Abstract
CCR5 was first characterized as a receptor for MIP-1, MIP-1β, and RANTES, and was rapidly shown to be the main coreceptor for M-tropic human immunodeficiency virus (HIV)-1 strains and simian immunodeficiency virus (SIV). Chemokines constitute a rapidly growing family of proteins and receptor-chemokine interactions are known to be promiscuous and redundant. We have therefore tested whether other CC-chemokines could bind to and activate CCR5. All CC-chemokines currently available were tested for their ability to compete with [125I]-MIP-1β binding on a stable cell line expressing recombinant CCR5, and/or to induce a functional response in these cells. We found that in addition to MIP-1β, MIP-1, and RANTES, five other CC-chemokines could compete for [125I]-MIP-1β binding: MCP-2, MCP-3, MCP-4, MCP-1, and eotaxin binding was characterized by IC50 values of 0.22, 2.14, 5.89, 29.9, and 21.7 nmol/L, respectively. Among these ligands, MCP-3 had the remarkable property of binding CCR5 with high affinity without eliciting a functional response, MCP-3 could also inhibit the activation of CCR5 by MIP-1β and may therefore be considered as a natural antagonist for CCR5. It was unable to induce significant endocytosis of the receptor. Chemokines that could compete with high affinity for MIP-1β binding could also compete for monomeric gp120 binding, although with variable potencies; maximal gp120 binding inhibition was 80% for MCP-2, but only 30% for MIP-1β. MCP-3 could compete efficiently for gp120 binding but was, however, found to be a weak inhibitor of HIV infection, probably as a consequence of its inability to downregulate the receptor.
CCR5 WAS CHARACTERIZED originally as a receptor responding functionally to the CC-chemokines MIP-1α, MIP-1β, and RANTES.1 CCR5 was further described as a major coreceptor for human immunodeficiency virus (HIV), after the demonstration that its three ligands constitute major HIV-suppressive factors produced by CD8+ lymphocytes.2 Cellular entry of primate lentivirus (HIV and simian immunodeficiency virus [SIV]) is initiated by the interaction between the virus membrane glycoprotein (gp120) and CD4. CD4 binding triggers conformational changes in gp120 that enable it to interact with a coreceptor, ultimately resulting in membrane fusion and viral entry.3HIV tropism can be largely explained by coreceptor usage. CCR5 is the major coreceptor for macrophage (M)-tropic HIV-1, HIV-2, and SIV strains (therefore referred to as R5 strains), whereas CXCR4 is the major coreceptor for T-tropic strains (now referred to as X4 strains). Direct interaction between gp120 and CCR5 was shown to involve conserved and variable regions of gp120.5-7 The strong resistance to HIV-1 infection of individuals homozygous for a deletion in the CCR5 coding region (CCR5Δ32), resulting in the production of a truncated receptor not expressed at the cell surface,8-10and HIV-1 inhibitory activities of MIP-1α, MIP-1β, and RANTES,2-11 have highlighted the essential role of CCR5 in HIV-1 pathogenesis.
Two mechanisms have been proposed to account for the ability of chemokines to inhibit HIV-1 infection. Firstly, the partial overlap of chemokine and gp120 binding sites allows a direct competition for access to the coreceptor.12 Secondly, receptor activation by full or partial agonists results in desensitization and internalization of the receptor, and therefore in the reduction of coreceptor surface expression.13 14 These mechanisms are likely to be complementary, although their relative contributions remain to be clarified in each case. The chemokine family has grown rapidly over the recent years, and more than 20 CC-chemokines have been reported to date. For most of the recently described proteins, receptors functionally responding to them have been identified. Some CC-chemokines however, such as LEC/ILINK and PARC/DC-CK1, have not yet been shown to act through one of the currently characterized chemokine receptors or related orphan receptors. Chemokine-receptor interactions are well known to be promiscuous and redundant, because most receptors are functionally activated by several chemokines, and most chemokines can bind and activate more than one receptor. It was therefore likely that some of the newly described chemokines would act through CCR5. Defining CCR5 pharmacology could help to understand the relationship between structural, functional, and anti-HIV properties of CC-chemokines.
In this work, we have tested all currently available CC-chemokines for their ability to bind to CCR5, to activate the receptor, to promote its downregulation, and to inhibit CCR5-mediated HIV-1 entry. We have shown that in addition to MIP-1β, MIP-1α, and RANTES, five other CC chemokines could efficiently compete for MIP-1β binding on CCR5. These were, with decreasing affinities: MCP-2, MCP-3, MCP-4, eotaxin, and MCP-1. Among these, MCP-2 and MCP-4 were full agonists, whereas MCP-3 could bind CCR5 with high affinity without eliciting a functional response and had the ability to inhibit functional response to MIP-1β. MCP-3 induced no CCR5 endocytosis, could also compete for gp120 binding, but was a weak inhibitor of HIV infection.
MATERIALS AND METHODS
Chemokines.
Recombinant human MIP-1α, MIP-1β, RANTES, MCP-1, MCP-2, MCP-3, MCP-4, eotaxin, MIP-3α, MIP-3β, SLC, TARC, MDC, I309, and MPIF-2 were obtained from R&D Systems (London, UK). Recombinant TECK, MPIF-1, PARC, and LEC were obtained from Peprotech (London, UK). HCC1, HCC2, and HCC3 were kindly provided by W.G. Forssmann (IPF, Hannover, Germany). [125I]-MIP-1β (specific activity: 2200 Ci/mmol) was obtained from New England Nuclear (Cambridge, MA). The lyophilized chemokines were dissolved as 10 μmol/L solutions in sterile phosphate-buffered saline (PBS) and stored at −20° C in aliquots. They were diluted to the working concentrations immediately before use.
Cell culture.
PM-1 were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum (Life Technologies, Merelbeke, Belgium), 100 units/mL penicillin, and 100 g/mL streptomycin (Life Technologies). CHO-K1 cells were cultured using HAM’s F12 medium supplemented with 10% fetal calf serum (Life Technologies), 100 units/mL penicillin, and 100 μg/mL streptomycin (Life Technologies).
Expression of mutant receptors in CHO-K1 cells.
A plasmid encoding apoaequorin and Gα16 under control of the SR promoter15 was transfected into CHO-K1 cells, using Fugene 6 (Boerhinger Mannheim, Mannheim, Germany). Zeocin (250 μg/mL; Invitrogen, Carlsbad, CA) selection of transfectants was initiated 2 days after transfection. Individual clones were isolated 3 weeks later, and the most responding clone was selected on the basis of its functional response (luminescence signal) to ionomycin A (100 nmol/L) and ATP (10 μmol/L). A construct encoding wild-type CCR5 was further transfected using Fugene 6 in this apoaequorin and Gα16-expressing cell line. Selection of transfected cells was made for 14 days with 400 μg/mL G418 (Life Technologies), and a clonal cell population expressing high CCR5 level was used for binding and functional studies. The level of receptor expression was measured by flow cytometry using antibodies directed against the second extracellular loop (2D7) of CCR5 (Pharmingen, San Diego, CA).
[125I]-MIP-1β binding assays.
CCR5-expressing CHO-K1 cells were collected from plates with Ca2+ and Mg2+-free PBS supplemented with 5 mmol/L EDTA, gently pelleted for 2 minutes at 1000g, and resuspended in binding buffer (50 mmol/L Hepes pH 7.4, 1 mmol/L CaCl2, 5 mmol/L MgCl2, 0.5% BSA). Competition binding assays were performed in Minisorb tubes (Nunc, Roskilde, Denmark), using 0.08 nmol/L 125I-MIP-1β (2200 Ci/mmol, New England Nuclear) as tracer, variable concentrations of competitors, and 40,000 cells in a final volume of 0.1 mL. Total binding was measured in the absence of competitor and nonspecific binding was measured with a 100-fold excess of unlabelled ligand. Samples were incubated for 90 minutes at 27°C, then bound tracer was separated by filtration through GF/B filters presoaked in 1% BSA. Filters were counted in a β-scintillation counter. Binding parameters were determined with the PRISM software (Graphpad Software, San Diego, CA) using nonlinear regression applied to a one-site competition model.
Functional assays.
Functional response to chemokines was analyzed by measuring the luminescence of aequorin as described.16 CCR5-, apoaequorin-, and G16-expressing cells were collected from plates with Ca2+ and Mg2+-free Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5 mmol/L EDTA, pelleted for 2 minutes at 1000g, resuspended in DMEM at a density of 5 × 106 cells/mL and incubated for 2 hours in the dark in the presence of 5 μmol/L coelenterazine H (Molecular Probes, Eugene, OR). Cells were diluted 7.5-fold before use. Agonists in a volume of 50 L DMEM were added to 50 μL of cell suspension (33,000 cells) and luminescence was measured for 1 minute in a Packard luminometer (Downers Grove, IL). For assaying antagonistic activities, chemokines were added to cell suspensions 1 minute before measuring the functional response to 1 nmol/L MIP-1β.
[125I]-gp120 binding assays.
Soluble JRFL gp120 was iodinated using Iodogen (Pierce, Rockford, IL) to a specific activity of 1000 Ci/mmol by using 5 μg protein with 500 μCi. Na125I radiolabeled proteins were purified from free Na125I by separation through a 0.3 mL Dowex column prepared in a 1 mL syringe and pre-equilibrated in Env binding buffer (50 mmol/L Hepes, pH 7.4, 5 mmol/L MgCl2, 1 mmol/L CaCl2) containing 1% BSA and 150 mmol/L NaCl. Env binding assays were performed by resuspending cells in 75 μL of Env binding buffer containing 5% BSA. 0.5 nmol/L of labeled protein, saturating amounts of sCD4 (100 nmol/L), and the indicated concentrations of chemokines were added to cells in 25 μL of binding buffer for a total volume of 100 μL. 2 × 105 293T cells transfected with CCR5 were incubated at room temperature for 1 hour unless specified. Unbound radioactivity was removed by filtering cells through 25 mm Whatman GF/C filters (Maidstone, UK) presoaked in 0.2% polyethylenimine (Sigma, St Louis, MO), and washing two times with 4 mL wash buffer (50 mmol/L Hepes pH 7.4, 500 mmol/L NaCl, 5 mmol/L MgCl2, 1 mmol/L CaCl2). Filters were counted in a Wallac 1470 Wizard gamma counter (Wallac, Turku, Finland).
Infection assays.
Plasmids encoding the HIV-1 ADA and BaL envs were provided by John Moore (Aaron Diamond AIDS Research Center, New York, NY). The NL4-3 luciferase virus backbone (pNL-Luc-E-R-) was provided by Ned Landau (Aaron Diamond AIDS Research Center). Luciferase reporter viruses were prepared as previously described by cotransfecting 293T cells with the indicated env construct and the NL4-3 luciferase virus. Virus supernatants were used to infect PM-1 cells, a T-cell line naturally expressing both CD4 and CCR5. Inhibition of infection by chemokines was assayed by adding 1 μg/mL of the chemokine at the time of virus infection. Incubation was performed at 37°C, and 4-days postinfection, the cells were lysed with 0.5% triton X-100 in PBS, and the lysate was analyzed for luciferase activity.
CCR5 endocytosis assay.
Chemokine-induced CCR5 endocytosis was performed as described.13 Briefly, CHO-K1 cells stably expresssing human CCR5 were collected from plates with 5 mmol/L EDTA in PBS, washed with PBS, and incubated for 2 hours at 37°C with various chemokines at a 500 nmol/L concentration. Cells were washed two times with 3 mL of cold PBS supplemented with 0.1% sodium azide and 0.1% BSA, and incubated for 30 minutes at 4°C with phycoerythrin-conjugated anti-CCR5 2D7 Mab (Pharmingen). Cells were further washed, resuspended, and their cell fluorescence was analyzed by fluorescence-activated cell sorter (FACS).
RESULTS
Promiscuous binding of CC-chemokines to CCR5.
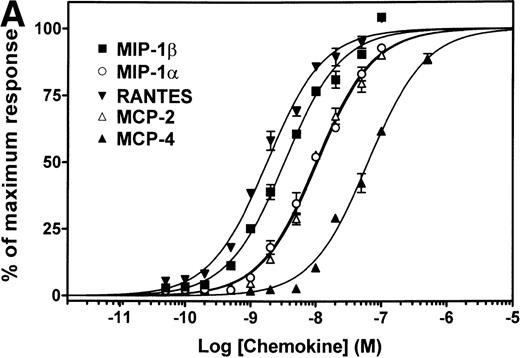
We investigated the ability of all CC-chemokines available to date to compete for [125I]-MIP-1β binding on CCR5 by using a CHO-K1 cell line stably expressing the receptor. We first screened chemokines for their ability to inhibit [125I]-MIP-1β binding at a concentration of 200 nmol/L. MIP-3α, MIP-3β, MIP-4, HCC-1, HCC-2, HCC-3, MPIF-1, MPIF-2, TARC, TECK, SLC, LEC, MDC, and I-309 had no significant effects on MIP-1β binding (data not shown). In addition to the three classical ligands of CCR5, MIP-1α, MIP-1β, and RANTES, five additional chemokines (MCP-1, MCP-2, MCP-3, MCP-4, and eotaxin) did compete for the binding of the tracer and inhibited 80% to 100% of MIP-1β binding at 200 nmol/L. For these ligands, competition binding curves were established (Fig1), allowing the determination of binding affinity parameters (Table 1). On this basis, the CCR5 ligands could be subdivided into high-affinity ligands (IC50 < 1 nmol/L, RANTES = MCP-2 MIP-1β > MIP-1α), intermediate affinity ligands (1 nmol/L < IC50 < 10 nmol/L, MCP-3 > MCP4) and low-affinity ligands (IC50 > 10 nmol/L, MCP-1 = eotaxin). Since for some G-protein–coupled receptors, the apparent affinity of ligands can vary depending on the nature of the tracer,17 we have performed similar competition binding assays, using three other iodinated chemokines as tracers: RANTES, MIP-1α, and MCP-2. The order of ligand affinities were not significantly different as compared with the results obtained with MIP-1β as tracer. As an example, the pIC50 obtained in one experiment using [125I]-MCP-2 as tracer were 9.61 ± 0.09 for MCP-2, 9.17 ± 0.21 for MIP-1β, 9.14 ± 0.15 for RANTES, 9.03 ± 0.13 for MIP-1α, and 7.82 ± 0.14 for MCP-3.
Binding of CC-chemokines to CCR5. A CHO-K1 cell line stably expressing human CCR5 and apoaequorin was established, and characterized by saturation-binding assay as expressing 2 pmoles receptor per mg protein. CC-chemokines were first tested for their ability to compete with [125I]-MIP-1β binding at high concentration (200 nmol/L), and competition curves were further established for chemokines displaying CCR5-binding activity. Chemokines that did not compete significantly at 200 nmol/L included MIP-3, MIP-3β, MIP-4, HCC-1, HCC-2, HCC-3, MPIF-1, MPIF-2, TARC, TECK, SLC, LEC, MDC, and I-309. The results were analyzed by the Graphpad Prism software, using a single-site model, and the data were normalized for the nonspecific binding (0%) and the specific binding in the absence of competitor (100%). All points were run in triplicate (error bars: S.E.M.). The presented curves are representative of at least two independent experiments. Table 1 presents the averaged values from the various experiments.
Binding of CC-chemokines to CCR5. A CHO-K1 cell line stably expressing human CCR5 and apoaequorin was established, and characterized by saturation-binding assay as expressing 2 pmoles receptor per mg protein. CC-chemokines were first tested for their ability to compete with [125I]-MIP-1β binding at high concentration (200 nmol/L), and competition curves were further established for chemokines displaying CCR5-binding activity. Chemokines that did not compete significantly at 200 nmol/L included MIP-3, MIP-3β, MIP-4, HCC-1, HCC-2, HCC-3, MPIF-1, MPIF-2, TARC, TECK, SLC, LEC, MDC, and I-309. The results were analyzed by the Graphpad Prism software, using a single-site model, and the data were normalized for the nonspecific binding (0%) and the specific binding in the absence of competitor (100%). All points were run in triplicate (error bars: S.E.M.). The presented curves are representative of at least two independent experiments. Table 1 presents the averaged values from the various experiments.
Summary of Binding and Functional Activities of CCR5 Ligands
| CC-Chemokines . | pIC50 (−Log M) ± s.e.m. . | pEC50 (−Log M) ± s.e.m. . |
|---|---|---|
| RANTES | 9.74 ± 0.19 | 8.87 ± 0.37 |
| MIP-1α | 9.05 ± 0.24 | 8.49 ± 0.47 |
| MIP-1β | 9.30 ± 0.24 | 8.47 ± 0.40 |
| MCP-2 | 9.40 ± 0.35 | 8.44 ± 0.32 |
| MCP-3 | 8.59 ± 0.11 | <6.3 |
| MCP-4 | 8.03 ± 0.28 | 7.04 ± 0.27 |
| Eotaxin | 7.72 ± 0.10 | <6.3 |
| MCP-1 | 7.34 ± 0.25 | <6.3 |
| CC-Chemokines . | pIC50 (−Log M) ± s.e.m. . | pEC50 (−Log M) ± s.e.m. . |
|---|---|---|
| RANTES | 9.74 ± 0.19 | 8.87 ± 0.37 |
| MIP-1α | 9.05 ± 0.24 | 8.49 ± 0.47 |
| MIP-1β | 9.30 ± 0.24 | 8.47 ± 0.40 |
| MCP-2 | 9.40 ± 0.35 | 8.44 ± 0.32 |
| MCP-3 | 8.59 ± 0.11 | <6.3 |
| MCP-4 | 8.03 ± 0.28 | 7.04 ± 0.27 |
| Eotaxin | 7.72 ± 0.10 | <6.3 |
| MCP-1 | 7.34 ± 0.25 | <6.3 |
The pIC50 (−log M) values were obtained from competition binding experiments, using [125I]-MIP-1β as a tracer (as displayed in Fig 1). The pEC50 (−log M) values were obtained from functional dose-response curves using the aequorin assay (as displayed in Fig 2A). Values represent the mean and s.e.m. of at least two (pIC50) or three (pEC50) independent determinations. One representative experiment is displayed in Fig 1.
Functional activity of CCR5 ligands.
The same set of chemokines was tested for their ability to activate CCR5, using a sensitive assay based on the use of apoaequorin as a reporter system for intracellular calcium release. Activation of chemokine receptors, including CCR5, is known to result in calcium signalling. The release of calcium was coupled to the production of a luminescent signal by coexpressing apoaequorin in the CCR5 cell line and incubating the cells with coelenterazin before the assay to reconstitute an active calcium-dependent enzyme complex.16G16 was also coexpressed in the CCR5/apoaequorin cell line, although this additional G protein is not necessary for the efficient coupling of the receptor to the production of luminescence in this assay (data not shown).
Chemokines that did not compete for MIP-1β binding on CCR5 at 200 nmol/L (MIP-3α, MIP-3β, MIP-4, HCC-1, HCC-2, HCC-3, MPIF-1, MPIF-2, TARC, TECK, SLC, LEC, MDC, and I-309) were also unable to promote intracellular calcium release in CCR5 transfected cells at all concentrations tested (up to 100 nmol/L). The chemokines binding CCR5 with high affinities (IC50 < 1 nmol/L) activated the receptor with high potency (Fig 2A). RANTES appeared as a slightly better agonist (EC50 of 1.3 nmol/L), whereas MIP-1α (EC50: 3.2 nmol/L), MCP-2 (EC50: 3.6 nmol/L), and MIP-1β (EC50: 3.4 nmol/L) were less potent. MCP-4 activated CCR5 with a potency (EC50: 103 nmol/L) in relation to its binding affinity. For the low-affinity ligands MCP-1 and eotaxin, although mild stimulation of the receptor was observed at the highest concentration tested (500 nM), a full dose response could not be established, and the EC50 was estimated as in excess of 500 nmol/L (Table 1).
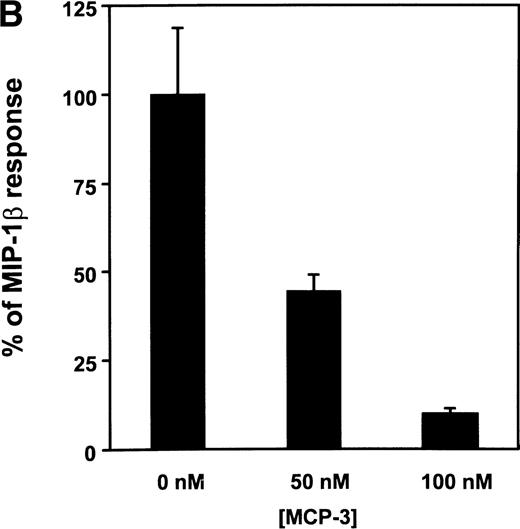
(A) Functional response of CCR5 to various CC-chemokines. The functional activity of chemokines able to bind to CCR5 was assayed by using a cell line coexpressing the receptor together with Gα16 and apoaequorin. Light emission resulting from the activation of the apoaequorin-coelenterazine complex in the presence of intracellular calcium was recorded in a luminometer. The results were analyzed by the Graphpad Prism software, using a single-site model, and the data were normalized for basal luminescence (0%) and maximal luminescence in the presence of 200 nmol/L MIP-1β (100%). All points were run in duplicate (error bars: S.E.M.). The displayed curves represent a typical experiment out of three performed independently. (B) Inhibition of the MIP-1β functional response by MCP-3. The antagonistic activity of MCP-3 was measured by preincubating the cells for 1 minute with MCP-3, before the addition of 1 nmol/L MIP-1β, and recording of luminescence. All points were run in triplicate (error bars: S.E.M.), and the results are representative of two independent experiments.
(A) Functional response of CCR5 to various CC-chemokines. The functional activity of chemokines able to bind to CCR5 was assayed by using a cell line coexpressing the receptor together with Gα16 and apoaequorin. Light emission resulting from the activation of the apoaequorin-coelenterazine complex in the presence of intracellular calcium was recorded in a luminometer. The results were analyzed by the Graphpad Prism software, using a single-site model, and the data were normalized for basal luminescence (0%) and maximal luminescence in the presence of 200 nmol/L MIP-1β (100%). All points were run in duplicate (error bars: S.E.M.). The displayed curves represent a typical experiment out of three performed independently. (B) Inhibition of the MIP-1β functional response by MCP-3. The antagonistic activity of MCP-3 was measured by preincubating the cells for 1 minute with MCP-3, before the addition of 1 nmol/L MIP-1β, and recording of luminescence. All points were run in triplicate (error bars: S.E.M.), and the results are representative of two independent experiments.
Interestingly, MCP-3 that is characterized by a better affinity for CCR5 than MCP-4 (IC50: 2.1 nmol/L versus 5.8 nmol/L) did not result in intracellular calcium release up to 100 nmol/L, a concentration that fully competed [125I]-MIP-1β binding on the CCR5 cell line. We further tested whether MCP-3 could inhibit the signaling mediated by MIP-1β. Different concentrations of MCP-3 were added on the CCR5 cell line 1 minute before the addition of MIP-1β (1 nmol/L). MIP-1β–induced signalling decreased with increasing MCP-3 concentrations (Fig 2B); 50 nmol/L MCP-3 reduced the luminescent signal by 50%, whereas 100 nmol/L MCP-3 totally blunted the functional response to MIP-1β. From these results, MCP-3 can be considered as a natural antagonist of CCR5.
Inhibition of gp120 binding and inhibition of HIV infection by CC-chemokines.
The ability of high-affinity (MIP-1α, MIP-1β, RANTES, and MCP-2) and intermediate-affinity (MCP-3 and MCP-4) CCR5 ligands to compete for HIV-1 gp120 binding was investigated. Recombinant gp120 from the M-tropic HIV-1 strain JR-FL was produced in 293T cells, purified, iodinated, and used as tracer in competition-binding assays using the CCR5-transfected HEK cells. As shown in Fig3, chemokines that competed efficiently for MIP-1β binding also inhibited gp120 binding, although with variable efficacy, in accordance with previously reported data, using YU2 competition binding.6 The IC50 observed for MIP-1α, MIP-1β, RANTES, and MCP-2 were found to be similar, although the maximal inhibition varied for different chemokines. MCP-2 (600 nmol/L) inhibited over 80% of gp120 binding whereas MIP-1β did not inhibit more than 50% at the same concentration. MCP-3 competed for gp120 binding with an efficiency similar to that of MCP-2. In contrast MCP-4 competed for gp120 binding with a much lower potency.
Inhibition of gp120 binding by CC-chemokines. CC-chemokines were tested for their ability to compete with the binding of [125I]-gp120 from the M-tropic HIV-1 strain JR-FL to CCR5-expressing 293 cells, in the presence of soluble CD4 (100 nmol/L). The displayed curves are representative of two independent experiments. The data were normalized for the nonspecific binding (0%) and the specific binding in the absence of competitor (100%). The data shown are the mean and S.E.M. derived from two independent experiments. ▩ 0 nmol/L, ▨ 20 nmol/L, □ 100 nmol/L, ▧ 600 nmol/L.
Inhibition of gp120 binding by CC-chemokines. CC-chemokines were tested for their ability to compete with the binding of [125I]-gp120 from the M-tropic HIV-1 strain JR-FL to CCR5-expressing 293 cells, in the presence of soluble CD4 (100 nmol/L). The displayed curves are representative of two independent experiments. The data were normalized for the nonspecific binding (0%) and the specific binding in the absence of competitor (100%). The data shown are the mean and S.E.M. derived from two independent experiments. ▩ 0 nmol/L, ▨ 20 nmol/L, □ 100 nmol/L, ▧ 600 nmol/L.
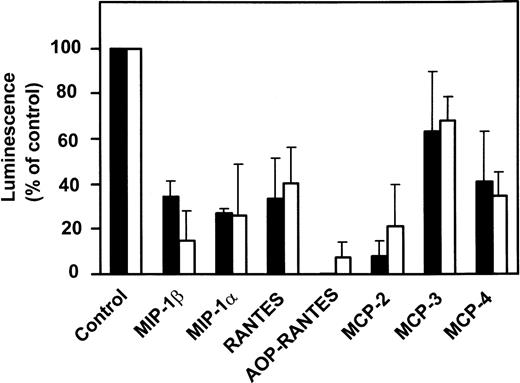
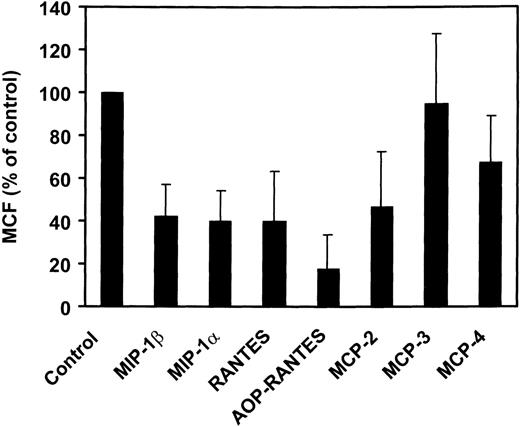
The various CC-chemokines were tested for their ability to inhibit infection of PM-1 cells, a human T-cell line naturally expressing CD4 and CCR5, by using luciferase reporter viruses pseudotyped with the Env proteins of two HIV-1 (ADA and BaL) R5 strains (Fig4). All high-affinity CCR5 ligands (MIP-1α, MIP-1β, RANTES, and MCP-2) displayed potent anti–HIV-1 inhibitory activities (60% to 80% inhibition at 1 μg/mL). Interestingly, MCP-2 displayed a greater inhibitory activity than MIP-1α and MIP-1β. In contrast, MCP-3 exhibited no significant inhibitory effects (20% inhibition), at concentrations competing efficiently for gp120 binding (1 g/mL). This suggested that competition for binding was not sufficient to inhibit viral entry. We therefore tested the ability of the various CCR5 ligands to promote receptor internalization. The reduction of cell surface CCR5 immunoreactivity was measured by FACS analysis, in response to 500 nmol/L chemokines, using the 2D7 monoclonal antibody (MoAb). Possibly in line with its lack of significant agonistic activity, MCP-3 did not reduce surface expression of CCR5, as compared with 30% reduction for MCP-4, 50% to 60% for MIP-1α, MIP-1β, MCP-2, and RANTES, and over 80% for AOP-RANTES (Fig 5).
Inhibition of HIV infection by CC-chemokines. Inhibition of viral entry by chemokines was assayed by infecting PM-1 cells with viruses pseudotyped with the env protein of the M-tropic HIV-1 strains ADA (▩) and BaL (□). Chemokines were used at a 1 μg/mL concentration, and the luciferase activity resulting from viral infection was measured. The data were normalized for basal luciferase activity (0%) and maximal activity in the absence of chemokines (100%). Each condition was run in triplicates, and the displayed results represent the mean of two independent experiments (error bars: S.E.M.).
Inhibition of HIV infection by CC-chemokines. Inhibition of viral entry by chemokines was assayed by infecting PM-1 cells with viruses pseudotyped with the env protein of the M-tropic HIV-1 strains ADA (▩) and BaL (□). Chemokines were used at a 1 μg/mL concentration, and the luciferase activity resulting from viral infection was measured. The data were normalized for basal luciferase activity (0%) and maximal activity in the absence of chemokines (100%). Each condition was run in triplicates, and the displayed results represent the mean of two independent experiments (error bars: S.E.M.).
Chemokine-induced internalization of CCR5. Internalization of CCR5 in the presence of various chemokines was estimated by FACS analysis of CHO-K1 cells expressing CCR5, using the 2D7 MoAb. The cells were incubated for 2 hours with the chemokines (500 nmol/L) before the test. The data (mean fluorescence) were normalized for the fluorescence of untransfected CHO-K1 cells (0%) and maximal fluorescence in the absence of chemokines (100%), and the displayed results represent the mean of three independent experiments (error bars: S.E.M.).
Chemokine-induced internalization of CCR5. Internalization of CCR5 in the presence of various chemokines was estimated by FACS analysis of CHO-K1 cells expressing CCR5, using the 2D7 MoAb. The cells were incubated for 2 hours with the chemokines (500 nmol/L) before the test. The data (mean fluorescence) were normalized for the fluorescence of untransfected CHO-K1 cells (0%) and maximal fluorescence in the absence of chemokines (100%), and the displayed results represent the mean of three independent experiments (error bars: S.E.M.).
DISCUSSION
CCR5 has been characterized originally as a receptor responding to the three CC-chemokines MIP-1α, MIP-1β, and RANTES.1 CCR5 was further identified as a coreceptor for HIV-1, HIV-2, and SIV strains.3 Most M-tropic strains that are found early in infection are strictly dependent on CCR5 for entry, and the essential role of CCR5 in HIV transmission was shown by the strong resistance to HIV infection of individuals homozygous for a 32 bp deletion in the CCR5 coding region.8 9 The absence of obvious phenotype associated with this defective CCR5 variant and the potent in vitro and ex vivo HIV-suppressive activities of CCR5 ligands and antibodies makes CCR5 an ideal target for therapeutic intervention.
Since we first characterized the pharmacology of CCR5, many new CC-chemokines have been identified or made available. Some of these chemokines have been shown to interact with receptors sharing high sequence similarities and/or common ligands with CCR5. To evaluate whether the range of CCR5 ligands had to be expanded in the context of the known redundancy between chemokine receptors, we tested all CC-chemokines described to date for their ability to bind and/or activate the human CCR5 receptor expressed in recombinant cell lines.
We have shown that, in addition to MIP-1α, MIP-1β, and RANTES, CCR5 could bind five other CC-chemokines at physiological concentration. MCP-2 interacted with the receptor with a high affinity (IC50 < 1 nmol/L) comparable to that of MIP-1α, MPI-1β, and RANTES. MCP-3 and MCP-4 displayed intermediate binding affinities (1 nmol/L < IC50 < 10 nmol/L), whereas MCP-1 and eotaxin were characterized by relatively low affinities (IC50 of 20 to 30 nmol/L). In functional assays, all high-affinity ligands were shown to be potent agonists (EC50 < 10 nmol/L). MCP-4 exhibited an agonistic activity in relation to its binding affinity. By contrast, MCP-3 did not show detectable functional activity at concentrations that completely competed for [125I]-MIP-1β binding, but could inhibit MIP-1β–induced signaling of CCR5, showing its ability to function as a natural CCR5 antagonist. MCP-1 and eotaxin displayed modest functional activities at high concentrations (above 50 nmol/L), suggesting that these two chemokines are unlikely to mediate biological activities through CCR5 in vivo. While this work was in progress, MCP-2 and MCP-4 have been described as agonists of CCR5.18,19 Our results show that full-length natural chemokines can bind to receptors without activating it. Truncated chemokines such as truncated RANTES, have also been shown to act as antagonists.20 MCP-3 appears, to our knowledge, as the first full-length natural chemokine displaying antagonistic activity on CCR5. Interestingly, MCP-3 was described as adopting preferentially a CXC-like dimer conformation,21 in contrast to other CC-chemokines. Whether this structural difference may be correlated with the antagonist activity of the chemokine on CCR5 is unclear at this stage.
CCR5, which is able to bind seven chemokines, appears therefore as a fairly promiscuous receptor. With the exception of MIP-1β, which so far does not activate other known receptors at low nmol/L concentrations, all other CC-chemokines binding to CCR5 also act through one or several other receptors. MIP-1α is the most potent agonist of CCR1 (IC50: 5 to 10 nmol/L). RANTES binds and activates CCR1 and CCR3. MCP-2 has a wide spectrum of receptor usage as it activates also CCR1, CCR2, and CCR3. It was, however, shown recently that the biological activity of MCP-2 on activated T cells was mediated essentially through CCR5, although these cells express other functional receptors for MCP-2.19 MCP-4 binds to CCR2 and CCR3, in addition to CCR5, with similar affinities for all three receptors. MCP-3 is an agonist for CCR1, CCR2, and CCR3. Noteworthy, its binding affinity for CCR1 and CCR2 (around 10 nmol/L) is comparable to that of CCR5.22 The antagonistic activities of MCP-3 on CCR5, therefore, takes place at concentrations at which MCP-3 activates other receptors, and may therefore have a functional relevance in vivo. MCP-1 and eotaxin are respectively active on CCR1 and CCR3.
The physiological significance of these overlapping activities of inflammatory chemokines is unclear. This lack of specificity could simply reflect the recent amplification of receptor and chemokine genes, as suggested by their genomic clustering, that have not evolved yet into nonredundant systems. It may also play a role in the coordinated recruitment of leukocyte subsets to orchestrate various types of immune responses. By contrast, constitutive chemokines, involved in the trafficking of leukocyte populations to specific compartments of lymphoid organs, appear as more specific in their interactions with receptors. TARC and MDC bind exclusively to CCR4, ELC and SLC to CCR7.
Primary sequence similarity among chemokines does not appear to correlate with their receptor usage. For example HCC-1 is much more similar to MIP-1β than MCP-2, but did not bind to or activate CCR5. Understanding the structural determinants that underlie the binding affinity and specificity of chemokines for receptors, as well as their agonistic properties could provide great insight for the design of chemokine analogs of higher affinity and efficacy. The N-terminal region of CC-chemokines has been shown to be important for receptor activation as well as receptor specificity.23-26 The sequence comparison of chemokines acting on a single receptor could allow to expand the understanding of structure-function relationships of chemokines.
High-affinity chemokine analogs could be valuable in the frame of inflammatory diseases and HIV infection. The blockade of CCR5 coreceptor function by different approaches was shown to potently inhibit HIV infection in vitro and ex vivo.2,11,27,28Natural or chemically modified chemokines as well as anti-CCR5 MoAb have been described as CCR5 antagonists with HIV-suppressive activity. The ability of additional chemokines acting on CCR5 to compete for HIV gp120 binding was therefore tested, as well as their HIV inhibitory properties in vitro. CCR5 high-affinity agonists (MIP-1α, MIP-1β, MCP-2, and RANTES) as well as ligands with intermediate affinity (MCP-3 and MCP-4) were able to compete for M-tropic HIV-1 gp120 binding on a CCR5 cell line although with variable efficacy. Interestingly, the maximal inhibition of gp120 binding was higher for MCP-2 (85% of gp120 total binding) than for other chemokines such as MIP-1β (50% of gp120 total binding). This result may suggest that gp120 can bind to CCR5 conformations not equally accessible for all chemokines. The same mechanism has been proposed earlier to explain the greater potency of AOP-RANTES to mediate HIV inhibition of infection.12
Inhibition of infection by the various chemokines was tested using pseudotyped viruses. MIP-1α, MIP-1β, and RANTES inhibited HIV (ADA, BaL) infection with similar efficacies. MCP-2 exhibited a greater potency to inhibit infection by HIV ADA. Interestingly, whereas MCP-3 could compete for HIV gp120 binding on CCR5 with high affinity, it did not inhibit HIV infection significantly. This may be due to the absence of agonistic properties of this ligand. When tested in a downregulation assay based on FACS analysis of cell-surface CCR5, MCP-3 did not induce downmodulation of the receptor, as compared with 60% for agonists such as MIP-1β, or over 80% for AOP-RANTES. Receptor endocytosis is classically linked to the phosphorylation of its agonist-bound active conformation,29 again showing the pure antagonistic nature of this ligand. It was suggested that potency of a chemokine or its derivatives to mediate inhibition of HIV infection not only depended on the competition for the virus-binding site,12 but could also be correlated to the downmodulation of coreceptor surface expression.13 The low efficacy of MCP-3 in an infection inhibition assay could be due to its lack of downmodulation effect on CCR5. Other agents, such as AOP-RANTES and truncated RANTES, described as CCR5 functional antagonists in chemotaxis assay but presenting partial agonistic properties in other functional assays (calcium flux, microphysiometer), have been described as potent HIV suppressors and efficient downregulators of receptor surface expression.
In conclusion, we have analyzed the pharmacology of CCR5 using binding and functional assays. A number of new chemokines able to interact with CCR5 were described, including MCP-2, MCP-3, and MCP-4. Among them, MCP-3 displayed the original property of acting as a natural antagonist of CCR5, at concentrations similar to those necessary to activate CCR1, CCR2, and CCR3. We also showed that inhibition of HIV infection required efficient activation of the coreceptor, in agreement with the hypothesis that receptor downregulation is the main mechanism of chemokine-mediated HIV suppressive activity.
ACKNOWLEDGMENT
Expert technical assistance was provided by M.J. Simons.
Supported by the Actions de Recherche Concertées of the Communauté Française de Belgique, the French Agence Nationale de Recherche sur le SIDA, the Belgian programme on Interuniversity, Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming, the BIOMED and BIOTECH programmes of the European Community (grants BIO4-CT98-0543 and BMH4-CT98-2343), the Fonds de la Recherche Scientifique Médicale of Belgium, and the Fondation Médicale Reine Elisabeth. The scientific responsibility is assumed by the authors. C.B. is Aspirant of the Belgian Fonds National de la Recherche Scientifique and C.G. is a fellow of the Fonds pour la Recherche dans l’Industrie et l’Agriculture. R.W.D. was supported by NIH NIAID R01-40880.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 1. Binding of CC-chemokines to CCR5. A CHO-K1 cell line stably expressing human CCR5 and apoaequorin was established, and characterized by saturation-binding assay as expressing 2 pmoles receptor per mg protein. CC-chemokines were first tested for their ability to compete with [125I]-MIP-1β binding at high concentration (200 nmol/L), and competition curves were further established for chemokines displaying CCR5-binding activity. Chemokines that did not compete significantly at 200 nmol/L included MIP-3, MIP-3β, MIP-4, HCC-1, HCC-2, HCC-3, MPIF-1, MPIF-2, TARC, TECK, SLC, LEC, MDC, and I-309. The results were analyzed by the Graphpad Prism software, using a single-site model, and the data were normalized for the nonspecific binding (0%) and the specific binding in the absence of competitor (100%). All points were run in triplicate (error bars: S.E.M.). The presented curves are representative of at least two independent experiments. Table 1 presents the averaged values from the various experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1899/5/m_blod41831001x.jpeg?Expires=1769161907&Signature=Z2HpKt0Yphy-WLWrLk20nZYkeDf-1YTpI3wBF1xLtEoPlZxMjqPWGuo8P4l47EHBZSrvioxEtdgom1c5TiSiZB~1fF5M8I3a6oew0dJKxuooFdCzuZHdnTEuKo9Wpwxa0X8Z3dLbVOaX8jGKS4soayWW5zBv08FkOG8HigOv21cnu~zxBXTDC8c9PF8bR6wvKkEVRVOuttzKMROd8w9IJt36kL3-P4OW6CvVuLcMVojD8ykZBzZ5mhnFcQxerXSl~PFmVtWO9BsWxZIASb~q7muS~EsTo9UaMDsqemmty8gd3gdZqT2qbeA1DdWPNPDPaWILYrvfdsD0rwvmO3G7nA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Inhibition of gp120 binding by CC-chemokines. CC-chemokines were tested for their ability to compete with the binding of [125I]-gp120 from the M-tropic HIV-1 strain JR-FL to CCR5-expressing 293 cells, in the presence of soluble CD4 (100 nmol/L). The displayed curves are representative of two independent experiments. The data were normalized for the nonspecific binding (0%) and the specific binding in the absence of competitor (100%). The data shown are the mean and S.E.M. derived from two independent experiments. ▩ 0 nmol/L, ▨ 20 nmol/L, □ 100 nmol/L, ▧ 600 nmol/L.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1899/5/m_blod41831003x.jpeg?Expires=1769161907&Signature=WXdO8Hx2x255NohFAu-N5GTW~3IiIEJyPB8Oyj1huTgqHmMxM3Zbc079KI29k~uRte0S2ndt3IFTOB9Sa1XkKVhH~S8vgrZFdv1SbOuFqtP9lIf6iFkVx24LVmBICp83QeevbMY76WGkhP1IqOuZSysFWGW4540XzcwsMYTx-IZY5mSQkLWp2XLa32iLMPG~mhhhSNz6YUOelZJU0A6mGA6rQ2MZvvIRAmg8-023XD84H5RipX-d4ON9wafVK2HXyTGywO8-HueUGILN3X7x2JrHsjLZXTJkeUQL0eHL7T5duwUKfjqhay20NOaXOBM6mUG788o1mOHiAVD6VIvsXA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal