Abstract
Thrombopoietin (TPO), the specific cytokine that regulates platelet production, is expressed in human bone marrow (BM), kidney, and liver. There appears to be no regulation of TPO in the kidney and liver, but TPO messenger RNA (mRNA) expression can be modulated in the stromal cells of the BM. In this study, we used primary human BM stromal cells as a model to study the regulation of TPO mRNA expression in response to various platelet -granular proteins. We showed that platelet-derived growth factor (PDGF) BB and fibroblast growth factor (FGF) 2 stimulated TPO mRNA expression in both a dose-dependent and time-dependent manner. The addition of 50 ng/mL of PDGF and 20 ng/mL of FGF resulted in maximal induction of TPO mRNA expression in 4 hours. We also found that platelet factor 4 (PF4), thrombospondin (TSP), and transforming growth factor-beta (TGF-β) are negative modulators of megakaryocytopoiesis. We observed suppression in TPO mRNA expression with 1 μg/mL of both PF4 and TSP and 50 ng/mL of TGF-β, with maximal suppression occurring 4 hours after the addition of these proteins. Finally, the addition of whole-platelet lysate produced a dose-dependent inhibition of TPO expression. On the basis of these findings, we propose that the platelet -granular proteins studied may regulate TPO gene expression in BM stromal cells by means of a feedback mechanism.
Thrombopoietin (TPO), the ligand for c-Mpl, supports megakaryocyte colony formation, increases megakaryocyte size and ploidy, and is the most important regulator of platelet production.1-4 TPO is produced in the liver3,5and, to a lesser extent, in the kidney, spleen, and bone marrow (BM).5,6 Because TPO is the physiologic regulator of platelet production, circulating levels of TPO would be expected to vary inversely with changes in platelet demand. At least 2 possible mechanisms of TPO regulation by platelets have been suggested. One proposed mechanism is that TPO gene expression is constant and that serum levels are controlled by the platelet mass through uptake and metabolism by means of binding to the Mpl receptor.7-11Megakaryocyte mass may also be an important regulator of serum TPO levels, since mice lacking the erythroid transcription factor NF-E2 have profound thrombocytopenia without concomitant elevated serum TPO levels.12,13 Nagata et al14 also found a relation between serum TPO concentration and megakaryocyte numbers in BM and spleen in mice with acute thrombocytopenia. Alternatively, TPO gene expression may be regulated by feedback control at the cellular level and TPO messenger RNA (mRNA) levels may vary accordingly. McCarty et al15 showed that administering antiplatelet antibodies to mice resulted in increased TPO mRNA expression in the BM but no noticeable differences in the kidney or liver. In addition, we previously observed an increase in TPO mRNA expression in stromal cells from patients with aplastic anemia and idiopathic thrombocytopenic purpura.6
The BM microenvironment plays a critical role in the regulation of hematopoiesis through direct cell-cell interactions and the release of hematopoietic growth factors.16-18 The stroma is composed of a mixed population of fibroblasts, endothelial cells, macrophages, and adipocytes.17,19 These cells and their cell-derived matrices provide attachment sites for hematopoietic progenitor cells.20 The establishment of long-term BM culture (LTBMC) systems, which are thought to mimic hematopoiesis in vivo, has greatly facilitated analysis of this microenvironment.21 LTBMC were initially generated from murine BM by Dexter et al,16 and the system was later adapted to human BM cells by Gartner and Kaplan.22 Stromal cells constitutively produce a variety of cytokines, including interleukin (IL) 1β, IL-3, IL-6, IL-7, IL-8, IL-11, granulocyte-macrophage colony-stimulating factor, granulocyte colony-stimulating factor (G-CSF), stem cell factor, platelet-derived growth factor (PDGF), fibroblast growth factor (FGF) 2, flt-3 ligand, and transforming growth factor β (TGF-β).23-25Studies have confirmed ubiquitous expression of TPO in BM stromal cells.26 27
The peptide growth factors PDGF and FGF are prime candidates for cytokines that may affect the production of hematopoietic growth factors. Both have a broad specificity for a number of cells, including fibroblasts, microvascular endothelial cells, and smooth muscle cells, and they induced the synthesis and release of macrophage colony-stimulating factor (M-CSF) in murine stromal cells.28 PDGF also stimulated the expression of collagenase in human skin fibroblasts.29 However, the effect of these growth factors on TPO mRNA expression in human BM stromal cells has not been investigated. Besides positive effectors, inhibitory influences may also regulate TPO expression. Platelet factor 4 (PF4), thrombospondin (TSP), and TGF-β are negative modulators of hematopoiesis, particularly megakaryocytopoiesis.30,31 PF4 has a high affinity for heparin and inhibits bone resorption and angiogenesis.32 TSP is synthesized and secreted by normal hematopoietic cells as well as by leukemic cell lines and can act as an adhesive ligand for developing hematopoietic cells.33TGF-β is also a potent regulator of extracellular matrix production and differentiation in a variety of cell types, including osteoblasts and vascular smooth-muscle cells.32 PF4, TSP, and TGF-β are all released by the α granules of platelets and inhibit megakaryocyte colony growth in vitro.30,31,34,35 It has been suggested that α-granular proteins may function in regulating thrombopoiesis by means of a feedback mechanism to the BM.32 However, the nature of this feedback mechanism is unknown.
In this study, we provide evidence that platelet α-granular proteins may regulate thrombopoiesis by modulating TPO mRNA expression in BM stroma. We found that PDGF and FGF stimulated TPO gene expression but PF4, TSP, and TGF-β suppressed it. TPO mRNA expression was also inhibited by whole-platelet lysate, an effect that was partly reversed by an antibody specific to PF4.
Materials and methods
Stimulating and inhibiting agents and antibodies
Human recombinant PDGF-BB (0.1 μg/μL) was purchased from Genzyme (Cambridge, MA) and human FGF-2 (0.1 μg/μL) from Gibco BRL (Gaithersburg, MD). Human PF4 (1 mg/mL) and TSP-1 (0.495 mg/mL) were purified from platelets.36 37 Recombinant human TGF-β1 (2 μg/mL) and the phorbol ester 12-myristate 13-acetate (PMA) were purchased from Sigma (St Louis, MO). The PMA was first dissolved in 100% ethanol and then further diluted to a final concentration of 50 ng/mL in culture medium. A polyclonal antibody against human PF4 (1 mg/mL) was purchased from Chemicon (Temecula, CA). A monoclonal anti-TSP antibody (0.5 mg/mL) was a gift from Dr Philip Hogg of the Centre for Thrombosis and Vascular Research.
BM specimens
Human BM samples were obtained with informed consent from adult patients with normal platelet counts who were undergoing BM aspiration for diagnostic purposes. The samples were essentially of normal BM obtained, for example, from patients undergoing staging procedures after diagnosis of non-Hodgkin lymphoma in which no BM involvement was observed. Aspirates were collected in minimum essential medium (MEM) (Sigma) supplemented with 20 U/mL preservative-free heparin (Fisons, Sydney, Australia). The aspirate was layered onto a Ficoll-Paque density gradient (Pharmacia, Uppsala, Sweden) and centrifuged for 35 minutes at 1400g. Mononuclear cells were collected from the interphase (density < 1.077 g/mL), washed several times in MEM, and resuspended in culture medium. The mononuclear cells represented 10% of total nucleated cells in the aspirate, so that a typical 10-mL sample of BM yielded about 10 million mononuclear cells. Cell viability was estimated by trypan blue exclusion.
Initiation and maintenance of culture
Freshly isolated mononuclear cells (8-10 × 106/mL) were seeded in 25-cm2tissue-culture flasks (Costar, Cambridge, MA) in an 8-mL total volume of LTBMC medium. This consisted of MEM supplemented with 12.5% fetal bovine serum (FBS) (Trace Biosciences, Victoria, Australia), 12.5% horse serum (Multicell; Trace Biosciences), 10−6mol/L hydrocortisone hemisuccinate (Sigma), and 1% glutamine solution with penicillin-streptomycin (10 000 U penicillin and 10 mg/mL streptomycin; Sigma).19 22 The cultures were incubated at 37°C in a 5% carbon dioxide (CO2) humidified atmosphere. A layer of adherent cells was obtained in 3 to 4 weeks. Each week, half of the medium was replaced with fresh medium. The adherent layers were subjected to passage at confluence by trypsinization (0.4% trypsin, Trace Biosciences) and cells were analyzed at low passage numbers (between passage 2 and 3).
In situ hybridization for mRNA regulation studies
A quantitative in situ hybridization (QISH) procedure enabling hybridization to be carried out directly on cells in tissue-culture plates was adapted from previous work.38-40 The adherent confluent cell layer was detached, and the cells were resuspended in complete LTBMC medium and seeded into 24-well flat-bottomed plates (Nunc, Naperville, IL) at a cell density of 5 × 104cells/mL. The plates were incubated at 37°C and 5% CO2in a humidified atmosphere. Preliminary studies carried out to optimize the conditions for maximal TPO mRNA stimulation and suppression in these cells found that PDGF and FGF had stimulatory effects, whereas PF4, TSP, and TGF-β had inhibitory effects.
In experiments to investigate the up-regulation of stromal cell TPO mRNA expression, the cells were grown to subconfluence and then allowed to become quiescent by placing them in MEM with 1% serum (0.5% FBS and 0.5% horse serum) for 24 hours.28 41 After 24 hours, either PDGF or FGF was added to the growth-arrested cells (time 0). As a control experiment, IL-3 mRNA expression was also measured in cells exposed to PDGF. In experiments to investigate the inhibition of TPO gene expression by PF4, TSP, or TGF-β, the confluent cells were left in complete medium containing 25% serum for 24 hours before these platelet proteins were added.
Time-course and dose-response assay
At time 0, PDGF, FGF, or PMA (as a positive control) was added to the growth-arrested cells at the appropriate concentrations. The assay was terminated at 2, 4, 7, 22, and 24 hours after the addition of the growth factors or PMA by gently aspirating the medium, washing the cells 3 times with ice-cold sterile phosphate-buffered saline (PBS), and fixing the cells in formalin. PF4, TSP, or TGF-β at the appropriate concentrations was added to cells in medium containing 25% serum. The assay was terminated at 0.25, 0.5, 2, 4, and 8 hours after the addition of the platelet proteins by washing with sterile PBS and fixing the cells in formalin as described previously. To investigate dose response, PDGF or FGF was added to the cells (at time 0) at increasing concentrations in the presence of medium containing 1% serum, whereas PF4, TSP, or TGF-β was added to cells (at time 0) in complete medium. The assay was terminated after 4 hours, and the cells were washed and fixed in formalin.
Fixation and pretreatment of cells
After termination of the assay, the cells were fixed by immersion of plates in 10% (vol/vol) analytical-grade formalin (BDH, Sydney, Australia) in PBS (pH 7.0) for 30 minutes at room temperature. The cells were then treated to render the cell membrane permeable for probe penetration by washing twice with PBS containing 0.25% (vol/vol) Triton X-100 (Sigma) and 0.25% (vol/vol) Nonidet P40 (Sigma) for 5 minutes for each wash. The plates were rinsed quickly in fresh PBS and placed in 20% (vol/vol) acetic acid in water for 30 seconds. Plates were then washed rapidly in 3 changes of sterile distilled water and placed in 100% ethanol for 5 minutes, the ethanol was decanted, and the plates were dried in a fan-forced nonhumidified oven at 42°C before the probes were added.
Complementary DNA (cDNA) probes and labeling
A 1.5-kb HindIII–BamHI fragment corresponding to human TPO cDNA (a gift from Dr J. Rasko and Prof D. Metcalf of the Walter and Eliza Hall Institute of Medical Research, Victoria, Australia) was labeled with biotin by using Photobiotin (Bresatec, Adelaide, South Australia) according to the manufacturer's instructions. The plasmid vector pBR322 was also labeled with biotin and used as a negative control. cDNA of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a positive control.
Hybridization conditions
Posthybridization and detection
The hybridization buffer was decanted from the plates and nonhomologous bound sequences were removed by incubating the plates in 2 × trisodium citrate (SSC) containing 50% (vol/vol) formamide for 15 minutes at 42°C. This was followed by 2 sequential washings with 2 × SSC–0.1% (wt/vol) sodium dodecyl sulfate (SDS) and 0.4 × SSC–0.1% (wt/vol) SDS at 42°C for 15 minutes each. The plates were then washed with 0.4 × SSC to remove the SDS. Biotin was detected by using an in situ hybridization detection kit (Dakopatts, Carpinteria, CA) with a slight modification of the manufacturer's instructions.
The following reagents were applied sequentially for 10 minutes each: streptavidin (1:100 dilution), biotinylated alkaline phosphatase (1:100 dilution), streptavidin (1:500 dilution), and biotinylated alkaline phosphatase (1:100).38 Plates were washed 4 times between each step in Tris-buffered saline (0.5 mol/L sodium chloride [NaCl], 0.1 mol/L Tris, and 10 mmol/L magnesium chloride [MgCl2]; pH 7.6). The alkaline phosphatase was detected with theP-nitrophenyl phosphate (P-NPP) conversion method. Five hundred microliters of P-NPP solution (prepared as 5-mg tablets of P-NPP Σ in 5 mL diethanolamine buffer) was added to each well.38 The plates were incubated in the dark at 37°C for 20 minutes. The yellow color development was stopped by adding 500 μL of 0.5 mol/L sodium hydroxide (NaOH) to each well. The contents were mixed and the absorbance was determined by using a plate reader at 405 nm (Multiskan MS; Labsystems, Helsinki, Finland).
RNA extraction and reverse transcription
Total cellular RNA was extracted from the treated stromal cells by using Trizol reagent (Gibco BRL). First-strand cDNA was generated by using the Superscript II reverse transcriptase (RT) enzyme (Gibco BRL). Briefly, 1 to 5 μg of total RNA and 1 μL oligo-d(T)15primer (0.5 mg/mL) in a volume of 12 μL were heated to 70°C for 10 minutes. Then, 1 × RT buffer (250 mmol/L Tris-hydrochloric acid [HCl] at pH 8.3, 375 mmol/L potassium chloride [KCl], and 15 mmol/L MgCl2), 10 mmol/L diethylnitrophenyl thiophosphate (dNTP) mix, 0.1 mol/L dithiothreitol, and 2500 U RNase inhibitor were added and incubated at 42°C for 2 minutes. Two hundred units of Superscript II RT enzyme was added and the reaction was continued at 42°C for 60 minutes and then 95°C for 5 minutes.
Semiquantitative polymerase chain reaction (PCR)
Sense and antisense primers for human TPO and GAPDH were used as follows: TPO forward, 319-5′ CTGCTTCGTGACTCCCATGTC 3′-340; TPO reverse, 695-5′ CGCACCTTTCCTCGGAGCAG 3′-714; GAPDH forward, 280-5′ ATCACCATCTTCCAGGAGCG 3′-300; and GAPDH reverse, 565-5′ GGTATCGTGGAAGGACTCATG 3′-585. Before the quantitative analysis, PCR was carried out at increasing cycle numbers and the intensity of the PCR products on the autoradiograph was measured. The relative band intensity was then plotted against the number of cycles to obtain a linear relation. The optimal cycle number was chosen in the midlinear region of the curve to avoid the plateau region of the reaction. The final conditions for the PCR reactions were 1 × PCR buffer (10 mmol/L Tris-HCl [pH 8.3] and 50 mmol/L KCl), 0.4 mmol/L dNTP, 10 μmol/L of each primer for each set, 1.5 mmol/L MgCl2, 2.5 U Taq DNA polymerase (Sigma), and 2 μL of first-strand cDNA. The mixture was heated to 95°C for 30 seconds, 61°C (TPO primers) for 1 minute, and 72°C for 1 minute for a total of 31 cycles, dependent followed by a final extension at 72°C for 3 minutes. The amplification procedure for the GAPDH primers consisted of denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds, and extension at 72°C for 90 seconds for a total of 19 cycles. A negative control that consisted of all PCR components except cDNA template was included in each reaction.
PCR products (367-base-pair [bp] TPO and 400-bp GAPDH) were separated by electrophoresis in a 2% agarose–Tris-acetate-EDTA gel and visualized by staining with ethidium bromide (0.1 μg/mL). The PCR products were transferred to a Hybond nitrogen membrane (Amersham, Buckinghamshire, United Kingdom) by capillary action using 15 × SSC. The blots were probed with phosphorus 32–labeled TPO or GAPDH cDNAs at about 8 to 10 × 106 cpm/blot. Hybridization was performed overnight at 42°C in 3 × SSC, 1% SDS, 50% formamide, 1 mol/L HEPES (pH 7.5), 1 × Denhardt solution (0.1% polyvinylpyrrolidone, 0.1% Ficoll type 400, and 0.1% bovine serum albumin [BSA]), denatured herring sperm DNA (200 μg/mL), and yeast transfer RNA (150 μg/mL). The blots were washed in 2 sequential washes of 2 × SSC–0.1% SDS and 1 × SSC–0.1% SDS for 30 minutes each at 42°C. The membranes were exposed to x-ray film (Amersham) at −80°C with an intensifying screen, and the film was developed in an automated developer (Curix-60; Agfa, Australia).
Cell-proliferation assay
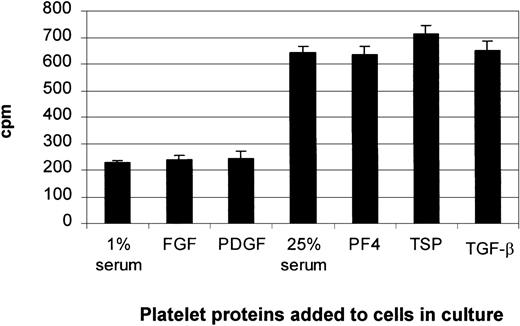
Tritium-thymidine incorporation into the stromal cells was used as a measure of DNA synthesis. The stromal cells were seeded at a density of 1.5 × 104 cells/well in a 96-well microtiter plate and grown to subconfluence. To investigate the stimulatory effects of PDGF and FGF, the cells were made quiescent in medium containing 1% serum for 24 hours. To study the inhibitory effects of PF4, TSP, and TGF-β, cells were left in complete medium. The cells were then incubated in the absence or presence of PDGF, FGF, PF4, TSP, or TGF-β for 18 hours, then pulsed for 6 hours with 100 000 cpm (24.8 × 1010 Bq/mmol) tritium-thymidine (NEN Life Sciences, Boston, MA). The assay was terminated by gently aspirating the medium and washing the cells with ice-cold sterile PBS. The PBS solution was carefully removed, and ice-cold 5% trichloroacetic acid was added to the cells to precipitate proteins and nucleic acids. Cells were solubilized by adding 0.1 mol/L NaOH, and isotope uptake was determined by liquid scintillation counting.42
The CellTiter 96 aqueous assay (Promega Corp, Madison, WI) was also used to measure changes in cell numbers.43 This assay is a nonradioactive alternative to tritium-thymidine incorporation. The system measures the conversion of a tetrazolium salt compound (MTS) into a soluble formazan product by the mitochondria of living cells.43 Cells seeded at a density of 5 × 104 cells/well were grown to subconfluence and incubated with the various platelet proteins for 4 hours at 37°C. The MTS reagent (prepared according to manufacturers' instructions) was added to the wells, including the control wells, which contained the same volume of culture medium but no cells. After an additional 4-hour incubation period, samples were read in a plate reader (Multiskan MS; Labsystems) at 492 nm.
Preparation of whole-platelet lysate
One unit of fresh blood (500 mL) was obtained with informed consent from patients undergoing regular venesection for hemachromatosis at a time when the patients' serum iron and ferritin levels were normal. The blood was collected in acid citrate dextrose and centrifuged for 10 minutes at 1200g to obtain platelet-rich plasma. The plasma was centrifuged for 15 minutes at 3000g to obtain a platelet pellet that was washed 3 times in PBS–1% EDTA and the platelets counted. The platelets were then resuspended at a concentration of 10 × 109 platelets/mL in PBS-EDTA. After 5 cycles of freezing and thawing, the platelet lysate was centrifuged for 30 minutes at 13 000g to remove cell debris. The protein concentration in the supernatant was determined by using bicinchoninic acid (BCA) protein assay reagents (Pierce, Rockford, IL) with BSA as reference. Serial dilutions of the platelet lysate equivalent to 1 × 109, 1 × 108, 1 × 107, and 1 × 106platelets/mL were added to the stromal cells and incubated for 4 hours. Anti-PF4 antibody (50 μg/mL) or the nonimmune IgG control (50 μg/mL), together with lysate equivalent to 1 × 109 platelets/mL, was also added to the cells and incubated for 4 hours. TPO mRNA expression levels were then measured with the QISH assay.
Identification of PF4 and TSP proteins in the platelet lysate
Equivalent amounts of platelet protein were subjected to SDS–polyacrylamide gel electrophoresis44 using either a 15% linear gel or a 4% to 15% gradient gel. Proteins were electrophoretically transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA), and Western blot analysis was carried out. Nonspecific binding sites were blocked with 1% skim milk in PBS—Tween 20 for 30 minutes at room temperature before the membranes were probed with a rabbit antihuman polyclonal anti-PF4 (2 μg/mL) and mouse monoclonal anti-TSP antibody (2 μg/mL). Immunostained proteins were detected with a horseradish peroxidase–labeled secondary antibody by using the enhanced chemiluminescence reagent (NEN Life Sciences, Boston, MA) according to the manufacturer's instructions.
TPO enzyme-linked immunosorbent assay (ELISA)
The supernatants from stromal cell cultures exposed to PDGF, PF4, and TSP were collected for protein measurement. A solid-phase sandwich ELISA was used to measure the TPO in the culture supernatants.45
Results
Stimulation of TPO mRNA expression
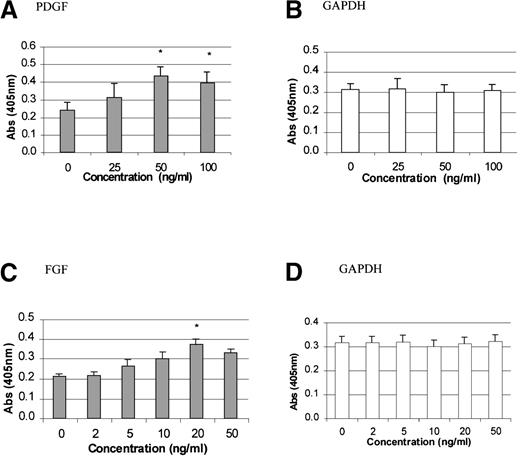
To study the stimulatory effects of PDGF-BB and FGF-2 on TPO mRNA expression, time-course and dose-response experiments were carried out. PDGF-BB (50 ng/mL) or FGF-2 (20 ng/mL) was added to growth-quiescent stromal cells and incubated for 2, 4, 7, 22, or 24 hours. The maximum poststimulatory peak in TPO mRNA expression was observed 4 hours after the addition of either growth factor (Figure1A and 1B). There was at least a 2-fold induction in TPO expression with the addition of PDGF and a 1.5-fold induction with the addition of FGF-2 (P < .05 compared with controls). Because PMA is a potent pharmacologic agent capable of inducing differentiation in a variety of cell lines in vitro by triggering growth-factor–dependent signaling pathways,46we used it as a positive control in our system. Figure 1C shows that TPO mRNA expression was induced in a time-dependent fashion in response to the addition of PMA. As expected, GAPDH mRNA levels remained unchanged in cells exposed to PDGF (Figure 1D), FGF, or PMA (data not shown). Figure 2 shows that TPO mRNA expression was specifically up-regulated in response to PDGF (50 ng/mL), whereas IL-3 expression was not altered. The dose-response curve for the effect of PDGF and FGF is shown in Figure3. Quiescent cells were exposed to increasing concentrations of PDGF (0, 25, 50, and 100 ng/mL) or FGF (0, 2, 5, 10, 20, and 50 ng/mL) and incubated for the optimal time of 4 hours. TPO mRNA expression was significantly stimulated with 50 ng/mL PDGF (Figure 3A) and 20 ng/ml FGF (Figure 3C) (P < .05 compared with controls).
Time-dependent stimulation of thrombopoietin (TPO) messenger RNA (mRNA) expression by platelet-derived growth factor (PDGF), fibroblast growth factor (FGF), and the phorbol ester 12-myristate 13-acetate (PMA) in human bone marrow (BM) stromal cells.
TPO mRNA expression levels were determined by using a quantitative in situ hybridization (QISH) assay at various time points after the addition of 50 ng/mL PDGF (A), 20 ng/mL FGF (B), or 50 ng/mL PMA (C). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels in cells exposed to PDGF are also shown (D). The results shown are the mean ± SD values from 5 separate experiments carried out in triplicate. *P < .05 compared with controls (time 0).
Time-dependent stimulation of thrombopoietin (TPO) messenger RNA (mRNA) expression by platelet-derived growth factor (PDGF), fibroblast growth factor (FGF), and the phorbol ester 12-myristate 13-acetate (PMA) in human bone marrow (BM) stromal cells.
TPO mRNA expression levels were determined by using a quantitative in situ hybridization (QISH) assay at various time points after the addition of 50 ng/mL PDGF (A), 20 ng/mL FGF (B), or 50 ng/mL PMA (C). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels in cells exposed to PDGF are also shown (D). The results shown are the mean ± SD values from 5 separate experiments carried out in triplicate. *P < .05 compared with controls (time 0).
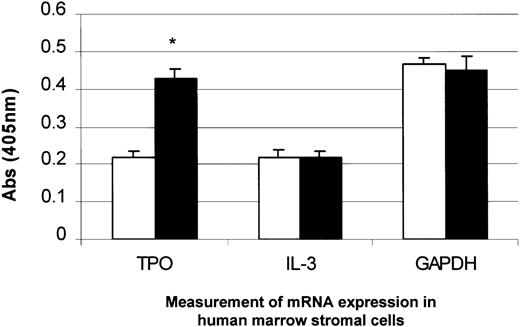
Measurement of TPO and interleukin 3 (IL-3) mRNA expression in BM stromal cells with the addition of PDGF.
PDGF (50 ng/mL) was added to BM stromal cells in culture and incubated for 4 hours. TPO mRNA expression was specifically up-regulated, whereas IL-3 and GAPDH mRNA remained unchanged, as measured by the QISH assay. The results shown are the mean ± SD values from 3 separate experiments carried out in triplicate. *P < .05 compared with controls (time 0). □, 1% serum; ▪, 1% serum plus PDGF.
Measurement of TPO and interleukin 3 (IL-3) mRNA expression in BM stromal cells with the addition of PDGF.
PDGF (50 ng/mL) was added to BM stromal cells in culture and incubated for 4 hours. TPO mRNA expression was specifically up-regulated, whereas IL-3 and GAPDH mRNA remained unchanged, as measured by the QISH assay. The results shown are the mean ± SD values from 3 separate experiments carried out in triplicate. *P < .05 compared with controls (time 0). □, 1% serum; ▪, 1% serum plus PDGF.
Dose effect of PDGF-BB or FGF-2 on TPO mRNA expression in BM stromal cells.
The levels of TPO mRNA expression in the BM stromal cell cultures were measured 4 hours after treatment with increasing concentrations of PDGF (A) or FGF (C). GAPDH mRNA levels remained unchanged in cells exposed to PDGF (B) or FGF (D). The results shown are the mean ± SD values from 5 separate experiments carried out in triplicate. *P < .05 compared with controls.
Dose effect of PDGF-BB or FGF-2 on TPO mRNA expression in BM stromal cells.
The levels of TPO mRNA expression in the BM stromal cell cultures were measured 4 hours after treatment with increasing concentrations of PDGF (A) or FGF (C). GAPDH mRNA levels remained unchanged in cells exposed to PDGF (B) or FGF (D). The results shown are the mean ± SD values from 5 separate experiments carried out in triplicate. *P < .05 compared with controls.
Suppression of TPO mRNA expression
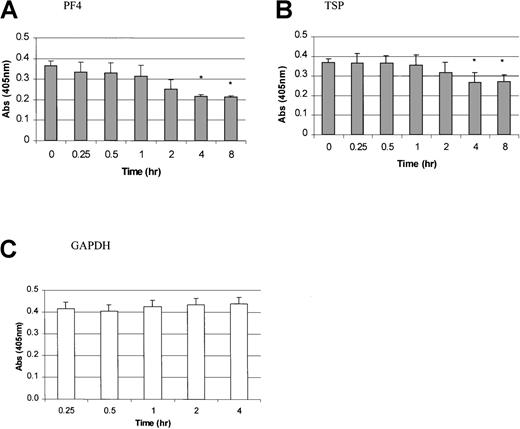
In contrast to the results with PDGF, FGF, and PMA, TPO mRNA expression was inhibited by the addition of PF4, TSP, or TGF-β. PF4 and TSP were added to the stromal cell cultures and incubated for 0.25, 0.5, 1, 2, 4, and 8 hours (Figure 4A and 4B). A significant inhibition of TPO mRNA expression was observed 4 hours after the addition of these platelet proteins. Figure5 shows the dose-dependent effects of PF4, TSP, and TGF-β. A significant inhibition in TPO mRNA expression was found with a PF4 or TSP dose ranging from 1 to 2 μg/mL and a TGF-β dose of 50 to 100 ng/mL (P < .05 compared with controls). GAPDH mRNA expression did not vary in cells exposed to PF4 (Figure 4D), TSP, or TGF-β (data not shown).
Time-dependent inhibition of TPO mRNA expression by platelet factor 4 (PF4) and thrombospondin (TSP) in BM stromal cells.
TPO mRNA expression levels were determined by using a QISH assay at various time points after the addition of PF4 (A) and TSP (B). GAPDH mRNA levels in cells exposed to PF4 are also shown (C). The results shown are the mean ± SD values from 4 separate experiments performed in triplicate. *P < .05 compared with controls (time 0).
Time-dependent inhibition of TPO mRNA expression by platelet factor 4 (PF4) and thrombospondin (TSP) in BM stromal cells.
TPO mRNA expression levels were determined by using a QISH assay at various time points after the addition of PF4 (A) and TSP (B). GAPDH mRNA levels in cells exposed to PF4 are also shown (C). The results shown are the mean ± SD values from 4 separate experiments performed in triplicate. *P < .05 compared with controls (time 0).
Dose effect of PF4, TSP, and transforming growth factor β (TGF-β) on TPO mRNA expression.
PF4 (A), TSP (B), and TGF-β (C) were added to BM stromal cell cultures at increasing concentrations, resulting in inhibition of TPO mRNA expression. GAPDH level in cells exposed to PF4 is also shown (D). The results shown are the mean ± SD values from 4 separate experiments performed in triplicate. *P < .05 compared with controls.
Dose effect of PF4, TSP, and transforming growth factor β (TGF-β) on TPO mRNA expression.
PF4 (A), TSP (B), and TGF-β (C) were added to BM stromal cell cultures at increasing concentrations, resulting in inhibition of TPO mRNA expression. GAPDH level in cells exposed to PF4 is also shown (D). The results shown are the mean ± SD values from 4 separate experiments performed in triplicate. *P < .05 compared with controls.
Confirmation of modulation in TPO mRNA expression by semiquantitative RT-PCR
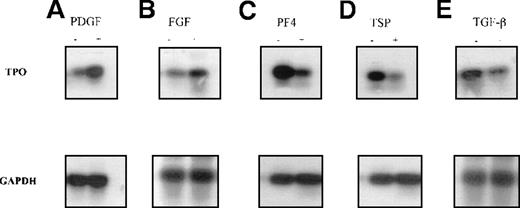
Semiquantitative RT-PCR was used to verify the QISH results. Total RNA was extracted from untreated stromal cells or cells treated with PDGF, FGF, PF4, TSP, or TGF-β, and RT-PCR was performed. First, the optimal numbers of PCR cycles to allow quantitation of TPO and GAPDH transcripts were determined. The relative band intensity of each PCR product was plotted against the increasing cycle numbers (Figure6A and 6B). The optimal cycle number for TPO transcripts was determined to be 31; that for GAPDH transcripts was found to 19. Second, PCR with GAPDH primers was carried out to ascertain whether equivalent amounts of high-quality cDNA were used in the experiments. We confirmed that TPO mRNA expression could be up-regulated by PDGF and FGF (Figure 7A and 7B). We also confirmed a significant inhibition in TPO expression with PF4, TSP, or TGF-β (Figure 7C, 7D, and 7E).
Optimization of quantitative polymerase chain reaction (PCR) analysis.
Autoradiograph showing PCR carried out with an increasing number of cycles and with the products subjected to Southern hybridization with a gene-specific probe. The optimal cycle number was determined to be in the midlinear region of the curve: 31 cycles for TPO transcripts (A) and 18 cycles for GAPDH (B).
Optimization of quantitative polymerase chain reaction (PCR) analysis.
Autoradiograph showing PCR carried out with an increasing number of cycles and with the products subjected to Southern hybridization with a gene-specific probe. The optimal cycle number was determined to be in the midlinear region of the curve: 31 cycles for TPO transcripts (A) and 18 cycles for GAPDH (B).
Changes in TPO mRNA expression in human BM stromal cells measured by semiquantitative reverse transcriptase (RT)-PCR.
Semiquantitative RT-PCR analyses used RNA isolated from primary BM stromal cells. The PCR products were separated by agarose-gel electrophoresis and analyzed by Southern hybridization with a gene-specific probe. It was confirmed that TPO mRNA expression was up-regulated after 4 hours of incubation with 50 ng/mL PDGF (A) or 20 ng/mL FGF (B) and down-regulated with 1 μg/mL PF4 (C), 1 μg/mL TSP (D), and 50 ng/mL TGF-β.
Changes in TPO mRNA expression in human BM stromal cells measured by semiquantitative reverse transcriptase (RT)-PCR.
Semiquantitative RT-PCR analyses used RNA isolated from primary BM stromal cells. The PCR products were separated by agarose-gel electrophoresis and analyzed by Southern hybridization with a gene-specific probe. It was confirmed that TPO mRNA expression was up-regulated after 4 hours of incubation with 50 ng/mL PDGF (A) or 20 ng/mL FGF (B) and down-regulated with 1 μg/mL PF4 (C), 1 μg/mL TSP (D), and 50 ng/mL TGF-β.
Changes in mRNA expression not a consequence of cell proliferation
Our thymidine-incorporation study and MTS assay (data not shown) demonstrated that the addition of individual platelet proteins to either quiescent cells or cells cultured in 25% serum had no significant effects on cell growth (Figure8). Trypan blue exclusion and morphologic assessment showed no decrease in cell viability and no toxicity changes. Therefore, the increase in TPO expression with the addition of PDGF or FGF was not associated with an increase in cell number, and the changes in TPO mRNA expression observed with the addition of the negative regulators (PF4, TSP, and TGF-β) were not a result of cell toxicity.
Changes in TPO mRNA expression not due to cell proliferation.
In experiments to investigate the up-regulation of TPO mRNA expression, BM stromal cells were allowed to become quiescent by placing them in 1% serum for 24 hours. The increase in TPO mRNA expression with the addition of PDGF or FGF was not due to an increase in cell proliferation, as shown by thymidine incorporation. In experiments to investigate the inhibition of TPO gene expression, the confluent cells were left in complete medium containing 25% serum for 24 hours. Suppression of TPO mRNA expression by PF4, TSP, or TGF-β was not a result of cell toxicity. The results shown are the mean ± SD values from 3 separate experiments carried out in triplicate.
Changes in TPO mRNA expression not due to cell proliferation.
In experiments to investigate the up-regulation of TPO mRNA expression, BM stromal cells were allowed to become quiescent by placing them in 1% serum for 24 hours. The increase in TPO mRNA expression with the addition of PDGF or FGF was not due to an increase in cell proliferation, as shown by thymidine incorporation. In experiments to investigate the inhibition of TPO gene expression, the confluent cells were left in complete medium containing 25% serum for 24 hours. Suppression of TPO mRNA expression by PF4, TSP, or TGF-β was not a result of cell toxicity. The results shown are the mean ± SD values from 3 separate experiments carried out in triplicate.
Suppression of TPO mRNA expression by whole-platelet lysate
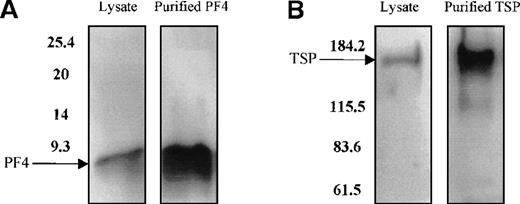
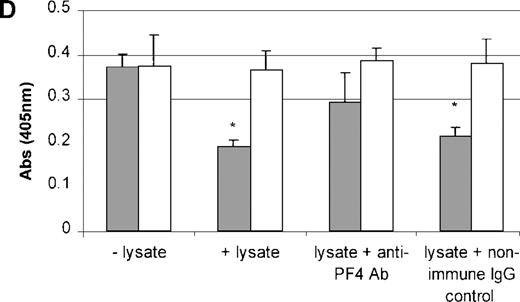
PDGF, FGF, PF4, TSP, and TGF-β do not act individually in vivo because they are stored and released together by the α granules of platelets. Therefore, to study the effect of the combination of these growth factors on TPO mRNA expression, platelets were freeze-thawed to release their contents. Using Western blot analysis, we confirmed the presence of PF4 and TSP proteins in the platelet lysate (Figure9A and 9B). The addition of the platelet lysate to the cultured cells resulted in inhibition of TPO mRNA expression in a dose-dependent manner. Lysate from 1 × 109 platelets/mL suppressed TPO mRNA expression levels by 50% (Figure 9C). Anti-PF4 antibody, when incubated together with the lysate from 1 × 109 platelets, partly reversed the inhibitory effect of the lysate (Figure 9D), whereas the nonimmune IgG control had no effect. Surprisingly, the antibody to TSP failed to rescue lysate inhibition of TPO expression (data not shown). This may have been because of the nonneutralizing nature of the anti-TSP antibody. GAPDH mRNA levels remained unchanged in response to incubation of the cells with the platelet lysate and antibodies (Figure9C and 9D).
Effect of whole-platelet lysate on TPO mRNA expression in BM stromal cells.
Equal amounts of lysate from freeze-thawed platelets were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and the presence of PF4 and TSP was confirmed by Western blot analysis using anti-PF4 (2 μg/mL) (A) and anti-TSP (2 μg/mL) antibodies (B). Serial dilutions of lysate from 1 × 109platelets/mL were added to the stromal cell cultures, resulting in a dose-dependent inhibition of TPO mRNA expression measured with the QISH assay (C). The concentration of lysate added is expressed as the amount of platelet granular constituents in increasing numbers of platelets per milliliter. GAPDH mRNA expression remained unchanged. The lysate inhibition of TPO mRNA was partly relieved with the incubation of an antibody specific to PF4, whereas the nonimmune IgG control had no effect; GAPDH mRNA expression remained unchanged in cells exposed to lysate or antibody (D). The results shown are the mean ± SD values from 3 separate experiments performed in triplicate. ▪, TPO; □, GAPDH. * P < 0.05 compared with the control.
Effect of whole-platelet lysate on TPO mRNA expression in BM stromal cells.
Equal amounts of lysate from freeze-thawed platelets were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and the presence of PF4 and TSP was confirmed by Western blot analysis using anti-PF4 (2 μg/mL) (A) and anti-TSP (2 μg/mL) antibodies (B). Serial dilutions of lysate from 1 × 109platelets/mL were added to the stromal cell cultures, resulting in a dose-dependent inhibition of TPO mRNA expression measured with the QISH assay (C). The concentration of lysate added is expressed as the amount of platelet granular constituents in increasing numbers of platelets per milliliter. GAPDH mRNA expression remained unchanged. The lysate inhibition of TPO mRNA was partly relieved with the incubation of an antibody specific to PF4, whereas the nonimmune IgG control had no effect; GAPDH mRNA expression remained unchanged in cells exposed to lysate or antibody (D). The results shown are the mean ± SD values from 3 separate experiments performed in triplicate. ▪, TPO; □, GAPDH. * P < 0.05 compared with the control.
TPO ELISA
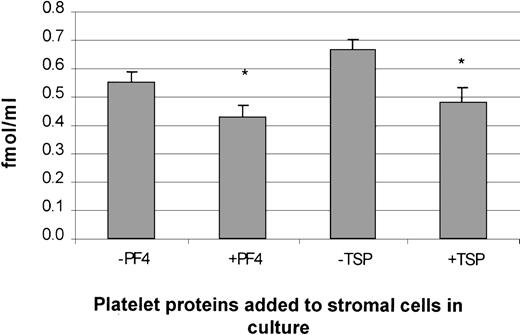
The protein concentration of TPO was measured in supernatants of stromal cells treated with and without PDGF, PF4, or TSP. A significant inhibition in TPO protein levels was observed in cells treated with PF4 or TSP (Figure 10). These data are consistent with the suppression of TPO mRNA in stromal cells treated with either PF4 or TSP. Unfortunately, results with supernatant collected from cells treated with PDGF were not significant because the basal levels of TPO protein expression (in growth-quiescent cells) were below the detection limit of the assay (data not shown).
Suppression of TPO protein expression in human BM stromal cells by PF4 and TSP.
The addition of PF4 and TSP to stromal cells in culture suppressed TPO protein expression, as measured by an enzyme-linked immunosorbent assay. The results shown are the mean ± SD values from 3 separate experiments carried out in triplicate. *P < .05 compared with controls.
Suppression of TPO protein expression in human BM stromal cells by PF4 and TSP.
The addition of PF4 and TSP to stromal cells in culture suppressed TPO protein expression, as measured by an enzyme-linked immunosorbent assay. The results shown are the mean ± SD values from 3 separate experiments carried out in triplicate. *P < .05 compared with controls.
Discussion
The current study demonstrated that TPO mRNA expression in primary BM stromal cells may be regulated both positively and negatively. We confirmed these results with 2 independent methods. The first method involved a QISH assay. This assay was previously applied in studies of the expression of Fc IgG receptors38,47 and of connective-tissue proteins in bone cells derived from humans.39 The quantitative aspects of this assay were demonstrated by a study of the effect of increasing cell numbers compared with the absorbance obtained.39 The QISH assay provides a quantitative determination of changes in gene expression where a direct log-linear relation between absorbance values and mRNA expression has been found.38-40 The second method we used was semiquantitative RT-PCR. Results obtained with this well-established technique12 15 were consistent with the QISH data, indicating that TPO gene expression by the stromal cells of the BM could indeed be modulated.
First, we showed that the peptide growth factors PDGF-BB and FGF-2 markedly stimulates the expression of TPO mRNA in both a time-dependent and dose-dependent manner. PDGF was previously shown to induce IL-1, IL-6, M-CSF, and G-CSF transcripts in human and murine stromal cells.28 41 Our data here showed at least a 2-fold induction in TPO mRNA expression with the addition of 50 ng/mL PDGF. A significant increase in TPO expression was also observed with 20 ng/mL FGF. We also used the phorbol ester PMA, which is capable of inducing differentiation in a variety of cell lines. PMA stimulated TPO mRNA levels in a time-dependent manner. However, the mechanism whereby TPO mRNA was induced by these growth factors has not been determined. Measurement of another mRNA species, IL-3, showed no change in response to PDGF. This further supports the idea that TPO mRNA expression was specifically up-regulated by PDGF. The increase in TPO mRNA expression observed was not due to increased cell proliferation, as demonstrated by thymidine uptake. In addition, measurements of GAPDH mRNA levels as a positive control were not affected by any of the regulatory molecules, suggesting that there were no changes in cell metabolism.
Second, we showed that TPO mRNA expression in the BM stromal cells may also be regulated by negative influences. PF4, TSP, and TGF-β are known inhibitors of megakaryocytopoiesis and hematopoiesis.30,31,35 On the basis of these observations, we hypothesized that these platelet granular proteins may play a role in the negative regulation of TPO mRNA expression in BM stromal cells. Using 2 independent methods, we showed that these molecules suppress TPO expression in both a dose-dependent and time-dependent manner. A significant inhibition was found with a PF4 or TSP dose ranging from 1 to 2 μg/mL. A significant inhibition was also observed with the addition of 50 to 100 ng/mL TGF-β. Furthermore, TPO protein secretion by stromal cells in culture was inhibited by PF4 and TSP, consistent with the effect of these proteins on TPO mRNA expression. In studies supporting these data, Lebeurier et al48 showed that parental administration of PF4 in mice resulted in significant reduction in circulating platelets, BM megakaryocytes, and colony-forming units–megakaryocytes.
Finally, it is well known that the regulatory molecules PDGF, FGF, PF4, TSP, and TGF-β are stored and released by platelet α granules.24,28,30,31 49 Because these molecules do not act as separate entities in vivo, we studied the combined effect of whole-platelet lysate on TPO mRNA expression. The addition of the platelet lysate to the cells in culture resulted in a dose-dependent inhibition of TPO mRNA expression, whereas lysate from 1 × 109 platelets/mL resulted in a 50% suppression of TPO expression. Interestingly, preincubation of the lysate with an antibody specific to PF4 partly reversed the inhibitory effect of the lysate on TPO expression. This suggests that PF4 may be one of the components in the platelet lysate that inhibits TPO mRNA expression. Preincubation of the platelet lysate with an antibody specific to TSP did not affect the degree of inhibition of TPO mRNA. This may have been due to the limitations in the ability of the antibody to neutralize the protein. Although these platelet α-granular proteins may individually up-regulate or down-regulate TPO gene expression in BM stromal cells, the combined effect of these proteins may in fact be inhibitory.
Our results suggest that PF4 and possibly TSP may have a greater impact than the other molecules, since these proteins are present at higher concentrations in the platelet granules; reported values are 20 μg PF4/109 platelets, 25 μg TSP/109 platelets, and 10 ng PDGF/109 platelets.33,49,50 Although plasma concentrations of PF4 and TSP are usually nanograms per milliliter, the local concentration of the platelet constituents released in the BM may be much higher than the concentration in the peripheral blood.51 52 Therefore, it is possible that the platelet constituents play a negative role in regulating TPO gene expression. We hypothesize a paracrine regulatory loop in which these molecules, when released from the platelet/megakaryocyte α granules, would modulate TPO gene expression in BM stromal cells. It is likely that this regulatory loop, acting as a negative feedback mechanism, becomes operative only in situations of increased megakaryocytopoiesis and platelet production. Conversely, in situations in which megakaryocytes and platelets are decreased, the negative modulation on TPO gene expression is lifted, allowing constitutive expression of TPO, together with stimulation by PDGF or FGF (secreted by cells in the surrounding microenvironment), to proceed unimpeded.
In summary, the results of this study extend those of previous studies6 15 suggesting that thrombopoiesis is regulated at the level of TPO mRNA expression in BM stroma. These data may provide further insights into the possible feedback mechanism that occurs locally in the BM microenvironment. Our findings with respect to this mechanism, however, do not exclude the presence of a passive mechanism in which plasma TPO levels are regulated by platelet/megakaryocyte mass through uptake and metabolism. In fact, the 2 mechanisms may coexist.
Acknowledgments
We thank Drs Tee Beng Kang, Aseem Lal, and Daniel Owens for their efforts in collecting the bone marrow samples, Dr Linda Bendall (ICPMR, Westmead Hospital, Sydney Australia) for technical advice on the LTBMC techniques, and Dr Melissa Holmes for helpful discussions and critical evaluation of the manuscript.
Funded in part by a program grant from the National Health and Medical Research Council of Australia and an infrastructural grant from the New South Wales state government.
Reprints:B.H. Chong, Department of Haematology, Prince of Wales Hospital, High Street, Randwick, NSW 2031, Australia; e-mail:b.h.chong@unsw.edu.au.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal