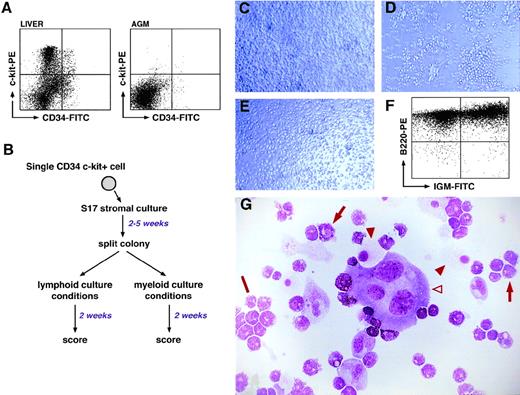
In vitro differentiation potential of CD34+c-kit+ cells from day 11 AGM and fetal liver. (A) FACSorting of CD34-FITC/c-kit-PE staining of day 11 fetal liver and AGM cells. Sorting gates were set using isotype-specific control antibodies, and dead cells were excluded using propidium iodide staining. The percentage of double-positive cells is 2.1% (fetal liver) and 0.1% (AGM). (B) Summary of the 2-step culture assay to test the differentiation potential of single CD34+/c-kit+ cells. (C) Photomicrograph of clone 1G12 after 13 days in culture under “multipotent conditions.” (D) Photomicrograph of clone 1G12 after 13 days of culture under lymphoid differentiation conditions. (E) Photomicrograph of clone 1G12 cultured for 13 days under myeloid conditions. (F) FACSscan analysis of the cells in panel D using anti-B220 and anti-IgM antibodies. (G) May-Grünwald-Giemsa staining of the cells in panel E; note the multimyeloid nature of the clone. The open arrowhead indicates a megakaryocyte; filled arrowheads indicate macrophages; arrows indicate myeloid cells of the granulocytic series; and the bar indicates a cluster of basophilic myeloid cells.
In vitro differentiation potential of CD34+c-kit+ cells from day 11 AGM and fetal liver. (A) FACSorting of CD34-FITC/c-kit-PE staining of day 11 fetal liver and AGM cells. Sorting gates were set using isotype-specific control antibodies, and dead cells were excluded using propidium iodide staining. The percentage of double-positive cells is 2.1% (fetal liver) and 0.1% (AGM). (B) Summary of the 2-step culture assay to test the differentiation potential of single CD34+/c-kit+ cells. (C) Photomicrograph of clone 1G12 after 13 days in culture under “multipotent conditions.” (D) Photomicrograph of clone 1G12 after 13 days of culture under lymphoid differentiation conditions. (E) Photomicrograph of clone 1G12 cultured for 13 days under myeloid conditions. (F) FACSscan analysis of the cells in panel D using anti-B220 and anti-IgM antibodies. (G) May-Grünwald-Giemsa staining of the cells in panel E; note the multimyeloid nature of the clone. The open arrowhead indicates a megakaryocyte; filled arrowheads indicate macrophages; arrows indicate myeloid cells of the granulocytic series; and the bar indicates a cluster of basophilic myeloid cells.
The functional potential of individual CD34+c-kit+ cells obtained in this manner was assessed using a previously reported 2-step in vitro differentiation assay15 (Fig 1B). This assay has been shown to allow multipotent cells to develop and subsequently differentiate into all hematopoietic lineages.25 26 In the first step of the assay, single cells are cultured on irradiated stromal S17 cell monolayers and expanded in the presence of IL-7, 2-Me, and c-kit ligand. The second step of the assay occurs after 14 to 28 days when growing clones are harvested and split. Half of the cells of the clone are cultured under conditions that promote the growth and differentiation of B lymphocytes (namely IL-7 and 2-Me), while the remaining half are cultured under conditions that promote myeloid cell development (in the presence of c-kit ligand and WEHI-conditioned medium). Single CD34+c-kit+ cells from day 11 AGM were assayed using this procedure; CD34+c-kit+ cells from day 11 fetal liver and adult bone marrow served as controls.
CD34+c-kit+ AGM cells are multipotent.
Under the conditions already described, the cloning efficiency was 22% (∼1 cell in 5) for AGM cells. However, when corrected for the number of empty wells observed in the sorted AGM populations, the cloning efficiency may be calculated at 37%.
Three types of clones derived from CD34+c-kit+AGM cells were identified: (1) clones with a restricted myeloid potential that were able to differentiate into either macrophages and megakaryocytes or macrophages only; none of these clones were able to produce B cells; (2) multimyeloid clones that showed the ability to differentiate into at least 3 myeloid lineages (mainly macrophages, megakaryocytes, and granular polymorphonuclear cells) but were not able to differentiate into B lymphocytes; and (3) multipotent clones that displayed a multimyeloid potentiality and the ability to yield B cells. An example of such a clone is shown in Fig 1C to G: panel C shows the clone after 2 weeks of culture in multipotent conditions, and panels D and E show the clone grown under lymphoid and myeloid conditions, respectively. Verification of the lymphoid and multimyeloid nature of the resulting cells was performed by either immunophenotyping of lymphoid cells (Fig 1F) or May-Grünwald-Giemsa staining of myeloid cells (Fig 1G). Taking all our experiments together, these lymphomyeloid stem cells represented 38% of the total clonogenic population, while cells with multimyeloid potential but lacking B-lymphoid potential represented 49%, and bilineage and unilineage restricted precursors accounted for approximately 13%.
To compare our data with previously reported stem-cell activity in the embryo, we also studied the differentiation potential of CD34+c-kit+ cells from day 11 fetal liver and from adult bone marrow. Table 1 shows the clone types in the different samples. Even when taking the cloning efficiency into account, the results show that the CD34+c-kit+ population from AGM is mainly composed of multipotent hematopoietic progenitors, while they represent a smaller fraction of the same population from fetal liver at the same stage of development. No multipotent progenitors could be detected in the CD34+c-kit+ population from bone marrow, presumably reflecting their low frequency in this population.
Differentiation Potential of CD34+c-kit+Cells
| Parameter . | Day 11 AGM . | Day 11 Liver . | Adult Bone Marrow . |
|---|---|---|---|
| Total clones (n) | 94 | 76 | 124 |
| Bilineage- or unilineage-restricted (%) | 13 | 71 | 100 |
| Multimyeloid (%) | 49 | 9 | 0 |
| Multimyeloid + B-cell potential (%) | 38 | 20 | 0 |
| Parameter . | Day 11 AGM . | Day 11 Liver . | Adult Bone Marrow . |
|---|---|---|---|
| Total clones (n) | 94 | 76 | 124 |
| Bilineage- or unilineage-restricted (%) | 13 | 71 | 100 |
| Multimyeloid (%) | 49 | 9 | 0 |
| Multimyeloid + B-cell potential (%) | 38 | 20 | 0 |
Clones developing from single CD34+c-kit+cells isolated from the AGM and fetal liver of day 11 mouse embryos and from the bone marrow of 3-week-old animals were analyzed for the capacity to differentiate into different hematopoientic lineages using the culture system shown in Fig 1B.
The high frequency of unilineage and bilineage committed precursors in the fetal liver and bone marrow samples suggested that the S17 culture system was capable of supporting the growth and development of less primitive progenitor cells. This, in turn, suggested that the high frequency of multipotent and multimyeloid colonies found in the AGM samples is a true reflection of the developmental potential of the CD34+c-kit+ population in the AGM and is not a result of a multipotent/multimyeloid bias in the culture system. To explore this further, we directly plated AGM CD34+c-kit+ cells into S17 cultures supplemented with myeloid growth factors including EPO. Consistent with the data obtained in the 2-step S17 culture assay, AGM cells primarily yielded multimyeloid colonies in this 1-step culture system (lymphopoiesis is not supported by these myeloid growth factor conditions), with few unilineage and bilineage colonies observed. These results are also consistent with experiments recently performed by Cumano et al (personal communication, September 1998), who have also observed that few EPO-dependent progenitors are present in the AGM at day 11. Those that do arise occur with a frequency similar to that found in cultures derived from embryo remnants, and are thus presumed to result from contamination due to embryonic blood circulation occurring at this stage of development.
Thus, although several thousand CD34+c-kit+ AGM cells are required for long-term reconstitution of adult recipients, a large fraction (8% or 14% when corrected for empty wells) have lympho-myeloid differentiation capacity as shown by these in vitro assays. This population may thus provide a tractable model system for the study of stem-cell fate, and we therefore sought to further characterize it at the molecular level. Since molecular analysis at the population level obscures any heterogeneity that may exist between individual cells, these studies were performed at the single-cell level using single-cell RT-PCR.
Single-cell RT-PCR.
To perform single-cell RT-PCR analysis, we used a method developed in our laboratory.17 This method uses gene-specific primers for reverse transcription, followed by 2 rounds of PCR with fully nested primer sets. The resulting high sensitivity of this method requires that appropriate controls be performed to ensure confidence in the specificity of detected expression. In control experiments with unilineage committed erythroid and myeloid cell lines, we were able to detect erythroid-affiliated gene expression (for example, β-globin and GATA-1) only in erythroid-committed cells, not in myeloid-committed cells. Similarly, we could only detect myeloid-associated gene expression (for example, MPO) in myeloid-committed cells, not in erythroid-committed cells. Expression of these hematopoietic genes was not detected in nonhematopoietic cell lines, nor was expression of nonhematopoietic genes (for example, EGF receptor) observed in any of the hematopoietic cells we tested.17 Data such as these encouraged confidence in the specificity of this method, although its sensitivity restricts its utility to qualitative as opposed to quantitative analysis.
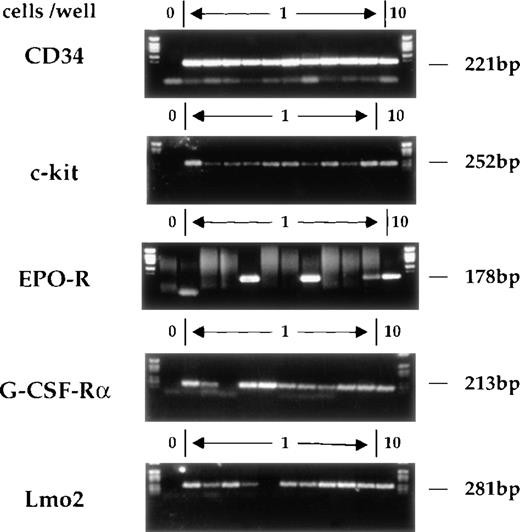
The single-cell RT-PCR method has already been described in detail.17 Briefly, single CD34+c-kit+ cells were obtained by FACSorting and lysed. Reverse transcription was performed using gene-specific primers prior to a multiplex first-round PCR. Aliquots of this reaction were used in separate second-round PCRs using fully nested primers for the individual genes of interest. Figure 2shows typical amplification products obtained on single sorted AGM cells. The analyses of CD34 and c-kit mRNAs serve as controls for the RT-PCR procedure and for the fidelity of FACSorting. Consistent with our previous studies of multipotent cell lines and primary multipotent cells freshly isolated from bone marrow, most CD34+c-kit+ AGM cells express G-CSF-R, whereas few express EPO receptor (EPO-R).
Gene expression in individual CD34+c-kit+ AGM cells. CD34+c-kit+ cells from AGM were individually sorted into 0.2-mL microtubes containing lysis buffer. Multiplex single-cell RT-PCR analysis was performed. The gel shows analysis of CD34, c-kit, EPO-R, G-CSF-R, and Lmo2. In each case, position 1 does not contain any cells and serves as a negative control, whereas position 12 contains 10 cells. The remaining positions (2-11) represent the amplification reactions of single cells. The expected migration position and molecular weight of appropriate RT-PCR products is indicated.
Gene expression in individual CD34+c-kit+ AGM cells. CD34+c-kit+ cells from AGM were individually sorted into 0.2-mL microtubes containing lysis buffer. Multiplex single-cell RT-PCR analysis was performed. The gel shows analysis of CD34, c-kit, EPO-R, G-CSF-R, and Lmo2. In each case, position 1 does not contain any cells and serves as a negative control, whereas position 12 contains 10 cells. The remaining positions (2-11) represent the amplification reactions of single cells. The expected migration position and molecular weight of appropriate RT-PCR products is indicated.
We next examined the expression patterns of Lmo2, AML-1, PU.1, and GATA-2, since these transcription factors have been implicated in the early stages of hematopoietic development by virtue of the panhematopoietic or multilineage deficits observed in the corresponding knockout mice. Expression of these factors may therefore be intimately linked to the determination of hematopoietic or stem-cell potential. Mice lacking Lmo2 function are bloodless and die at about embryonic day 10 (E10).27 The importance of this gene in the differentiation along the nonerythroid pathways is still unclear, but most lineages seem to be affected.28 The analysis of AML-1 knockout mice suggests a block in the development of all definitive hematopoietic lineages leading to fetal death by E12.5, but primitive hematopoiesis is not affected.29 Mice lacking PU.1 die 1 to 3 days before birth.30 The principal hematopoietic defects are the absence of monocytes, granulocytes, and T and B lymphocytes; erythroid precursors are present and β-globin gene expression is unaffected. Mice lacking GATA-2 function die at about E10.5. Although all hematopoietic lineages are affected, stem cells are present but appear to be deficient in their self-renewal/proliferative capacities.31 32
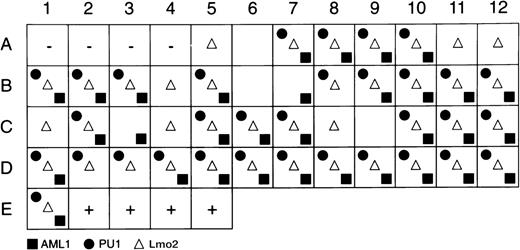
Single cells were sorted and a multiplex first-round PCR was performed using specific primers for CD34, c-kit, PU.1, AML-1, and Lmo2. Individual second rounds of amplification were performed using nested primers. Figure 2 shows representative data from Lmo2 reactions, and Fig 3 summarizes PU.1, AML-1, and Lmo2 expression patterns for the cells that were PCR-positive for CD34 and c-kit. In averaging 2 independent experiments, 89% of AGM cells expressed Lmo2, 73% expressed PU.1, and 71% were positive for AML-1; GATA-2 was expressed by an average of 60% of cells (data not shown). These frequencies of coexpression suggest that AGM cells are reasonably homogeneous with respect to expression of these factors, which therefore presumably do not define subpopulations of committed or lineage-restricted cells within the CD34+c-kit+AGM compartment.
Summary of Lmo2, AML-1, and PU.1 gene-expression programs of individual CD34+c-kit+ cells. The first 4 positions (A1-A4) contain no cells and serve as negative controls, and the last 4 positions (E2-E5) contain 10 cells each and serve as positive controls. The remaining positions represent the amplification reactions of single cells, with the presence of a symbol indicating positive amplification for the gene product indicated.
Summary of Lmo2, AML-1, and PU.1 gene-expression programs of individual CD34+c-kit+ cells. The first 4 positions (A1-A4) contain no cells and serve as negative controls, and the last 4 positions (E2-E5) contain 10 cells each and serve as positive controls. The remaining positions represent the amplification reactions of single cells, with the presence of a symbol indicating positive amplification for the gene product indicated.
Coactivation of erythroid and myeloid programs.
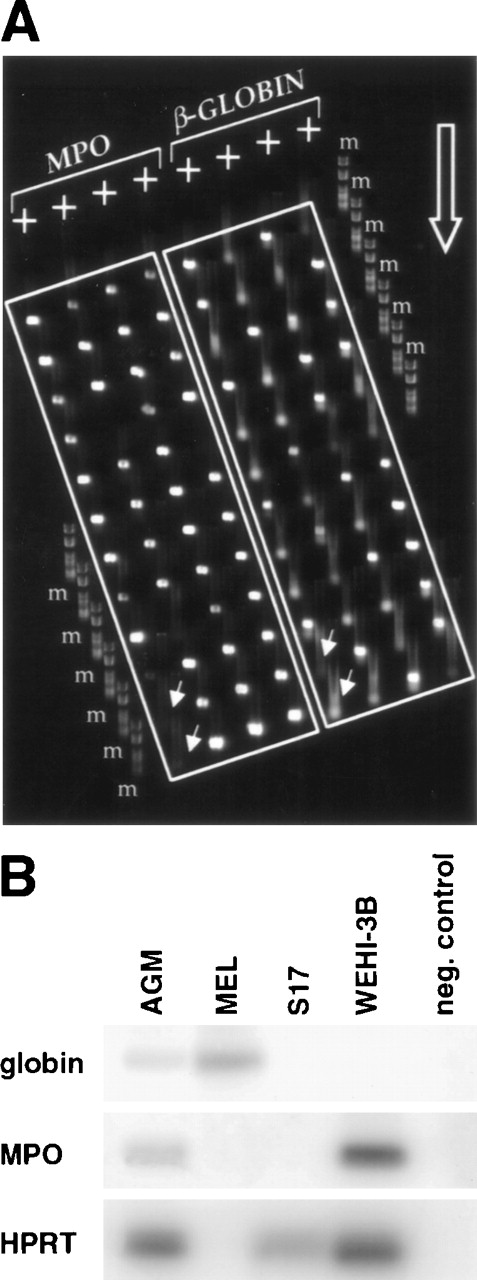
Previous analyses of single adult multipotent cells and cell lines have led to the hypothesis that individual multipotential cells may prime more than 1 lineage-affiliated program of gene expression before exclusive commitment and differentiation to a single hematopoietic lineage.17 For example, genes such as β-globin and MPO that encode erythroid- and myeloid-affiliated lineage-specific functions33 were found to be expressed, at least at low level, ahead of an exclusive unilineage commitment decision to either erythroid or myeloid cell fates. We therefore studied the expression of some key lineage-affiliated genes in day 11 AGM cells. Hu et al17 showed that the neutrophil-specific MPO gene is expressed in a large percentage of FDCP-mix and is often coexpressed with β-globin.17 We found that over 90% of day 11 CD34+c-kit+ AGM or liver cells express MPO and that about 50% of them coexpress β-globin. This is illustrated in Fig 4A, showing an agarose MadgeBio gel (see Materials and Methods) run of RT-PCR products from a representative experiment. The first 4 lanes are amplification products for MPO on single CD34+c-kit+ AGM cells, while the last 4 lanes show products for β-globin on the same cells. Analysis of single positive cells (CD34+c-kit− or CD34−c-kit+) from AGM seldom showed coexpression of MPO and β-globin. In all experiments performed, we never observed coexpression of MPO and β-globin in CD34−c-kit+ cells, while only 10% of CD34+c-kit− cells exhibited dual expression (data not shown).
Coexpression of MPO and β-globin in individual CD34+c-kit+ AGM cells. (A) Individual cells were subjected to multiplex 2-round RT-PCR using primers specific for CD34, c-kit, MPO, and β-globin. Second-round β-globin and MPO PCR products from CD34+c-kit+ cells were electrophoresed on an agarose MadgeBio gel, with MPO products on the left side and β-globin products on the right side. The migration direction of the gel is indicated by the large open arrow, and negative control reactions are indicated by the offset small arrows. The 2 most leftward wells of the first and fifth lanes do not contain any cells and serve as negative controls. Note the high frequency of cells that score positive for both MPO and β-globin. m, marker #6 (Roche Diagnostics, Lewes, UK). (B) Analysis of gene expression in sorted CD34+c-kit+ AGM cells compared with different cell lines. Several dilutions of each cDNA were amplified to ensure quantitation. The amount shown corresponded to nonsaturated PCR. MEL cells were used as a positive control for globin expression and WEHI-3B for MPO expression, while S17 was used as an example of a nonhematopoietic cell line. The clone of MEL cells used here is HPRT-negative, but the amount of cDNA used was controlled by OD to be equivalent to the other cDNAs amplified here.
Coexpression of MPO and β-globin in individual CD34+c-kit+ AGM cells. (A) Individual cells were subjected to multiplex 2-round RT-PCR using primers specific for CD34, c-kit, MPO, and β-globin. Second-round β-globin and MPO PCR products from CD34+c-kit+ cells were electrophoresed on an agarose MadgeBio gel, with MPO products on the left side and β-globin products on the right side. The migration direction of the gel is indicated by the large open arrow, and negative control reactions are indicated by the offset small arrows. The 2 most leftward wells of the first and fifth lanes do not contain any cells and serve as negative controls. Note the high frequency of cells that score positive for both MPO and β-globin. m, marker #6 (Roche Diagnostics, Lewes, UK). (B) Analysis of gene expression in sorted CD34+c-kit+ AGM cells compared with different cell lines. Several dilutions of each cDNA were amplified to ensure quantitation. The amount shown corresponded to nonsaturated PCR. MEL cells were used as a positive control for globin expression and WEHI-3B for MPO expression, while S17 was used as an example of a nonhematopoietic cell line. The clone of MEL cells used here is HPRT-negative, but the amount of cDNA used was controlled by OD to be equivalent to the other cDNAs amplified here.
As previously discussed, the single-cell method described here provides only qualitative data regarding the cell distribution of specific transcripts and provides little or no information with regard to the level. The level of expression of lineage-affiliated genes like β-globin or MPO is not a priori expected to be particularly high in multipotential cells, as expression in this compartment is not likely related to the function of these products in these cells, but is instead a reflection of the priming of these loci for future activity in appropriate unilineage committed cells (see Discussion). Indeed, using a different single-cell RT-PCR method, Brady et al34detected β-globin and MPO transcripts in unilineage committed cells, but not in multipotent cells. This may partly reflect different sensitivities in the methodologies used or different criteria in selection of the cells analyzed.
To gain an appreciation of the level of expression of MPO and β-globin in AGM cells, we prepared total cDNA from a population of sorted CD34+c-kit+ AGM cells. After reverse transcription and 1 round of PCR for β-globin, MPO, and HPRT, reaction products were fractionated on agarose gels, blotted, and hybridized with radiolabeled oligomers specific for the β-globin, MPO, and HPRT products, respectively. The results presented in Fig 4B are from cDNA dilutions that do not yield saturated signals by PCR and show that both β-globin and MPO can be easily detected in the CD34+c-kit+ population of AGM cells. In contrast, only β-globin is detected in the erythroid committed cell line MEL, and only MPO is detected in the myeloid committed cell line WEHI-3B. Neither β-globin nor MPO were detected in the nonhematopoietic stromal cell line S17. HPRT serves as a loading control for these experiments except in the case of MEL cells, which are mutant at the HPRT locus and do not express HPRT.
Analysis of lymphoid genes.
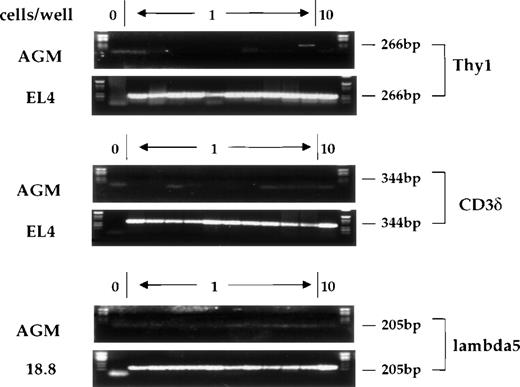
Expression of the lymphoid-specific genes Thy-1, CD3, and λ5 was also studied using single-cell RT-PCR. Thy-1 is a surface marker for thymocytes and T cells that is expressed very early in T-cell development, prior to CD3 expression.35 Thy-1 is also expressed at a low level in mouse (C57BL/Ka-Thy-1.1) bone marrow, where the Thy-1.1lo, Sca-1hi, lin−population has been shown to contain all of the multipotent progenitors.36 Additionally, Thy-1 is expressed outside of the hematopoietic system, most notably in cells of the nervous system.37 The CD3δ gene of the CD3 complex of the T-cell receptor is 1 of the earliest definitive markers of T-cell differentiation.27 λ5 is 1 of the 2 chains comprising the surrogate light chain of the pre-B receptor.38,39 Figure5 shows the signals obtained with single cells after amplification of the mRNAs for these genes from AGM cells, as well as from control cells known to express these different genes. We were not able to detect any cells expressing λ5 or CD3, and only 2% to 4% of CD34+c-kit+ AGM cells were positive for Thy-1 expression. This is consistent with data reported by Sanchez et al,6 who examined the surface expression of Thy-1.2 on AGM cells by flow cytometry. In these studies, while 9.5% of AGM cells were scored positive for Thy-1.2 expression, only 0.5% of c-kit+ AGM cells expressed Thy-1.2.
Expression of Thy-1, CD3δ, and λ5 in individual CD34+c-kit+ cells from AGM or from control cell lines. Cells were analyzed as previously described. Position 1 does not contain any cells and serves as a negative control, positions 2-11 contain single cells, and position 12 contains 10 cells. Product sizes are indicated.
Expression of Thy-1, CD3δ, and λ5 in individual CD34+c-kit+ cells from AGM or from control cell lines. Cells were analyzed as previously described. Position 1 does not contain any cells and serves as a negative control, positions 2-11 contain single cells, and position 12 contains 10 cells. Product sizes are indicated.
DISCUSSION
AGM cells as an in vitro model.
Previous studies have shown that the long-term reconstitution activity of AGM cells resides within the CD34+c-kit+population.6 Our present in vitro analysis of the CD34+c-kit+ population at the level of single cells has revealed the following. (1) The incidence of clonogenic cells is surprisingly high. This may relate to the cycling status of these cells: certainly, an actively cycling population would square with the hypothesis that the AGM is a site of stem-cell development or expansion in the early embryo.40 (2) A high percentage of AGM-derived clonogenic cells in our experiments retain both multimyeloid and B-lymphoid differentiation potential. Previous experiments have shown that all P-Sp clones with B-cell potential are able to differentiate into T lymphocytes25 and that this is also the case for all day 11 liver- or blood-derived clones.15 While we have not tested the T-lymphoid differentiation potential of CD34+c-kit+ AGM cells directly in the experiments described here, it seems likely that AGM cells with multimyeloid and B-lymphoid potential also possess T-lymphoid differentiation capacity.
Our data show that in the CD34+c-kit+population from day 11 AGM, 1 in 6 cells, at the very least, is multimyeloid and 1 in 12 is also able to differentiate into lymphocytes. This frequency is in agreement with the published frequency of cells able to form CFU-S40,41 but is much higher than the frequency of cells able to reconstitute irradiated adult recipients.6 This could indicate that the majority of cells that have a multipotent phenotype in vitro are not hematopoietic stem cells; alternatively, recent studies using conditioned newborn mice as recipients for reconstitution with embryonic cells seem to indicate that the discrepancy might be due to a failure of embryonic stem cells to engraft in adult recipients.5,7 11 Although AGM cells have not yet been tested under these conditions, it is possible that the frequency of reconstituting cells in newborns may better reflect the frequency of multipotent cells detected in our in vitro studies.
Our data thus suggest that cells from the AGM will provide a valuable model system in which to examine and experimentally manipulate hematopoietic lineage specification processes in vitro. In particular, the high frequency of multipotent cells predicts that daughter-cell experiments will prove fruitful. AGM cells should also prove uniquely valuable in the study of knockout mice with hematopoietic defects. The effects on hematopoiesis of otherwise lethal mutations can be studied in in vitro AGM cultures, as long as these cells themselves are generated and the embryos survive to at least day 10 to 11 of gestation.
Multilineage priming.
Previous studies have documented low-level transcription of genes with known lineage-affiliated function (such as β-globin) in adult multipotential cell lines (FDCP-mix A4), as well as multipotential cells freshly isolated from adult mouse bone marrow.17Importantly, transcripts characteristic of distinct lineages (eg, β-globin and MPO) did not segregate to different cells within the population, but were frequently found to be coexpressed in a large percentage of individual cells. Evidence provided in this report suggests that this coexpression of lineage-affiliated markers in multipotent progenitors is not unique to the adult stage of hematopoiesis, nor is it simply a function of continually forced self-renewal of multipotential cell lines in cell culture. Rather, such limited coactivation of lineage-affiliated genes such as β-globin and MPO appears to be an early event in the ontogeny of the hematopoietic system. Thus, the molecular process underlying the priming of these loci may be closely coupled or associated with the registration of hematopoietic potential.
While the specificity of the low-level transcription observed in these cells seems secure enough (ie, nonhematopoietic-associated transcripts are not detected), its precise significance remains speculative. Our results are consistent with the notion that the expression of genes like MPO or β-globin occurs in cells that have multipotent or at least multimyeloid potential. Certainly, it occurs within cells that coexpress transcription factors such as AML-1, Lmo2, and PU.1, since these encompass the vast majority of cells analyzed; RT-PCR experiments on single AGM cells have confirmed this (data not shown). Arguably, the cloning efficiency of the cells used in our experiments precludes formal assignment of a β-globin/MPO phenotype to a cell with proven multipotent potential. When our data are adjusted for the frequency of wells that contain no cells after sorting, we estimate that approximately 33% of AGM cells are capable of generating multilineage colonies in vitro. An additional 2% to 3% yield bilineage or unilineage colonies after 2-step culture. This leaves approximately 65% of the sorted population that is not clonogenic under these conditions and whose developmental potential therefore cannot be categorically determined. There is no a priori reason to assume that these cells are particularly different in their lineage potential versus those that do produce colonies in vitro. Certainly, we have found little evidence for more committed cells in this population using the 2-step culture system or in different culture systems for the growth and development of more committed progenitors. Given that at least 90% of the AGM cells analyzed express MPO, we can safely infer that MPO is expressed (or rather primed) in at least some multipotent AGM cells. Since 50% of CD34+c-kit+ AGM cells coexpress MPO and β-globin (note that coexpression of β-globin and MPO is seldom found in single CD34+ or c-kit+AGM cells), it is formally possible that these coexpressing cells lie specifically within the 65% that do not give rise to colonies. However, by applying Ockham’s razor, the simplest and most plausible explanation consistent with the current culture data and molecular analyses is that coexpression occurs within multilineage-potential hematopoietic cells, and this interpretation of the data agrees well with previous studies in this area.
Our favored interpretation with regard to the nature of this low-level or sporadic transcription is that it reflects the partially accessible nature, or “primed” configuration, of the chromatin structure at the loci in question. Evidence for this primed chromatin configuration has been obtained in multipotential cell line models such as FDCP-mix A4, where appropriate numbers of cells for chromatin and biochemical analyses are readily obtainable.42-46 Similar observations have also been made on human leukemic cells and cell lines.47 We have also speculated that priming may be achieved by so-called “stand-in” factors rather than by the set of transcription factors normally associated with high-level transcriptional output of lineage-affiliated loci in terminally differentiated unilineage-committed cells.42 Transcription factors such as Lmo2, AML-1, PU.1, and GATA-2, where gene targeting has revealed an early hematopoietic defect and which we show to be consistently coexpressed in multipotential AGM cells, may be prime candidates for this priming function. The transcripts we have observed may reflect low-level or sporadic transcription from authentic promoters, ie, a consequence of locus activation. Alternatively, they themselves may be active participants in the opening of a locus. In this regard, it is interesting to note the recent observations of Ashe et al,48 who detected intergenic transcription throughout the globin locus, which they suggested may function as part of the locus-opening machinery.
Finally, our studies of priming in AGM-derived stem cells have failed to find evidence of lymphoid-affiliated gene activation: neither CD3, λ5, nor Thy-1 transcripts were consistently detected by RT-PCR. This is seemingly at odds with our previous studies of FDCP-mix A4 cells, where the CD3δ enhancer was shown to be in a DNase I–hypersensitive configuration.49 Similarly, λ5 expression is detectable by RT-PCR in FDCP-mix, and these cells also express Thy-1 on their surface.17,35 These lymphoid characteristics of FDCP-mix probably reflect their forced self-renewal in tissue culture. Certainly, FDCP-mix cells are not considered entirely authentic stem cells, and despite displaying lymphoid features, they have not been successfully differentiated down the lymphoid pathway. However, the recent observations of Wang et al50 provide cogent evidence that T- and B-lymphoid gene loci are coactivated before unilineage T- or B-lymphoid commitment.
These considerations aside, we are intrigued by another possibility that relates to the evolutionary origins of hematopoietic stem cells. We suggest that the ground state of developmentally early stem cells may essentially be myeloid-disposed, reflecting (1) the evolutionary origins of hematopoiesis in monocytic/phagocytic cells and granulocytes and (2) the early evolutionary requirement for oxygen-carrying red blood cells. This evolutionary baggage may also underlie the essentially eythro-myeloid disposition of early yolk sac–derived progenitors and the later developmental appearance of lymphoid potential in intraembryonic progenitors of the paraaortic splanchnopleura and AGM. Further studies of lymphoid gene activation in early stem cells at the level of chromatin structure will be required to test this hypothesis.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Tariq Enver, PhD, Section of Gene Function and Regulation, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Rd, London SW3 6JB; e-mail:<tariq@icr.ac.uk>.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal