Abstract
Apoptosis and platelet activation share common morphological and biochemical features. Because caspases are essential mediators of apoptosis, we examined whether platelets contain these proteinases and use them during platelet activation. Human platelets contained caspase-9, caspase-3, and the caspase activators APAF-1 and cytochrome c as shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. Upon treatment with cytochrome c and dATP, platelet cytoplasmic extracts recapitulated apoptotic events, including sequential activation of procaspase-9 and procaspase-3 and subsequent proteolysis of caspase substrates. Calcium ionophore-stimulated platelets also recapitulated apoptotic events, including cell shrinkage, plasma membrane microvesiculation, phosphatidyl serine externalization, and proteolysis of procaspase-9, procaspase-3, gelsolin, and protein kinase C-δ. Strikingly, however, these events occurred without caspase activation or release of mitochondrial cytochrome c, suggesting a role for a noncaspase proteinase. Supporting this, inhibition of the calcium-dependent proteinase, calpain, prevented caspase proteolysis, ‘apoptotic’ substrate cleavage, and platelet microvesiculation. In vitro, purified calpain cleaved recombinant procaspase-9 and procaspase-3 without activating either caspase, confirming the inhibitor studies. These data implicate calpain as a potential regulator of caspases and suggest that calpain, not caspases, promotes apoptosis-like events during platelet activation.
PROFOUND ALTERATIONS in cellular architecture characterize apoptotic cell death. Regardless of the initiating signal, the cell shrinks, the plasma membrane blebs and vesiculates, and phosphatidylserine (PS) redistributes to the cell surface. Meanwhile, the chromatin condenses, the nucleus breaks up, and the DNA fragments into oligonucleosomal pieces. These events culminate in removal of the apoptotic cell by neighboring phagocytes without induction of an inflammatory response.1
Nuclear collapse and DNA fragmentation are dramatic and universal features of apoptosis. However, the nucleus itself is not required for apoptosis because apoptotic stimuli readily induce apoptotic morphological features in anucleate cytoplasts.2-4Furthermore, platelets, small anucleate cells derived from bone marrow megakaryocytes, undergo events similar to apoptosis upon activation with hemostatic agents like thrombin and collagen or stimulation with pharmacological agents like calcium ionophore.5-8Regardless of the stimulus, the platelet shrinks and extends filopodia, the plasma membrane blebs and microvesiculates, and PS redistributes to the platelet surface.5-8 These events promote hemostasis by inducing platelet aggregation, platelet adhesion to injured blood vessels, and activation of the coagulation cascade.
Caspase proteinases are centrally involved in apoptotic signaling and execution.1 Caspases are aspartate-specific cysteine proteinases that exist as latent zymogens, but once activated by apoptotic signals, they promote apoptosis by specific limited proteolysis of key cellular substrates. Death stimuli typically facilitate autoactivation of initiator caspases such as caspase-8 and caspase-9. The adapter molecule FADD initiates caspase-8 activation following ligation of death receptors such as Fas.9,10Chemotherapeutic agents, UV-irradiation, and other apoptotic stimuli cause release of mitochondrial cytochrome c into the cytosol.11,12 Cytochrome c then binds to apoptotic proteinase activating factor-1 (APAF-1) and this complex, along with adenine nucleotides, promotes caspase-9 autoactivation.13Active caspase-8 also causes release of mitochondrial cytochrome c, thereby linking the activation pathways.14,15 Once activated, both caspase-8 and caspase-9 activate caspase-3, which in turn cleaves other caspases, fodrin, gelsolin, protein kinase C-δ (PKCδ), and DNA fragmentation factor-45 (DFF45).1 16-22Proteolysis of these and other caspase substrates induces the hallmarks of apoptotic cell death.
Calpain, a calcium-dependent proteinase, may also function in apoptosis by cleaving cytoskeletal proteins such as fodrin, actin, and the pro-apoptotic protein Bax.18,23,24 It is unclear, however, whether calpain functions upstream or downstream of caspases in apoptosis.18,25-27 During platelet activation, calpain promotes platelet microvesiculation, clot retraction, and proteolysis of fodrin and other cytoskeletal components.28-31Therefore, calpain functions in both apoptosis and platelet activation. However, whether caspases participate in platelet activation is unknown.
In this report, we examine whether platelets contain caspases and use them during platelet activation. We found that human platelets contain caspase-9, caspase-3, and the caspase activators APAF-1 and cytochrome c. In vitro, these proteins recapitulate the cytoplasmic events of apoptosis. Surprisingly, however, we found that calpain, not caspases, promotes apoptosis-like events during platelet activation. Therefore, platelet activation is not equivalent to apoptosis and certain aspects of the apoptotic phenotype may occur without caspase activation.
MATERIALS AND METHODS
Materials.
Antibodies against the following proteins were purchased commercially: caspase-3 (Pharmingen, La Jolla, CA), cytochrome c (Pharmingen), fodrin (Chemicon International, Temecula, CA), PKCδ (Santa Cruz Biotechnology, Santa Cruz, CA), gelsolin (Sigma, St Louis, MO), CD3 (Sigma), and glycoprotein (gp) IIb (Southern Biotech, Birmingham, AL). Dr Xiadong Wang (University of Texas Southwestern Medical Center, Dallas, TX) provided anti-DFF45 polyclonal antibodies.22 Antisera against APAF-1 and caspase-9 was prepared by immunizing rabbits with an APAF-1 peptide (S38EEEKVRNEPTQQQR52) or with recombinant caspase-9. Horseradish peroxidase–conjugated secondary antibodies were from Amersham (Arlington Heights, IL). N-acetyl-Asp-Glu-Val-Asp-p-nitroanilide (DEVD-pNA), benzoyloxycarbonyl-Asp-Glu-Val-Asp-amino-4-trifluoro-methyl-coumarin (DEVD-AFC), N-acetyl-Tyr-Val-Ala-Asp-AFC (YVAD-AFC), N-acetyl-Val-Glu-Ile-Asp-AFC (VEID-AFC), N-acetyl-Leu-Glu-His-Asp-AFC (LEHD-AFC), succinyl-Leu-Leu-Val-Tyr-AFC (LLVY-AFC), and benzoyloxycarbonyl-Val-Ala-Asp(OMe)-fluoromethyl ketone (ZVAD-fmk) were from Enzyme Systems Products (Livermore, CA). Benzoyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone (ZVAD-fmk*) was from Bachem. All other reagents were from Sigma.
Proteinases.
Proteolytically active caspases and caspase zymogens were expressed inEscherichia coli and purified as described.16,32Active caspase-3 was active site titrated with ZVAD-fmk*.1635S-procaspase-9 was prepared using a procaspase-9 cDNA (provided by Dr Emad Alnemri, Thomas Jefferson University, Philadelphia, PA) and the TnT reticulocyte lysate system (Promega, Madison, WI).33 Porcine μ-calpain was purchased from Calbiochem (San Diego, CA) and active site titrated with calpeptin. Hereafter we will refer to μ-calpain simply as calpain. Granzyme B (13 U/μg) was from Enzyme Systems Products.
Platelets and cells.
Platelets were isolated from human platelet concentrates (San Diego Blood Bank, San Diego, CA) or from whole blood obtained from normal human donors using a protocol approved by the Review Board for Human Studies at the La Jolla Institute for Allergy and Immunology.34 A mixed population of lymphocytes and granulocytes was obtained from the buffy coat after hypotonic lysis of red blood cells.35 Jurkat cells were originally from American Type Culture Collection (ATCC; Rockville, MD) and maintained as described.36
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Cell-free apoptosis.
Cytosolic extracts from Jurkat cells and buffy coat cells were prepared as described.36,37 Extracts were prepared from platelets using the same method except that platelets were subjected to 3 freeze/thaw cycles before homogenization. Extract protein concentrations were determined with the Bradford dye reagent (BioRad). Cell-free apoptosis was initiated with cytochrome c and dATP as indicated in the figure legends. At the indicated times, aliquots were withdrawn for proteinase assay and Western blot analysis.37To detect caspase-dependent DNase activity, extracts were incubated with ≈106 rat liver nuclei and buffer, caspase-3 (100 nmol/L), or cytochrome c (10 μmol/L) and dATP (1 mmol/L). DNA fragmentation was visualized via agarose gel electrophoresis and ethidium bromide staining.36
Platelet activation.
Before activation, freshly isolated platelets were suspended at a concentration of 2 × 107/mL in buffer A (10 mmol/L HEPES pH 7.4, 137 mmol/L NaCl, 2.7 mmol/L KCl, 1.7 mmol/L MgCl2, 3 mmol/L CaCl2, 25 mmol/L glucose, 0.05% bovine serum albumin). Platelets were treated with buffer or calcium ionophore A23187 (2 μmol/L) and incubated at 37°C for the indicated times. Cell shrinkage (decreased forward scatter) and PS externalization were monitored by flow cytometry as described.4 In addition, whole-cell lysates were prepared and analyzed for caspase activity, caspase processing, and apoptotic substrate cleavage. For comparison, control and UV-treated (apoptotic) Jurkat cells were also analyzed.38
Platelet subcellular fractionation.
Platelets were preincubated for 30 minutes at room temperature in the absence or presence of ZVAD-fmk (25 μmol/L) or calpeptin (25 μmol/L) and then stimulated with ionophore A23187. After 15 minutes, subcellular fractions were prepared by differential centrifugation as decribed.28,29 This procedure yields a 15,000gpellet (P15) and supernatant (S15) as well as a 100,000g pellet (P100), which represent the mitochondrial, cytosolic, and microvesicular fractions, respectively. The P15 and S15 fractions were analyzed by SDS-PAGE and Western blotting with anti-cytochrome c antibodies to detect release of mitochondrial cytochrome c.11 The P100 fractions were suspended in 50 μL of caspase buffer (20 mmol/L PIPES, 100 mmol/L NaCl, 10 mmol/L dithiothreitol, 1 mmol/L EDTA, 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonic acid (CHAPS), 10% sucrose, pH 7.2). Aliquots (10 μL) of each sample were subjected to SDS-PAGE and Western blotting with anti-gp IIb antibodies to assess microvesiculation.28 29
Proteinase assays.
To detect caspase-3–like activity, samples (75 μg protein) from cell-free reactions were incubated at 37°C for 30 minutes with DEVD-pNA (100 μmol/L in caspase buffer). Substrate hydrolysis was measured by sample absorbance at 406 nm. A molar extinction coefficient of 10,000 M−1cm−1 was used to determine the concentration of pNA generated from substrate hydrolysis.
For studies involving platelet activation and apoptosis, we determined caspase activity using more sensitive AFC-based fluorogenic substrates. LEHD-AFC and DEVD-AFC were used to detect caspase-9–like and caspase-3–like activity, respectively.39 Whole-cell lysates (25 μg protein) were incubated with 100 μmol/L LEHD-AFC or DEVD-AFC in caspase buffer at 37°C. Initial rates of substrate hydrolysis were determined using a Tecan SpectroFluor fluorimeter in the kinetic mode. Excitation was at 400 nm and emission was at 500 nm (slit widths 10 nm). A standard curve of AFC concentration versus fluorescence was used to determine the concentration of AFC generated from substrate hydrolysis.
Inhibitor studies with purified proteinases.
Purified calpain or caspase-3, each at a final concentration of 100 nmol/L, were incubated with ZVAD-fmk (0 to 10 μmol/L) or calpeptin (0 to 10 μmol/L) for 20 minutes at room temperature. Residual enzyme activity was then measured fluorimetrically using LLVY-AFC (100 μmol/L) for calpain or DEVD-AFC (100 μmol/L) for caspase-3.39 40 Calpain reactions were conducted in calpain buffer (caspase buffer lacking EDTA and supplemented with 1 mmol/L CaCl2) and caspase-3 reactions were conducted in caspase buffer. Initial rates of substrate hydrolysis were determined at each inhibitor concentration. Results were plotted as percent maximal enzyme activity (determined in the absence of inhibitor) versus the logarithm of the inhibitor concentration.
Reaction of calpain with procaspase-9 and procaspase-3 in vitro.
Calpain was reacted with 35S-procaspase-9 or recombinant procaspase-3 for various times at 37°C in calpain buffer as detailed in the legend for Fig 6. Reaction products were analyzed by SDS-PAGE followed by autoradiography (35S-procaspase-9) or staining with Coomassie brilliant blue (procaspase-3).
To determine if calpain activates procaspase-3, procaspase-3 (0.5 μmol/L) was reacted with calpain (100 nmol/L) for 15 minutes at 37°C and then caspase-3 activity was measured by monitoring DEVD-AFC hydrolysis as described above. As a positive control, procaspase-3 was activated with granzyme B (1 U) under the same conditions. We also examined whether granzyme B could activate calpain-processed caspase-3 because preliminary studies showed that calpain-cleaved procaspase-3 was not active. To accomplish this, we treated procaspase-3 with calpain for 15 minutes and then with granzyme B (1 U) for an additional 15 minutes before DEVDase assay.
Identification of calpain cleavage sites in procaspase-3 and procaspase-9.
Fifty micrograms of each caspase zymogen was digested with 100 nmol/L calpain in calpain buffer for 15 minutes at 37°C. The active site mutant of caspase-9 was used because this allows purification of large amounts of the zymogen from E coli. Reaction products were separated by 8% to 18% SDS-PAGE, blotted to polyvinylidene fluoride membranes and the major fragments sequenced by Edman degradation using an Applied Biosystems 476A Protein Sequencer.
RESULTS
Identification of platelet caspases and caspase activators.
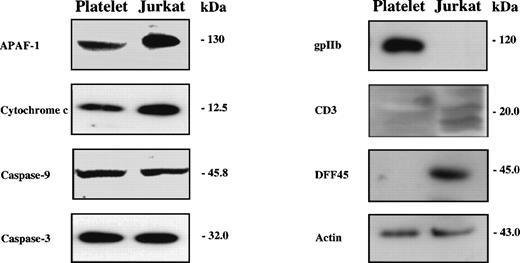
To determine whether human platelets contain caspases and their activators, we subjected platelet lysates to Western blotting with antibodies against these proteins. We analyzed equivalent amounts of Jurkat cell lysates for comparison. As shown in Fig 1, comparable amounts of APAF-1, cytochrome c, caspase-9, and caspase-3 were present in platelets and Jurkat cells. Both caspase-9 and caspase-3 were detected solely as the zymogen forms in both extracts. No APAF-1 or caspase-9 immunoreactivity was detected when preimmune rabbit serum was substituted for immune antiserum (not shown). Because platelets contain abundant amounts of adenine nucleotides,41 platelets contain all components necessary for mitochondria-dependent caspase activation.
Human platelets contain caspases and caspase activators. Whole-cell lysates (25 μg protein) were subjected to SDS-PAGE and Western blotting with antibodies against apoptotic proteins (APAF-1, cytochrome c, caspase-9, caspase-3, DFF45), a platelet marker (gpIIb), a T-cell marker (CD3), and actin as a loading control. The presented blots were performed with lysates from platelet concentrates. Comparable results were obtained with freshly isolated platelets.
Human platelets contain caspases and caspase activators. Whole-cell lysates (25 μg protein) were subjected to SDS-PAGE and Western blotting with antibodies against apoptotic proteins (APAF-1, cytochrome c, caspase-9, caspase-3, DFF45), a platelet marker (gpIIb), a T-cell marker (CD3), and actin as a loading control. The presented blots were performed with lysates from platelet concentrates. Comparable results were obtained with freshly isolated platelets.
To assess the purity of our platelet preparations, we subjected platelet lysates to Western blotting with antibodies against gpIIb, a platelet marker, and antibodies against CD3, a component of the T-cell receptor. Although the preparation showed immunoreactivity to gpIIb, the platelets contained no detectable CD3 immunoreactivity (Fig 1). Additionally, platelet extracts (50 μg protein) contained no detectable caspase-activated DNase activity in a cell-free assay and lacked DFF-45, a caspase substrate that must be cleaved before DNA fragmentation in nucleated cells (Fig 1 and data not shown).21 22 Since caspase-dependent DNA fragmentation was detectable in Jurkat cell extracts or buffy coat extracts with as little as 0.1 μg of extract (not shown), we conclude that our platelet preparations were not significantly contaminated with nucleated cells. Comparable results were obtained with freshly isolated platelets and platelets isolated from platelet concentrates.
Cytochrome c and dATP-dependent caspase activation.
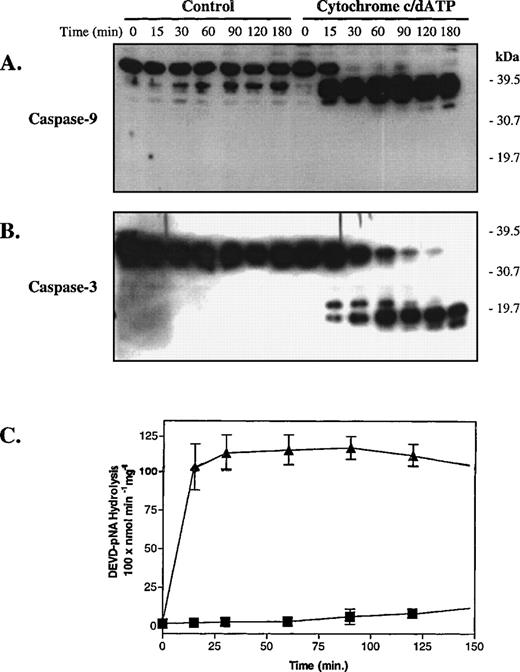
We next determined if cytochrome c and dATP would activate platelet caspase-9 and caspase-3 in vitro. To accomplish this, we incubated platelet cytosolic extracts with and without cytochrome c and dATP and then analyzed procaspase processing by SDS-PAGE and Western blotting. As shown in Fig 2, cytochrome c and dATP initiated rapid sequential processing of procaspase-9 and procaspase-3 to their proteolytically active fragments. Procaspase processing coincided with detection of caspase-3–like DEVDase activity in the cytochrome c and dATP-treated platelet extracts (Fig 2C).39No caspase activation occurred in the absence of cytochrome c and dATP. Thus, cytochrome c and dATP initiate sequential activation of caspase-9 and caspase-3 in platelet cytosolic extracts as in extracts from nucleated cells.33 37
Cytochrome c and dATP initiate sequential caspase-9 and caspase-3 activation in platelet cytosolic extracts. Platelet cytosolic extracts derived from hospital concentrates (final concentration, 5 mg protein/mL) were treated with buffer or cytochrome c (10 μmol/L) and dATP (1 mmol/L) and then analyzed by SDS-PAGE and Western blotting (35 μg protein/lane). (A) Procaspase-9 processing in response to cytochrome c and dATP. (B) The blot from (A) was stripped and reprobed with anti–caspase-3 antibodies. At the indicated times after incubation with buffer (▪) or cytochrome c and dATP (▴), aliquots (75 μg protein) of cell-free reactions were analyzed for activity against DEVD-pNA (C). (A) and (B) are representative of 3 independent experiments. In (C), error bars represent the SEM, N = 3.
Cytochrome c and dATP initiate sequential caspase-9 and caspase-3 activation in platelet cytosolic extracts. Platelet cytosolic extracts derived from hospital concentrates (final concentration, 5 mg protein/mL) were treated with buffer or cytochrome c (10 μmol/L) and dATP (1 mmol/L) and then analyzed by SDS-PAGE and Western blotting (35 μg protein/lane). (A) Procaspase-9 processing in response to cytochrome c and dATP. (B) The blot from (A) was stripped and reprobed with anti–caspase-3 antibodies. At the indicated times after incubation with buffer (▪) or cytochrome c and dATP (▴), aliquots (75 μg protein) of cell-free reactions were analyzed for activity against DEVD-pNA (C). (A) and (B) are representative of 3 independent experiments. In (C), error bars represent the SEM, N = 3.
Cytochrome c and dATP-dependent caspase activation promotes proteolysis of apoptotic substrates.
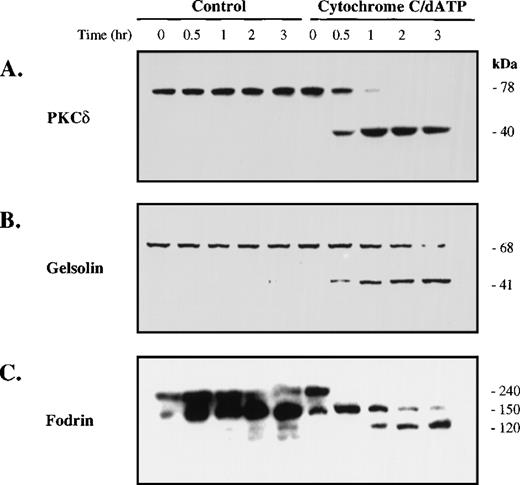
As shown in Fig 3, cytochrome c and dATP-dependent caspase activation in platelet extracts leads to proteolysis of gelsolin, PKCδ, and fodrin, proteins cleaved during apoptosis.17-20 Cleavage of these substrates was evident within 30 minutes and nearly complete by 3 hours. In the absence of cytochrome c and dATP, no substrate cleavage was observed, even at the latter time points. Thus, cytochrome c and dATP initiate caspase activation and subsequent cleavage of caspase substrates in platelet extracts. Therefore, platelet extracts recapitulate cytosolic events that occur in apoptotic cells and in cytosolic extracts from nucleated cells.
Cytochrome c and dATP-dependent caspase activation promotes proteolysis of apoptotic substrates. Cell-free reactions were prepared as described in the legend to Fig 2. At the indicated times, portions (50 μg protein) of the extract were removed for Western blotting with antibodies against PKCδ (A), gelsolin (B), or fodrin (C). Molecular weights (kD) are listed for the endogenous and cleaved forms.
Cytochrome c and dATP-dependent caspase activation promotes proteolysis of apoptotic substrates. Cell-free reactions were prepared as described in the legend to Fig 2. At the indicated times, portions (50 μg protein) of the extract were removed for Western blotting with antibodies against PKCδ (A), gelsolin (B), or fodrin (C). Molecular weights (kD) are listed for the endogenous and cleaved forms.
A comparison of platelet activation and apoptosis.
Because activated platelet extracts demonstrated caspase activity in vitro, we wished to determine whether caspases function during platelet activation. To accomplish this, we stimulated freshly isolated platelets with calcium ionophore A23187 and then examined caspase processing and activity. Naive and UV-irradiated Jurkat cells were also analyzed to compare apoptotic events with similar events during ionophore stimulation. We observed cell shrinkage (decreased forward scatter) and PS externalization by fluorescence-activated cell sorting (FACS) analysis in both ionophore-stimulated platelets and UV-treated (apoptotic) Jurkat cells (not shown). However, these events occurred within 15 minutes in ionophore-treated platelets, but required 6 hours to occur in apoptotic cells. In the studies presented in Fig 4, greater than 90% of A23187-treated platelets were PS positive, compared with 1.5% of untreated platelets. With Jurkat cells, 68% of UV-treated cells were PS positive, compared with 3% of untreated cells.
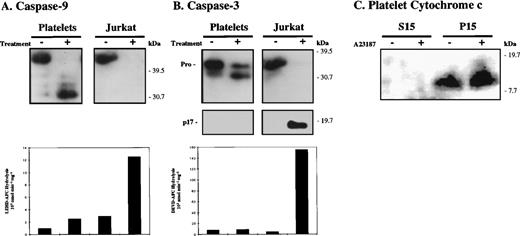
Caspase activation accompanies apoptosis but not platelet activation. Platelets were activated with calcium ionophore A23187 (2 μmol/L) for 15 minutes at 37°C. For comparison, control and apoptotic (6 hours post UVB-treatment) Jurkat cells were examined in parallel with platelets. Platelet activation and apoptosis were confirmed by monitoring cell shrinkage (decreased forward scatter) and PS externalization by FACS analysis as detailed in the text. Cell lysates were analyzed by SDS-PAGE and Western blotting (50 μg protein/lane) with antibodies against caspase-9 (A) and caspase-3 (B). Fifty micrograms of lysate were also assayed against LEHD-AFC and DEVD-AFC to detect caspase-9–like (A, bottom panel) and caspase-3–like activity (B, bottom panel). In (C), control and A2387-treated platelets were subjected to subcellular fractionation. The cytosolic (S15) and mitochondrial (P15) fractions (40 μg protein) were then subjected to SDS-PAGE and Western blotting with anti-cytochrome c antibodies. The presented data are representative of 3 independent experiments.
Caspase activation accompanies apoptosis but not platelet activation. Platelets were activated with calcium ionophore A23187 (2 μmol/L) for 15 minutes at 37°C. For comparison, control and apoptotic (6 hours post UVB-treatment) Jurkat cells were examined in parallel with platelets. Platelet activation and apoptosis were confirmed by monitoring cell shrinkage (decreased forward scatter) and PS externalization by FACS analysis as detailed in the text. Cell lysates were analyzed by SDS-PAGE and Western blotting (50 μg protein/lane) with antibodies against caspase-9 (A) and caspase-3 (B). Fifty micrograms of lysate were also assayed against LEHD-AFC and DEVD-AFC to detect caspase-9–like (A, bottom panel) and caspase-3–like activity (B, bottom panel). In (C), control and A2387-treated platelets were subjected to subcellular fractionation. The cytosolic (S15) and mitochondrial (P15) fractions (40 μg protein) were then subjected to SDS-PAGE and Western blotting with anti-cytochrome c antibodies. The presented data are representative of 3 independent experiments.
As shown in Fig 4, apoptosis correlated with processing of procaspase-9 and procaspase-3 and the onset of caspase activity (Fig 4A and B). After 6 hours, caspase-9 immunoreactivity was no longer detectable in the apoptotic Jurkat cells; however, a low level of caspase-9–like activity was detected using the substrate LEHD-AFC.39 The apoptotic Jurkat cells contained only the active form of caspase-3 and showed activity against the caspase-3 substrate DEVD-AFC.39Procaspase processing also occurred in the ionophore-stimulated platelets; however, no caspase activity was detected (Fig 4A and B). Procaspase-9 was cleaved to multiple fragments, primarily to an ≈30-kD form. This contrasts to the 35-kD active form of caspase-9 observed in response to cytochrome c and dATP (Fig 2). A slight increase in LEHDase activity was observed in the ionophore-stimulated platelets; however, this result was not statistically significant as assessed by the paired Student’st-test. Similarly, procaspase-3 was processed to an ≈30-kD form during platelet activation, which is distinct from the active p17 caspase-3 fragment observed during apoptosis and in response to cytochrome c and dATP (Figs 2 and 4B). No caspase-3–like DEVDase activity was detected after ionophore stimulation. Thus, procaspase processing occurs without detectable caspase activation during ionophore stimulation. Furthermore, cytochrome c did not translocate to the platelet cytosol (S15) during platelet activation (Fig 4C), eliminating the possibility that caspase processing requires cytochrome c release or APAF-1.
To confirm that the preceding findings were not limited to ionophore stimulation, we examined caspase processing and activity after treatment of platelets with physiological platelet agonists. We observed procaspase-3 processing to an ≈30-kD form after treatment of platelets with thromboxane agonist U46619, thrombin, or thrombin and collagen; however, no caspase-3–like DEVDase activity was detected (not shown). Procaspase-9 processing was not examined; however, none of the preceding treatments induced activity against the caspase-9 substrate LEHD-AFC. Furthermore, anti-Fas antibodies did not activate platelets or induce platelet caspase activation (not shown). Therefore, platelet activation apparently occurs independently of mitochondria– and death-receptor–dependent caspase pathways.
ZVAD-fmk and calpeptin inhibit microvesiculation, caspase processing, and substrate cleavage during platelet activation.
We next sought to determine whether calpain might promote apoptosis-like events during platelet activation or whether platelet activation proceeds with a very low level of caspase activity (below the assay detection level). To accomplish this, we examined the effect of ZVAD-fmk and calpeptin, 2 cell-permeable proteinase inhibitors, on platelet activation. ZVAD-fmk inhibits most caspases and calpeptin potently inhibits calpain.42,43 Calpeptin and other calpain inhibitors like MDL 28,170 therefore inhibit calpain-dependent platelet activation events.28 We incubated platelets with buffer, ZVAD-fmk, or calpeptin for 30 minutes and then stimulated them with ionophore A23187. After 15 minutes of activation, we assessed how the inhibitors affected various platelet activation parameters.
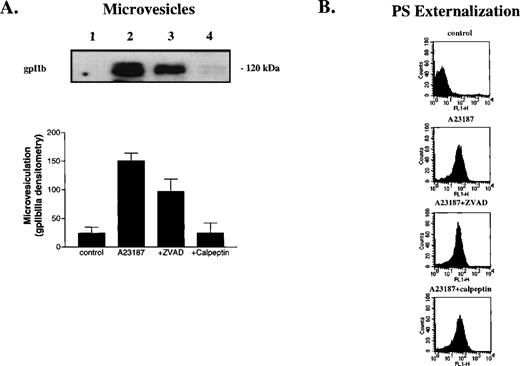
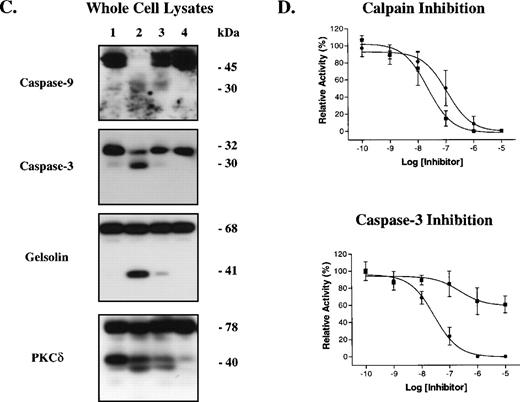
Ionophore A23187 induced platelet microvesiculation, PS externalization, caspase processing, and cleavage of the ‘apoptotic’ substrates, gelsolin and PKCδ (Fig 5). ZVAD-fmk and calpeptin attenuated microvesiculation, caspase processing, and substrate cleavage; however, calpeptin was a more potent inhibitor. By contrast, these inhibitors did not significantly alter PS externalization or cell shrinkage (Fig 5B and not shown). We observed no activity against the caspase substrates LEHD-AFC, DEVD-AFC, VEID-AFC, or YVAD-AFC in the absence or presence of inhibitors (Fig 4and not shown), suggesting that ZVAD-fmk inhibits a noncaspase proteinase(s) that promotes proteolysis of caspases, gelsolin and PKCδ.39 Because ZVAD-fmk inhibited the same events as calpeptin, the data suggest that ZVAD-fmk inhibits calpain and that calpain cleaves caspases and ‘apoptotic’ substrates.
Calpain inhibition prevents platelet microvesiculation, caspase processing, and apoptotic substrate cleavage, but not PS externalization. Platelets were preincubated with vehicle (lanes 1 and 2), ZVAD-fmk (25 μmol/L; lane 3) or calpeptin (25 μmol/L; lane 4) for 30 minutes at room temperature and then treated with buffer (lane 1) or A23187 (lanes 2 through 4) for 15 minutes at 37°C. Microvesicles were then isolated from platelet supernatants as described in Materials and Methods and analyzed by Western blotting (10-μL aliquots/lane) with anti-gpIIb antibodies (A). The graph in (A) shows the extent of microvesiculation as assessed by gpIIb densitometry (arbitrary units). In (B), plasma membrane PS externalization was determined by annexin V-FITC binding and FACS analysis. (C) Whole-cell lysates analyzed by Western blotting with antibodies against caspase-9, caspase-3, gelsolin, and PKCδ (50 μg protein/lane). (D) Inhibition of 100 nmol/L purified calpain and caspase-3 by ZVAD-fmk (•) and calpeptin (▪). Error bars represent the SEM, N = 4.
Calpain inhibition prevents platelet microvesiculation, caspase processing, and apoptotic substrate cleavage, but not PS externalization. Platelets were preincubated with vehicle (lanes 1 and 2), ZVAD-fmk (25 μmol/L; lane 3) or calpeptin (25 μmol/L; lane 4) for 30 minutes at room temperature and then treated with buffer (lane 1) or A23187 (lanes 2 through 4) for 15 minutes at 37°C. Microvesicles were then isolated from platelet supernatants as described in Materials and Methods and analyzed by Western blotting (10-μL aliquots/lane) with anti-gpIIb antibodies (A). The graph in (A) shows the extent of microvesiculation as assessed by gpIIb densitometry (arbitrary units). In (B), plasma membrane PS externalization was determined by annexin V-FITC binding and FACS analysis. (C) Whole-cell lysates analyzed by Western blotting with antibodies against caspase-9, caspase-3, gelsolin, and PKCδ (50 μg protein/lane). (D) Inhibition of 100 nmol/L purified calpain and caspase-3 by ZVAD-fmk (•) and calpeptin (▪). Error bars represent the SEM, N = 4.
To determine the relative potency of ZVAD-fmk and calpeptin for calpain and caspase-3, we incubated the purified proteinases with various concentrations of the inhibitors and then determined residual proteinase activity against fluorogenic substrates. Both ZVAD-fmk and calpeptin effectively inhibited calpain, with both inhibitors completely abolishing calpain activity by 1 μmol/L inhibitor (Fig5D). ZVAD-fmk potently inhibited caspase-3; however, calpeptin was a poor caspase-3 inhibitor (Fig 5D). Together with the preceding data, these results suggest that ZVAD-fmk and calpeptin inhibit calpain during platelet activation and this inhibition prevents caspase processing and ‘apoptotic’ substrate cleavage.
Reaction of purified calpain with procaspase-9 and procaspase-3 in vitro.
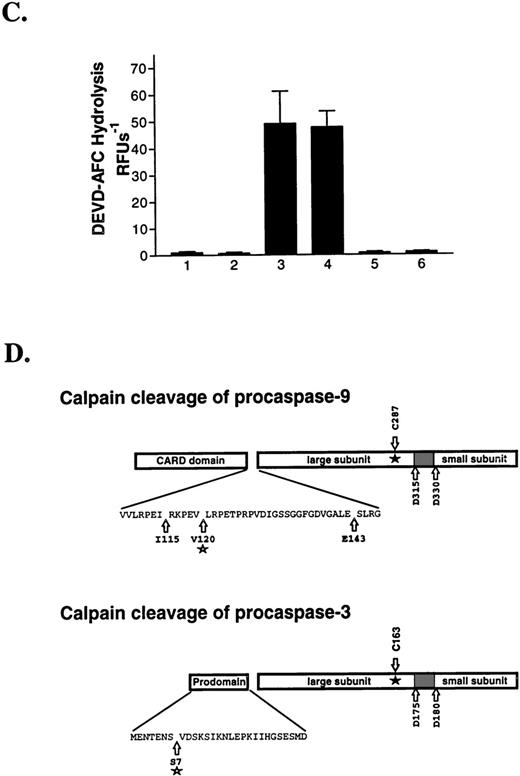
We next incubated purified calpain with 35S-procaspase-9 to determine if the proteinase directly processed the zymogen. As shown in Fig 6A, calpain rapidly processed the 45-kD procaspase-9 to several fragments. Major fragments of ≈30 to 34 kD and ≈11 kD were observed. Nearly complete procaspase-9 processing was observed within 10 minutes and no further proteolysis was observed after 1 hour of incubation. ZVAD-fmk and calpeptin abolished procaspase-9 processing (Fig 6A, lanes 8 and 9).
Purified calpain removes the prodomain of caspase-9 and caspase-3 via specific limited proteolysis. Calpain (100 nmol/L) was reacted with 35S-procaspase-9 (5 μL; A) or recombinant procaspase-3 (10 μmol/L; B) in the absence and presence of calpeptin (1 μmol/L) or ZVAD-fmk (1 μmol/L) and the products were analyzed by SDS-PAGE and autoradiography (A) or staining with Coomassie Blue (B). To determine whether calpain activates procaspase-3, we treated the procaspase with calpain and then measured caspase-3 activity by monitoring DEVD-AFC hydrolysis as shown in (C). The graph shows DEVD-AFC hydrolysis by procaspase-3 (1), procaspase-3 reacted with calpain (2), granzyme B (3), or calpain then granzyme B (4) (error bars represent the SEM, N = 3). Columns 5 and 6 show the DEVDase activity of calpain and granzyme B, respectively. In (D), purified procaspase-9 and procaspase-3 were digested with calpain and the products sequenced by Edman degradation. The insets show the calpain cleavage sites. C287 and C163 denote the caspase-9 and caspase-3 active site cysteines. The shaded areas between the caspase large and small subunits represent the inter-domain linker that is removed upon caspase activation.
Purified calpain removes the prodomain of caspase-9 and caspase-3 via specific limited proteolysis. Calpain (100 nmol/L) was reacted with 35S-procaspase-9 (5 μL; A) or recombinant procaspase-3 (10 μmol/L; B) in the absence and presence of calpeptin (1 μmol/L) or ZVAD-fmk (1 μmol/L) and the products were analyzed by SDS-PAGE and autoradiography (A) or staining with Coomassie Blue (B). To determine whether calpain activates procaspase-3, we treated the procaspase with calpain and then measured caspase-3 activity by monitoring DEVD-AFC hydrolysis as shown in (C). The graph shows DEVD-AFC hydrolysis by procaspase-3 (1), procaspase-3 reacted with calpain (2), granzyme B (3), or calpain then granzyme B (4) (error bars represent the SEM, N = 3). Columns 5 and 6 show the DEVDase activity of calpain and granzyme B, respectively. In (D), purified procaspase-9 and procaspase-3 were digested with calpain and the products sequenced by Edman degradation. The insets show the calpain cleavage sites. C287 and C163 denote the caspase-9 and caspase-3 active site cysteines. The shaded areas between the caspase large and small subunits represent the inter-domain linker that is removed upon caspase activation.
Calpain also showed activity against purified procaspase-3 (Fig 6B). Calpain processed the zymogen to an ≈30-kD fragment within 15 minutes at 37°C. ZVAD-fmk and calpeptin both attenuated procaspase-3 processing (lanes 5 and 6). The 16-kD band present in all lanes represents a contaminant, which does not promote procaspase-3 activation, as the preparation showed no activity against DEVD-AFC (Fig6C). Calpain-treated procaspase-3 showed no activity against DEVD-AFC, indicating that calpain cleavage does not activate procaspase-3 (Fig6C). By contrast, granzyme B, a potent caspase activator, activated procaspase-3 within 15 minutes at 37°C, as shown by the appearance of DEVDase activity.16 Granzyme B readily activated calpain-treated procaspase-3, indicating that calpain cleavage does not inactivate procaspase-3.
Identification of the calpain cleavage sites in procaspase-9 and procaspase-3.
To identify the calpain cleavage sites in procaspase-9 and procaspase-3, we digested each zymogen with calpain and sequenced the products by Edman degradation. Calpain cleaved procaspase-9 primarily at Val120 and to a lesser extent at Ile115 and Glu143 (Fig 6D). These cleavage sites are all located between the procaspase-9 prodomain and the large subunit.44Therefore, processing at any of these sites would not activate the caspase. Similarly, calpain removed a fragment of the procaspase-3 prodomain by cleaving after Ser7.45 This cleavage would not promote procaspase-3 activation, consistent with the preceding activation studies.
DISCUSSION
In this study, we found that human platelets contain functional pro-apoptotic caspases. Surprisingly, however, we found that calpain, not caspases, promotes events resembling apoptosis during platelet activation. This suggests that platelet activation is not equivalent to apoptosis and that certain aspects of the apoptotic phenotype may occur without caspase activation.
Platelets contain caspase-3, caspase-9, and the caspase activators APAF-1 and cytochrome c, at comparable levels to nucleated cells (Fig1). Furthermore, the extent and rate of caspase-3 activation and apoptotic substrate cleavage observed in platelet extracts is similar to that observed in extracts from nucleated cells, suggesting that the 2 extracts are functionally equivalent (Figs 2 and3).33,36,37 However, platelets lack DFF45 (Fig 1) and consequently contain no detectable caspase-dependent DNase (DFF40) activity. The platelet cell-free system should therefore be useful to dissect the role of DFF45, DFF40, and other molecules in nuclear apoptosis and DNA fragmentation. Furthermore, because megakaryocytes undergo apoptosis, study of megakaryocyte differentiation may provide insight into developmental regulation of caspases and DNases.46
During apoptosis, caspases mediate cell shrinkage, plasma membrane microvesiculation, PS externalization, and apoptotic substrate cleavage.1 We observed comparable events during platelet activation; surprisingly, however, we found that caspases do not participate in this process. Three lines of evidence support this finding. First, apoptosis, but not platelet activation, correlated with procaspase processing and activation (Fig 4). Second, calpain inhibition, not caspase inhibition, prevented apoptosis-like events during A23187 stimulation (Fig 5). Third, there was no release of mitochondrial cytochrome c during platelet activation, an event critical for caspase activation in many forms of apoptosis (Fig 4). Because we also failed to detect caspase activation after treatment of platelets with thrombin, thrombin and collagen, or a thromboxane analog, we conclude that platelet activation occurs independently of caspase activation.
Because platelet activation occurs irreversibly, the process results in platelet death. In this context, platelet activation might represent a nonapoptotic form of cell death that involves neither caspases nor other pro-apoptotic molecules. Supporting this, many platelet activation events including adhesion to endothelial cells, extension of filopodia, self-aggregation, and release of intracellular granules distinguish this process from apoptosis.5-7G-protein–coupled receptors, adhesion receptors, and second messenger systems involving protein phosphorylation, calcium influx, and metabolism of inositol polyphosphates and diacyl glycerol mediate these events; however, definitive roles for these signaling molecules in apoptosis have not been established.5-7 Furthermore, platelet activation proceeds much more rapidly than apoptosis and does not involve nuclear events. Thus, certain aspects of platelet activation are distinct from apoptosis and do not require caspases or other apoptotic signaling molecules.
Alternatively, platelet activation pathways may use pro-apoptotic molecules other than caspases. For example, Vanags et al8recently detected upregulation of Bax, a pro-apoptotic Bcl-2 family member, in activated platelets. Bax can induce caspase-independent cell death and this may contribute to platelet demise during activation.47,48 Additionally, noncaspase proteinases such as calpain might cleave caspase substrates to recapitulate apoptotic events during platelet activation. Cleavage of caspase substrates like gelsolin (Fig 6), PKCδ (Fig 6), fodrin, and pp125-focal adhesion kinase by calpain supports this hypothesis.17 49-51
Two surprising findings in this study concern the calcium-dependent proteinase, calpain. First, we found that the broad specificity caspase inhibitor, ZVAD-fmk, potently inhibits calpain (Fig 5). Although calpain prefers Tyr, Met, or Arg in the P1 position of peptide substrates, it cleaves procaspase-9 after a Glu residue (Fig 6) and glucagon after an Asp residue.40 Therefore, our finding that ZVAD-fmk inhibits calpain is not entirely unexpected, especially since the fluoromethyl ketone (fmk) group efficiently modifies the catalytic thiol of cysteine proteinases. Although this study focused on μ-calpain, m-calpain demonstrates similar substrate specificity and thus may also be susceptible to ZVAD-fmk.40 Because ZVAD-fmk inhibits caspase-3 and calpain, inhibitor studies alone cannot distinguish the biological activities of these proteinases. Caution should therefore be used when using ZVAD-fmk to study apoptotic events.
A second interesting finding was that calpain cleaves procaspases without activating them, both in vitro and during platelet activation (Figs 5 and 6). Proteolysis of procaspase-3 removed a portion of the caspase’s prodomain. However, proteolysis did not inactivate caspase-3 because granzyme B activated the calpain-processed zymogen, in agreement with studies we recently published concerning activation of caspase-3 mutants.16 Full or partial deletion of the procaspase-3 prodomain did not significantly alter procaspase-3 activation by caspase-8, caspase-10, or granzyme B. Furthermore, once activated, the mutant proteins displayed equal reactivity toward substrates and the baculovirus p35 caspase inhibitor. Together, these results suggest that limited proteolysis of procaspase-3 does not alter its interaction with other proteinases or inhibitors.
Calpain also removed the procaspase-9 prodomain during platelet activation and in vitro in a purified system. Because binding of caspase-9 to APAF-1 is necessary for its activity and because the caspase-9 prodomain mediates this interaction,13,32,33calpain-dependent removal of the prodomain could significantly abrogate caspase-9 activation. Since calpain also did not activate procaspase-3 (Fig 5), this finding suggests that calpain must function downstream or parallel to caspases during apoptosis. In fact, 2 recent reports support this hypothesis.26 27 Alternatively, if calpain acted on caspase-9 before an apoptotic stimulus, then removal of the prodomain might promote cell survival by preventing caspase-9 activation and apoptosis.
In summary, human platelets contain functional pro-apoptotic caspases; however, caspases do not function during platelet activation, even though this process resembles apoptosis. What role might caspases play in platelet biology? Because megakaryoctes can undergo apoptosis, caspases might simply have been transferred to the platelet during development. However, platelet production in vitro correlates with the onset of megakaryocyte apoptosis, suggesting a potential role for apoptosis and caspases in platelet production.46 By contrast, because caspases and apoptosis often mediate cell senescence, caspases might function during platelet senescence. Further study of platelets and megakaryocytes will provide answers to these questions and enhance our knowledge of the caspase system.
ACKNOWLEDGMENT
The authors thank Drs Xiadong Wang and Emad Alnemri for reagents.
B.B.W. was supported by a Mentored Clinical Scientist Development Award (CA75268-01). H.R.S. was supported by a grant from the Danish Natural Science Foundation (9600412). G.S.S. and D.R.G. received funding from the National Institutes of Health (NS37878 to G.S.S. and CA69831 and AI40646 to D.R.G.). This is publication no. 300 from the La Jolla Institute for Allergy and Immunology.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Beni B. Wolf, MD, PhD, La Jolla Institute for Allergy and Immunology, 10355 Science Center Dr, San Diego, CA 92121; e-mail: 102251.1444@compuserve.com.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal