Abstract
Smad family proteins are essential for transforming growth factor β (TGF-β) signal mediation downstream of a heteromeric complex of the type I and type II receptor serine/threonine kinases. A distant family member, Smad7, is expressed in most mammalian tissues and cells and prevents TGF-β signaling. In this study, we examined the physiologic role of Smad7 in mediating the effects of activin, a member of the TGF-β superfamily of peptides that functions in a number of processes, including blood-cell development. We report here that Smad7 expression is specifically absent in particular hematopoietic cells that respond to activin by differentiating into the erythroid lineage and that ectopic production of Smad7 causes mouse erythroid leukemia (F5-5) cells to become resistant to activin induction of erythroid differentiation. When coexpressed with type I activin receptor ActR-I or ActR-IB in concert with type II receptor ActR-II, Smad7 efficiently reduced an early transcriptional response mediated by ActR-I but had only a minimal effect on the response mediated by ActR-IB. In the presence of Smad7, overexpression of an activated form of ActR-IB, but not of an activated form of ActR-I, induced F5-5 cells to differentiate. These results suggest that Smad7 selectively interferes with the ActR-I pathway in activin signal transduction. The findings also indicate the existence of a novel activity of Smad7 that inhibits erythroid differentiation by blocking intracellular signaling of activin.
The cell fate determination of hematopoietic progenitors in both the embryonic and adult body is controlled by the action of many molecules, including various growth factors and cytokines. Members of the transforming growth factor β (TGF-β) family have also been suggested to play critical roles in the regulation of cell proliferation and differentiation during hematopoiesis. The TGF-β superfamily consists of a large number of related polypeptides in diverse multicellular organisms and includes activins, bone morphogenetic proteins (BMPs), TGF-βs, and many other secreted factors. The signals of these factors are involved in a wide array of biologic processes, including growth control, matrix production, immune response, and embryonic patterning. The factors transmit signals through their specific receptors composed of at least 2 distinct transmembrane serine and threonine kinases known as type I and type II receptors. According to currently proposed models, ligand-induced association of the type I and type II receptors results in the phosphorylation and consequent activation of the type I receptor, which is required for downstream signal cascades.1 2
Genetic and developmental studies in Drosophila fruit flies revealed that mad and medea genes are essential for intracellular signaling of Dpp (the decapentaplegic gene product), which is required for many differentiation events and is closely related to vertebrate BMP2/4.3,4 Several homologs of these genes, which are now referred to as Smads, have been observed in vertebrate cells.5,6 Smad transcripts are widely expressed in a variety of tissues and cells, indicating substantial involvement of the Smad proteins in signaling from various TGF-β ligands.7,8 Individual Smad proteins are dedicated to transducing the intracellular signals for a specific subclass of TGF-β ligands; for example, Smad1 mediates the BMP signal, whereas Smad2 and Smad3 transduce the TGF-β signal.9-11 In both of these types of signaling, however, it appears that Smad4, which is encoded by the tumor suppressor gene DPC4 (initially identified at a chromosome locus that is frequently mutated in human carcinomas12), is commonly used. In addition, it was shown that Smad5 is involved in the inhibitory effect of TGF-β on hematopoietic cell proliferation.13 Recently, complementary DNAs (cDNAs) encoding the distantly related family members Smad614,15 and Smad716-20 were identified. Ectopic expression of each of these cDNAs prevents the signaling of various TGF-β family ligands. However, the physiologic importance of these antagonistic Smads has not been clarified.
Activin was originally identified as an activity in porcine ovarian fluid that induces secretion of follicle-stimulating hormone from pituitary cells.21,22 In the TGF-β superfamily, activin is a unique and multifunctional factor that can stimulate hormone production in ovarian and placental cells; support neuronal cell survival; influence cell-cycle progress positively or negatively, depending on the cell type; and induce mesodermal differentiation in amphibian embryos.23-25 Moreover, erythroid differentiation factor, which was isolated from the conditioned medium of stimulated human monocytic cells, was found to be a human homolog of activin.26 Consistent with this observation, natural and recombinant activins promote the early stages of erythrocyte differentiation, accompanied by hemoglobin synthesis.27,28Activin first binds to a specific type II receptor, and then a type I receptor binds to this complex.29,30 The activin type I receptor cannot bind activin when produced alone; coexpression of type II receptor is required for binding of the ligand.31-33 The presence of a type I receptor is also necessary for signal transduction, which suggests that the functional form is a complex between activin and both types of receptors.31
Two activin type I receptors, ActR-I (also called as Tsk-7L or ALK-2) and ActR-IB (ALK-4),31-36 and 2 activin type II receptors, ActR-II and ActR-IIB,37-39 have been characterized in vertebrate cells. Although differences in the affinity to activin and the tissue distribution of ActR-II and ActR-IIB have been observed, the importance of the existence of the 2 type II receptors is unclear.38,39 On the other hand, activin has been suggested to elicit discrete signals, as well as common signals, from the 2 type I receptors, ActR-I and ActR-IB.31,33,40 41
The goal of this study was to elucidate the physiologic role of Smad7 in activin signaling. We found that Smad7 expression, which occurs normally in various tissues, is very low, specifically in cells from some leukemic origins that can start erythroid differentiation in response to agonist stimuli. Stable expression of Smad7 effectively prevents these cells from differentiating in response to activin. Our results suggest that Smad7 may exert control by selectively interfering with different activin signaling pathways.
Materials and methods
Cell culture
F5-5, K562, HEL, HL-60, and MEG01 cells were cultured in RPMI-1640 medium. CTLL-2 cells were cultured in RPMI-1640 with 100 U/mL recombinant murine interleukin 2. NMuMG, M1, P815, Sp2, Swiss-3T3, WEHI-3, and 293 cells were cultured in Dulbecco modified Eagle medium. ST2, C7, CHO-K1, and Mv1Lu cells were cultured in minimum essential medium α. All media were supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 0.1 mg/mL each of ampicillin and streptomycin, except where indicated otherwise. F5-5, Swiss-3T3, and Mv1Lu cells were obtained from the Riken Cell Bank (Tsukuba, Japan). K562, NMuMG,42 Sp2, and 293 cells were from the American Type Culture Collection (Manassas, VA). HEL, HL-60, and MEG01 cells were from Dr M. Matsuoka (Kumamoto University, Kumamoto, Japan). ST2 cells43 were obtained from Dr S. Nishikawa (Kyoto University, Kyoto, Japan), CHO-K1 cells from Dr M. Tsuneoka (Kurume University, Kurume, Japan), and C7, CTLL-2, M1, P815, and WEHI-3 cells from Dr T. Kunisada (Tottori University, Yonago, Japan). To assay erythroid differentiation, F5-5 cells (1 × 104cells/mL) were cultured in complete medium with or without activin A (1 nmol/L), a gift from Dr Y. Eto (Ajinomoto, Kawasaki, Japan). Five days after stimulation, 90 μL of the cell culture was added to 10 μL of fresh dianisidine solution (0.2% 3,3′-dimethoxybenzidine, 2% hydrogen peroxide, and 5% acetic acid in water). The blue-stained cells were counted with use of microscopy.
DNA clones
Degenerate polymerase chain reaction (PCR) primers were used to screen a mouse cDNA library (17-day embryo marathon-ready cDNA; Clontech, Palo Alto, CA) for identification of Smad family genes. On the basis of regions of sequence similarity between Smad protein members, 2 degenerate antisense primers were synthesized: gene-specific primer 1 (GSP1), which was 5′-GTT(A/C/G/T)A(A/G)GTG(A/C/G/T)AC(C/T)TC(A/C/G/T)A(G/T)CCAGCA(A/C/G/T)GG-3′, and gene-specific primer 2 (GSP2, which was 5′-GTA(A/C/G/T)(C/T)(A/C)(A/C/G/T)G(C/G)(A/C/G/T)- CCCCA(A/C/G/T)CC(C/T)TT(A/C/G/T)AC(A/G)AA-3′. The primer set for the first PCR was GSP1 and an adapter primer (AP1; Clontech). A second PCR was performed by using the first PCR product (1:500 dilution) as a template and GSP2 and a nested adapter primer (AP2, Clontech). The PCR products were screened by sequencing and used to isolate full-length clones of the corresponding cDNA from a mouse embryo cDNA library. A 3.6-kilobase (kb) mouse cDNA containing the putative entire coding regions of Smad7 was isolated and its nucleotide sequence determined. Sequence analyses revealed that mouse Smad7 cDNA comprised 2 closely related sequences that encoded polypeptides of 426 and 425 amino acids, respectively. We designated the latter sequence Smad7B to distinguish it from the former, Smad7. Compared with Smad7B, Smad7 has 1 additional amino acid residue in a linker region between the Mad homology 1 and Mad homology 2 domains, possibly because of use of an alternative RNA splicing site causing a 3-nucleotide insertion. The sequence data were submitted to the European Molecular Biology Laboratory database, GenBank, and the DNA Databank of Japan (accession numbers, AJ000550 and AJ000551).
Expression vectors were constructed with coding sequences that were obtained by PCR of the cDNA and entirely sequenced. The coding region of Smad7 was amplified with the following primers: upstream, 5′-CCGCTAGCACCATGTTCAGGACCAAACGATCTGCGCTCGTC-3′; and downstream, 5′-CCGGATCCTATCGCGAGTTGAAGATGACCTCCAGCCAGCACG-3′. For receptor cDNAs, the 5′ and 3′ primers used for PCR were synthesized according to known sequences.37,44,45 The cloned NheI-BamHI fragment of Smad7,XbaI-EcoRI fragments of ActR-I and ActR-IB, andXbaI-BamHI fragment of ActR-II were subcloned into the expression vector pactEF.46 ActR-I (Q207D) and ActR-IB (T206D), which contained an activating mutation and consequently had signaling activity in the absence of ligand,29,47 were generated by a PCR-based strategy. ActR-I and ActR-II were modified at their carboxy-terminals by the addition of a Flag tag and an HA tag, respectively. Provirus plasmid vector pGψ+ was obtained from the 5-kbEagI-BamHI backbone of pGD′48 by insertion of a 0.7-kb EagI-BamHI fragment of LXSN (Clontech). pGψ+neo was constructed by inserting a 1.1-kbXhoI-HincII fragment of the neo expression cassette from pMC1neo (Stratagene, La Jolla, CA) into a 5.7-kbXhoI-ClaI fragment of pGψ+. Similarly, pGψ+pur was made from the 5.7-kbXhoI-ClaI backbone of pGψ+ by insertion of a 0.4-kb XhoI-HindIII fragment containing the SV40 early promoter from LXSN and a 0.6-kb AlfII-NgoMI PCR fragment bearing the puromycin resistance gene (Clontech). TheNheI-BamHI fragment of Smad7 and theXbaI-BamHI fragment of Myc-Smad7 were subcloned into pGψ+neo and pGψ+pur. TheXbaI-EcoRI fragments of ActR-I, ActR-IB, and their mutants and the XbaI-BamHI fragment of ActR-II were inserted into pGψ+neo at the HpaI site. The reporter construct p3TP-Lux was provided by Dr J. Massagué(Memorial Sloan-Kettering Cancer Center, New York, NY). An internal control reporter, pRL-1PGK, was constructed by inserting a 0.5-kbEcoRI-PstI fragment from pKJ2,49 which includes the constitutive promoter of the mouse phosphoglycerate kinase 1 (Pgk-1) gene, between the BglII and NheI sites of the pRL-null vector (Promega, Madison, WI) containing a sea pansy luciferase reporter gene.
Retrovirus preparation
Highly transfectable 293 cells were transfected with DNA constructs by using the calcium phosphate coprecipitation procedure described previously.50 Typically, 20 μg of expression vector assembled with pGψ+ and 10 μg of the replication-defective packaging construct pMoψ−were cotransfected into 293 cells that were seeded at a density of 3 × 106 cells per 90-mm dish 24 to 30 hours earlier. The supernatants were harvested, supplemented with HEPES–sodium hydroxide (pH 7.1) to a concentration of 20 mmol/L, filtrated through 0.45-μm-pore filters, and stored at −80°C.
DNA transfection and luciferase assays
Cells (2 × 104/well) were placed in 24-well plates. The next day, they were transfected with p3TP-Lux (0.3 μg), the control pRL-1PGK (0.1 μg), and the indicated constructs by using FuGENE 6 (Boehringer Mannheim, Indianapolis, IN). Twenty-hours after transfection, cells were stimulated with 1 nmol/L activin A in medium containing 0.2% FBS. After 24 hours, the cells were lysed and the 2 luciferase activities were measured by using the Dual-luciferase Reporter Assay System (Promega).
Reverse transcriptase (RT)-PCR assays
cDNA was synthesized from the total RNA (1 μg) by using the SuperScript preamplification system (GIBCO BRL, Gaithersburg, MD), treated with RNase H, and subjected to PCR (KOD Dash; Toyobo, Japan). A fragment of Smad7 cDNA (207 bp) was amplified with the following primers (which can work as specific primers commonly for mouse, mink, and human Smad7 cDNAs): upstream, 5′-AAAGTGTTCCCTGGTTTCTCCATCAAGGC-3′; and downstream, 5′-CTACCGGCTGTTGAAGATGACCTCCAGCCAGCAC-3′. The PCR conditions were as follows: denaturation at 96°C for 2 minutes, followed by 28 cycles at 96°C for 15 seconds, 65°C for 10 seconds, and 70°C for 30 seconds. A fragment of G3PDH cDNA (452 bp) was amplified with the primers, 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′. A fragment of β-globin cDNA (244 bp) was amplified with the primers 5′-ACTCCGATGAAGTTGGTGGTG-3′ and 5′-GGATCCACATGCAGCTTGTCA-3′.51
Immunochemical procedures
To prepare Smad7 protein as an immunogen, the cDNA fragment corresponding to amino acid residues 91 to 426 was made by PCR and subcloned in pET3Δ (Novagen, Madison, WI) for expression inEscherichia coli. Specific antibody was purified from rabbit serum by immunoaffinity chromatography with an immobilized-antigen column prepared by using Affi-gel 15 (Bio-Rad, Hercules, CA). For immunoblotting, cells were lysed as described previously.46After determination of protein concentration with use of a MicroBCA system (Pierce, Rockford, IL), a fixed amount of protein was boiled again in the presence of 2% sodium dodecyl sulfate and 5% 2-mercaptoethanol, subjected to gel electrophoresis on a 10% polyacrylamide gel, and transferred to a nitrocellulose membrane (0.2-μm pore). After blocking with 3% bovine serum albumin in Tris-buffered saline (TBS), the blot was incubated with antimouse Smad7 antibody for 2 hours. The membrane was then incubated with horseradish peroxidase–conjugated donkey IgG against rabbit Ig (1:2000; Amersham, Buckinghamshire, UK) in 5% skim milk in TBS for 1 hour. This was followed by detection with a chemiluminescent substrate (SuperSignal Ultra, Pierce).
Results
Smad7 expression is specifically absent in erythroid leukemic cells
To study the biologic importance of Smad7, we surveyed different tissues and cells for the expression of Smad7. Northern blot analysis showed that the main transcript of 4.2 kb was contained ubiquitously in various organs of mice, with higher levels in the brain and kidney (Figure 1A). Further analysis with RT-PCR revealed Smad7 messenger RNA (mRNA) in a wide variety of cultured cells established from human and rodent tissues of epithelial, fibroblastic, and hematic origins (Figure 1B). These included cells known to be responsive to TGF-β (Mv1Lu lung epithelial cells and CTLL-2 T lymphocytes),52,53 activin (M1 myeloblast cells and HL-60 promyelocytic cells),54,55 and BMP (Swiss-3T3 fibroblast cells),56 thereby suggesting an important role for Smad7 in many biologic processes. Mouse F5-5 and human K562 and HEL hematopoietic malignant cells, however, had barely detectable levels of Smad7 mRNA (Figure 1B). Interestingly, these leukemia cells are known to start accumulating hemoglobin, an erythroid cell marker, in response to activin.27,28 57
Specific absence of Smad7 mRNA in particular hematopoietic cells.
(A) Northern blot analysis of mouse Smad7 mRNA. Each lane contained 2 μg of poly(A)+ RNA from the indicated mouse organs. A Smad7 probe (625-base pair [bp] complementary DNA [cDNA] fragment) revealed one major transcript in various tissues. Positions of RNA size markers (kilobase [kb]) are shown on the left. (B) RT-PCR analysis of Smad7 mRNA in different types of cells. The examined cell lines were mink Mv1Lu (lung epithelial), mouse Swiss-3T3 (embryonic fibroblast), F5-5 (erythroid leukemia), M1 (myeloblast), WEHI-3 (myelomonocyte), C7 (macrophage), P815 (mastocytoma), CTLL-2 (T lymphocyte), Sp2 (plasmacytoma), human HEL (erythroleukemia), K562 (chronic myelogenous leukemia), HL-60 (promyelocytic leukemia), and MEG-01 (megakaryoblast). cDNAs to 1 μg of RNA isolated from these cells were synthesized by reverse transcription with random primers. Equal aliquots from each reaction were subjected to PCR to amplify a Smad7 and a control G3PDH cDNA fragment. The PCR primer pairs and conditions were designed for amplification of a 207-bp Smad7 sequence in both rodent and human cells. The data indicate that F5-5, HEL, and K562 cells specifically lack Smad7 expression.
Specific absence of Smad7 mRNA in particular hematopoietic cells.
(A) Northern blot analysis of mouse Smad7 mRNA. Each lane contained 2 μg of poly(A)+ RNA from the indicated mouse organs. A Smad7 probe (625-base pair [bp] complementary DNA [cDNA] fragment) revealed one major transcript in various tissues. Positions of RNA size markers (kilobase [kb]) are shown on the left. (B) RT-PCR analysis of Smad7 mRNA in different types of cells. The examined cell lines were mink Mv1Lu (lung epithelial), mouse Swiss-3T3 (embryonic fibroblast), F5-5 (erythroid leukemia), M1 (myeloblast), WEHI-3 (myelomonocyte), C7 (macrophage), P815 (mastocytoma), CTLL-2 (T lymphocyte), Sp2 (plasmacytoma), human HEL (erythroleukemia), K562 (chronic myelogenous leukemia), HL-60 (promyelocytic leukemia), and MEG-01 (megakaryoblast). cDNAs to 1 μg of RNA isolated from these cells were synthesized by reverse transcription with random primers. Equal aliquots from each reaction were subjected to PCR to amplify a Smad7 and a control G3PDH cDNA fragment. The PCR primer pairs and conditions were designed for amplification of a 207-bp Smad7 sequence in both rodent and human cells. The data indicate that F5-5, HEL, and K562 cells specifically lack Smad7 expression.
Smad7 prevents induction of erythroid differentiation by activin
Activin acts as a hematopoietic growth factor that induces erythroid precursor cells to undergo differentiation to the erythroid lineage. The absence of Smad7 expression in the particular leukemic cells in our study but not in other activin-responsive cells could mean either that Smad7 is not required for activin signals in these leukemic cells or that it must be kept at low levels to allow erythroid differentiation to occur.
To investigate the function of Smad7 protein in cellular responses, Smad7 cDNA was subcloned into a retroviral vector because of its limited copy number of gene integration and very high transducing efficiency. In this vector, the Moloney long-terminal-repeat promoter drove Smad7 expression and was followed by an internal thymidine kinase promoter with neor as a selectable marker gene. The Smad7 or control vector construct was transiently cotransfected with a replication-defective ecotropic provirus onto 293 cells to generate a high titer of recombinant viruses,50 which typically transduced theneor marker into a major fraction of the target mouse fibroblast NIH3T3 cells (95%-100%) and erythroid F5-5 cells (85%-95%).
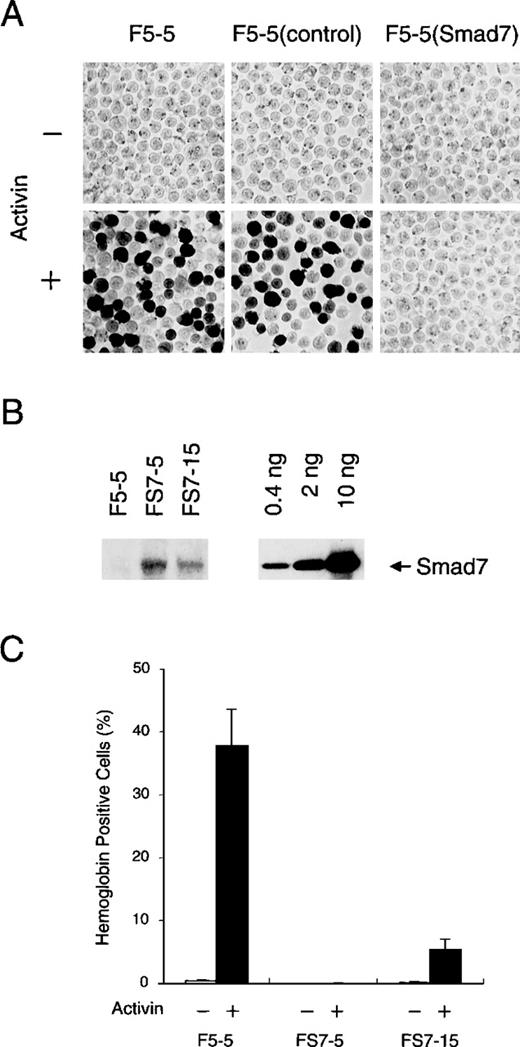
We generated F5-5 cells stably expressing Smad7 and cells containing the control vector by infection of the appropriate viruses and selection with G418. The cells were then treated with 1 nmol/L activin (recombinant human activin A) for 5 days and assessed for hemoglobin content with use of benzidine staining. As shown in Figure2A, activin efficiently increased the proportion of cells that contained hemoglobin, usually up to 40% to 50% in a population of G418-resistant control cells, as in parent F5-5 cells, whereas less than 0.1% of the untreated cells were stained positively. These results confirmed the previously reported effects of activin.27 28 In sharp contrast, the cells infected with the Smad7 vector were strongly prevented from having hemoglobin synthesis even after treatment with activin, suggesting that Smad7 protein suppressed activin-induced erythroid differentiation of these cells.
Inhibitory effect of Smad7 on erythroid differentiation induced by activin.
(A) Hemoglobin accumulation in F5-5 mouse erythroid leukemia cells and in derivatives. F5-5 cells were infected with control or Smad7 transducing virus and selected for acquiring G418 resistance. After 5 days of culture with (+) or without (−) 1 nmol/L activin, cells were stained with benzidine to reveal hemoglobin content. F5-5 (control) indicates G418-resistant control cells, and F5-5 (Smad7) indicates Smad7 virus-transduced cells. (B) Immunoblotting analysis of Smad7 stably expressed in clonal cells. FS7-5 and FS7-15 cells were cloned from single G418-resistant cells that had been infected with Smad7 expression vector. The whole-cell lysate of 2 × 104 cells (left) or the indicated amount (0.4, 2, or 10 ng) of the bacterially expressed, purified Smad7 protein (amino acids 91-426; right) was loaded into each lane and subjected to electrophoresis and blotting. Smad7 was detected with specific antibody against Smad7. Densitometric analysis indicated that 2 × 104 F5-5 parent cells, FS7-5 clonal cells, and FS7-15 clonal cells contained less than 4 pg, 100 pg, and 20 pg, respectively, of Smad7 protein. (C) Effect of Smad7 level on activin-induced hemoglobin synthesis. F5-5, FS7-5, and FS7-15 cells were cultured for 5 days in the absence (−) or presence (+) of 1 nmol/L activin and assessed for hemoglobin accumulation. The results shown are the percentages of total cells that contained hemoglobin. Values are the mean and SD from 3 separate experiments.
Inhibitory effect of Smad7 on erythroid differentiation induced by activin.
(A) Hemoglobin accumulation in F5-5 mouse erythroid leukemia cells and in derivatives. F5-5 cells were infected with control or Smad7 transducing virus and selected for acquiring G418 resistance. After 5 days of culture with (+) or without (−) 1 nmol/L activin, cells were stained with benzidine to reveal hemoglobin content. F5-5 (control) indicates G418-resistant control cells, and F5-5 (Smad7) indicates Smad7 virus-transduced cells. (B) Immunoblotting analysis of Smad7 stably expressed in clonal cells. FS7-5 and FS7-15 cells were cloned from single G418-resistant cells that had been infected with Smad7 expression vector. The whole-cell lysate of 2 × 104 cells (left) or the indicated amount (0.4, 2, or 10 ng) of the bacterially expressed, purified Smad7 protein (amino acids 91-426; right) was loaded into each lane and subjected to electrophoresis and blotting. Smad7 was detected with specific antibody against Smad7. Densitometric analysis indicated that 2 × 104 F5-5 parent cells, FS7-5 clonal cells, and FS7-15 clonal cells contained less than 4 pg, 100 pg, and 20 pg, respectively, of Smad7 protein. (C) Effect of Smad7 level on activin-induced hemoglobin synthesis. F5-5, FS7-5, and FS7-15 cells were cultured for 5 days in the absence (−) or presence (+) of 1 nmol/L activin and assessed for hemoglobin accumulation. The results shown are the percentages of total cells that contained hemoglobin. Values are the mean and SD from 3 separate experiments.
To confirm this possibility, we next established several cell lines from single infected F5-5 cells and assessed expression levels of Smad7 protein with quantitative immunoblotting using a specific antibody (Figure 2B). As expected from the lack of mRNA, the parent F5-5 cells did not contain a detectable level of endogenous Smad7. In subclonal FS7-5 cells, which produced a high level of transduced Smad7, activin treatment did not induce any hemoglobin synthesis (Figure 2C). Even at the lower level present in subclonal FS7-15 cells, Smad7 caused obvious repression of the increase in hemoglobin-containing cells in the culture treated with activin. These results clearly show that Smad7 inhibits the induction of hemoglobin synthesis by activin in F5-5 cells in a dose-dependent manner. In addition, the high propensity of F5-5 cells to undergo activin-induced erythroid differentiation may be related to the absence of Smad7.
Smad7 effectively inhibits ActR-I signaling but only inefficiently inhibits ActR-IB signaling
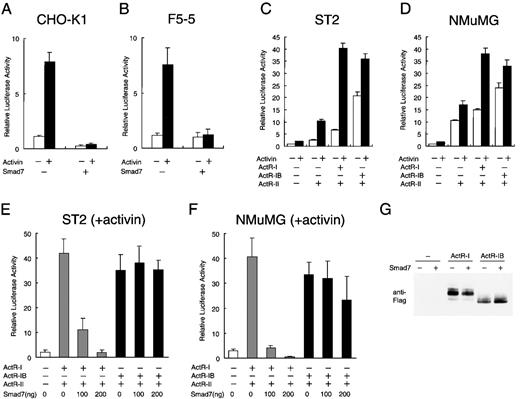
Several steps, including transcriptional alteration, are postulated to exist between activin-stimulated receptor assembly and the eventual phenotype expression in the cells. To examine the effects of Smad7 on activin-induced signaling, we used luciferase reporter assays with the p3TP-Lux plasmid,58 which contains a TGF-β–responsive element from the plasminogen activator inhibitor-1 gene promoter and 3 tetradecanoyl phorbol acetate–responsive elements from a collagenase gene enhancer. This promoter complex was shown to be transcriptionally activated by both TGF-β and activin through mediation of their respective receptors, when p3TP-Lux was transiently introduced into the responsive cells.31,58 We initially transfected p3TP-Lux, together with a control reporter driven by the constitutive promoter of the Pgk-1 gene,59 into CHO-K1 cells, which are responsive to activin and have high transfection efficiency60 and then continued the culture with or without activin. After 18 hours of treatment with 1 nmol/L activin, luciferase activity was elevated by 8-fold compared with that in untreated cells (Figure 3A). However, coexpression of Smad7 strongly precluded the reporter activity from being stimulated by activin. Transient transfection of the reporter into F5-5 cells yielded essentially similar results (Figure 3B). These results suggest that Smad7 acts as a potent negative regulator of the early transcriptional response initiated by activin.
Selective inhibition by Smad7 of signaling of type I activin receptor.
Chinese hamster ovary (CHO) K1 cells (A) and F5-5 mouse erythroid leukemia cells (B) were transfected with 300 ng of reporter construct p3TP-Lux, 100 ng of the internal control pRL-1PGK, and 200 ng of either control vector pactEF or Smad7 expression vector pactEF-Smad7 and then cultured in the absence or presence of 1 nmol/L activin. Transcriptional activity of the 3TP promoter was measured as firefly luciferase activity in the cell lysate. Values were normalized by comparison with values for sea pansy luciferase activity, which was driven by the constitutive promoter of the mouse phosphoglycerate kinase gene in pRL-1PGK. Results are expressed as the increase in induction (x-fold) compared with the value in the untreated vector control. Values are the mean and SD from at least 3 assays. (C) ST2 mouse fibroblast cells rendered highly responsive to activin by transfection of activin receptors in the p3TP-Lux assays. In parent cells, activin treatment produced only a weak activation of the 3TP promoter. ST2 cells were transiently transfected with 300 ng of p3TP-Lux, 100 ng of pRL-1PGK, and 50 ng each of expression vector for ActR-II (pactEF-ActR-II) and either ActR-I (pactEF-ActR-I) or ActR-IB (pactEF-ActR-IB) and then cultured in the absence or presence of activin. Luciferase activity was determined as described above. (D) Effects of transfection of activin receptors into NMuMG mouse mammary epithelial cells. Cells were treated and transcriptional activation was assessed as described in (C). (E and F) Efficient suppression by Smad7 of p3TP-Lux activation by activin in the cells producing ActR-I but only inefficient suppression in the cells producing ActR-IB. Increasing amounts of Smad7 vector (0-200 ng) were cotransfected with vectors for ActR-II and either ActR-I or ActR-IB (50 ng each) into ST2 cells (E) and NMuMG cells (F). After treatment with activin, the cells were harvested for the luciferase assay described above. (G) Immunoblot showing comparable levels of the type I receptor proteins, ActR-I and ActR-IB, in transiently transfected cells. NMuMG cells were transfected with carboxy-terminal Flag-tagged ActR-I or ActR-IB in the absence or presence of Smad7. A fixed amount (20 μg each) of total protein from the whole-cell lysate was analyzed by gel electrophoresis and subsequent immunoblotting with anti-Flag monoclonal antibody.
Selective inhibition by Smad7 of signaling of type I activin receptor.
Chinese hamster ovary (CHO) K1 cells (A) and F5-5 mouse erythroid leukemia cells (B) were transfected with 300 ng of reporter construct p3TP-Lux, 100 ng of the internal control pRL-1PGK, and 200 ng of either control vector pactEF or Smad7 expression vector pactEF-Smad7 and then cultured in the absence or presence of 1 nmol/L activin. Transcriptional activity of the 3TP promoter was measured as firefly luciferase activity in the cell lysate. Values were normalized by comparison with values for sea pansy luciferase activity, which was driven by the constitutive promoter of the mouse phosphoglycerate kinase gene in pRL-1PGK. Results are expressed as the increase in induction (x-fold) compared with the value in the untreated vector control. Values are the mean and SD from at least 3 assays. (C) ST2 mouse fibroblast cells rendered highly responsive to activin by transfection of activin receptors in the p3TP-Lux assays. In parent cells, activin treatment produced only a weak activation of the 3TP promoter. ST2 cells were transiently transfected with 300 ng of p3TP-Lux, 100 ng of pRL-1PGK, and 50 ng each of expression vector for ActR-II (pactEF-ActR-II) and either ActR-I (pactEF-ActR-I) or ActR-IB (pactEF-ActR-IB) and then cultured in the absence or presence of activin. Luciferase activity was determined as described above. (D) Effects of transfection of activin receptors into NMuMG mouse mammary epithelial cells. Cells were treated and transcriptional activation was assessed as described in (C). (E and F) Efficient suppression by Smad7 of p3TP-Lux activation by activin in the cells producing ActR-I but only inefficient suppression in the cells producing ActR-IB. Increasing amounts of Smad7 vector (0-200 ng) were cotransfected with vectors for ActR-II and either ActR-I or ActR-IB (50 ng each) into ST2 cells (E) and NMuMG cells (F). After treatment with activin, the cells were harvested for the luciferase assay described above. (G) Immunoblot showing comparable levels of the type I receptor proteins, ActR-I and ActR-IB, in transiently transfected cells. NMuMG cells were transfected with carboxy-terminal Flag-tagged ActR-I or ActR-IB in the absence or presence of Smad7. A fixed amount (20 μg each) of total protein from the whole-cell lysate was analyzed by gel electrophoresis and subsequent immunoblotting with anti-Flag monoclonal antibody.
We attempted to analyze whether Smad7 expression interrupts a particular receptor signaling of activin or blocks all signalings nonspecifically. It was previously shown that transfection of receptor cDNA can confer specific responsiveness to the corresponding ligand on certain cells that are otherwise less responsive.31,58,61Murine cells suitable for this analysis were screened by transfecting p3TP-Lux in combination with vectors containing cDNAs for various activin receptors. We found that in ST2 fibroblast cells, which were established from bone marrow stroma, activin treatment only slightly increased the activity of singly transfected p3TP-Lux. However, cotransfection of vectors for ActR-I and ActR-II, or for ActR-IB and Act-RII, dramatically elevated the reporter activity in these cells in the presence of added activin (Figure 3C), although some ligand-independent elevation of reporter activity was also observed, as previously reported.29,33,62 Coexpression of a kinase-deficient Act-RII mutant with either type I receptor failed to support the reporter activation by activin (data not shown), consistent with the absolute requirement of functional type II receptor kinase for activin signaling.31
Comparable reporter activation occurred in transfected cells expressing ActR-I and in those expressing ActR-IB, together with the wild-type ActR-II (Figure 3C), and similar levels of ActR-I and ActR-IB were detected by immunoblotting using antibody against the Flag tag at the carboxy-terminal of each receptor (Figure3G). These results showed that ActR-I and ActR-IB can act as functional type I receptors for activin in these cells and were consistent with previous observations in other cell systems.31-34 36 They also suggest that the intracellular signaling machinery in ST2 cells can effectively transmit activin stimulation into transcriptional regulation when sufficient amounts of activin receptors are supplied.
We then used this system to examine the effect of Smad7. p3TP-Lux activation by activin in the cells producing ActR-I was largely suppressed by coexpression of Smad7 in a dose-dependent manner, indicating that activin signals through ActR-I are sensitive to Smad7 (Figure 3E). In sharp contrast, Smad7 did not reduce reporter activation in the cells expressing ActR-IB (Figure 3E). Almost identical results were obtained in an entirely different cell type, NMuMG epithelial cells derived from the mouse mammary gland (Figures 3D and 3F). Hence, Smad7 very effectively suppresses the ActR-I–dependent transcriptional activation of activin but only inefficiently suppressed the ActR-IB–dependent activation in cells of various origins.
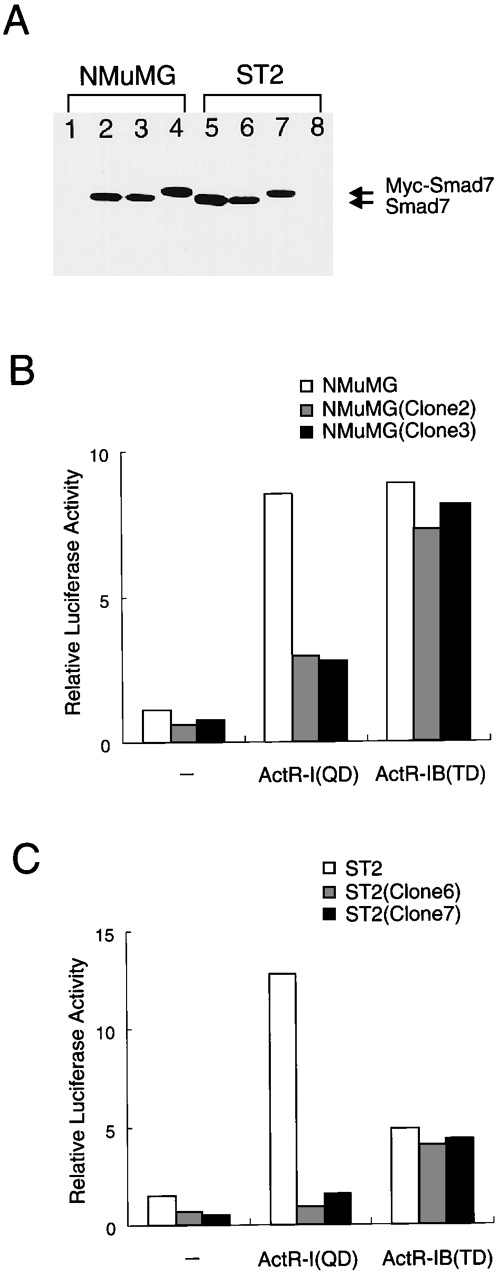
To investigate the effects described above under more physiologic conditions, we used single retrovector-infected cells to prepare cells that stably expressed moderate levels of Smad7. These subcloned and parent cells produced detectable levels and undetectable levels, respectively, of Smad7 (Figure 4A). We therefore tested the reporter response to coexpression with activated derivatives of type I receptors, ActR-I (QD) and ActR-IB (TD), which were known to have acquired constitutive signaling activity in the absence of ligand.47 In both types of cells, ActR-IB (TD) enhanced p3TP-Lux activity to an equal extent (8-fold; Figure 4B, right bars), indicating that stable expression of Smad7 barely affected Act-RIB signaling. In the parent NMuMG cells, ActR-I (QD) activated the reporter to about the same extent as did ActR-IB (TD), whereas in the cells producing Smad7 (clone 2 and clone 3), ActR-I (QD) caused only limited (2.5-fold) stimulation of reporter activity (Figure 4B). Equivalent results were obtained with ST2-derived clonal cells (Figure 4C). Thus, Smad7 preferentially blocked ActR-I signaling but had minimal effects on ActR-IB signaling. These results indicate that a moderate level of expression of Smad7 may cause selective mediation of intracellular activin signals.
Different signaling activities of 2 type I activin receptors in cells stably expressing Smad7.
(A) Immunoblot showing detectable levels of Smad7 (or Myc-Smad7) in stably infected cells and undetectable levels in parent cells. Clonal cells stably expressing Smad7 were established by retrovector infection and selection. A fixed amount (20 μg each) of total protein was analyzed with gel electrophoresis and subsequent immunoblotting using specific antibody against Smad7. Lane 1 shows results with parent NMuMG cells; lanes 2 and 3, results with clonal progeny cells infected with Smad7 virus and designated NMuMG (clone 2) and NMuMG (clone 3); lane 4, results with NMuMG cells infected with Myc-Smad7 virus and designated NMuMG (clone 4); lanes 5 and 6, results with ST2 progeny cells infected with Smad7 and designated ST2 (clone 5) and ST2 (clone 6); lane 7, results with ST2 progeny cells infected with Myc-Smad7 and designated ST2 (clone 7); and lane 8, results with parent ST2 cells. (B and C) In the cells stably producing Smad7, ActR-I (QD) induced only limited stimulation of the reporter, whereas ActR-IB (TD) enhanced p3TP-Lux activity to the same extent as in the parent cells. ST2 cells, NMuMG cells, and their derivatives were transfected with p3TP-Lux (300 ng), ActR-II vector, and either ActR-I (QD) or ActR-IB (TD) vector (50 ng each) and then cultured without activin treatment. Results are expressed as the increase in induction (x-fold) compared with the value in the vector control in parent cells. These data suggest that in cells expressing an increased amount of Smad7, ActR-IB signaling maintains its activity, whereas ActR-I signaling is prevented from causing the transcriptional response.
Different signaling activities of 2 type I activin receptors in cells stably expressing Smad7.
(A) Immunoblot showing detectable levels of Smad7 (or Myc-Smad7) in stably infected cells and undetectable levels in parent cells. Clonal cells stably expressing Smad7 were established by retrovector infection and selection. A fixed amount (20 μg each) of total protein was analyzed with gel electrophoresis and subsequent immunoblotting using specific antibody against Smad7. Lane 1 shows results with parent NMuMG cells; lanes 2 and 3, results with clonal progeny cells infected with Smad7 virus and designated NMuMG (clone 2) and NMuMG (clone 3); lane 4, results with NMuMG cells infected with Myc-Smad7 virus and designated NMuMG (clone 4); lanes 5 and 6, results with ST2 progeny cells infected with Smad7 and designated ST2 (clone 5) and ST2 (clone 6); lane 7, results with ST2 progeny cells infected with Myc-Smad7 and designated ST2 (clone 7); and lane 8, results with parent ST2 cells. (B and C) In the cells stably producing Smad7, ActR-I (QD) induced only limited stimulation of the reporter, whereas ActR-IB (TD) enhanced p3TP-Lux activity to the same extent as in the parent cells. ST2 cells, NMuMG cells, and their derivatives were transfected with p3TP-Lux (300 ng), ActR-II vector, and either ActR-I (QD) or ActR-IB (TD) vector (50 ng each) and then cultured without activin treatment. Results are expressed as the increase in induction (x-fold) compared with the value in the vector control in parent cells. These data suggest that in cells expressing an increased amount of Smad7, ActR-IB signaling maintains its activity, whereas ActR-I signaling is prevented from causing the transcriptional response.
Smad7 blocks erythroid differentiation induced by activated type I activin receptors with different efficiencies
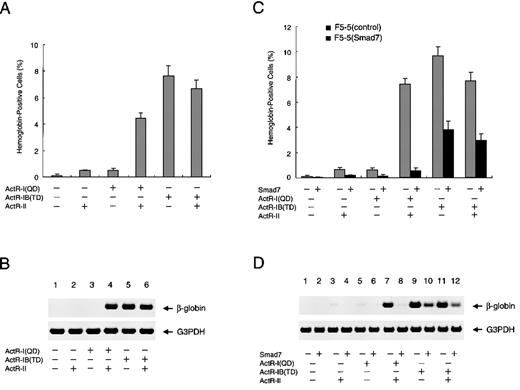
Finally, we examined whether erythroid differentiation of F5-5 cells could be mediated through the signaling of a particular activin receptor or receptors. RT-PCR analyses showed that these cells expressed detectable but limited levels of endogenous ActR-I, ActR-IB, and ActR-II (data not shown). The cultures were infected with neomycin-selectable retroviral constructs that expressed the activated mutants of activin type I receptors, either alone or in combination with the type II receptor. In singly infected F5-5 cells in the absence of activin, either ActR-II or ActR-I (QD) induced only limited elevation of hemoglobin synthesis, whereas transduced ActR-IB (TD) caused a significant increase in the proportion of hemoglobin-containing cells (up to about 8%) (Figure5A). Double infection with the combination of ActR-I (QD) and ActR-II, however, increased the proportion of hemoglobin-positive cells to a level comparable to that induced by ActR-IB (TD) and ActR-II. These results revealed that in concert with ActR-II, either one of the type I receptors can signal F5-5 cells to undergo erythroid differentiation, albeit with relatively lower efficiency than activin treatment. Analyses of β-globin mRNA in the infected cells verified these observations (Figure 5B). These findings indicate that in the presence of ActR-II, ActR-I and ActR-IB can induce the same final consequences in F5-5 cells. The results also implicate both the type I receptors and the type II receptor in the erythroid differentiation of F5-5 cells in response to activin.
Effect of Smad7 on erythroid differentiation induced by activated forms of type I activin receptors.
(A) Promotion of erythroid differentiation of F5-5 cells by expression of constitutively activated ActR-I (QD) or ActR-IB (TD). Cells were infected with various combinations of vector viruses expressing ActR-I (QD), ActR-IB (TD), and ActR-II. Infected cells were supplemented with medium to achieve the initial density of 3 × 104cells/mL and incubated for a week. At the end of the incubation period, a portion of the culture was examined by using benzidine staining for hemoglobin and the rest were collected for RNA extraction. The results shown are the percentages of total cells that contained hemoglobin. Values are the mean and SD from triplicate assays. (B) Total RNA harvested from the above cells was analyzed with RT-PCR for the presence of β-globin mRNA as a marker of erythroid differentiation. G3PDH served as a loading control. (C and D) Smad7 efficiently inhibits erythroid differentiation induced by ActR-I (QD) but only inefficiently inhibits that induced by ActR-IB (TD). F5-5 cells were infected with puromycin-selectable virus expressing Smad7 or with control virus. Stably transduced cells, F5-5 (Smad7) cells and F5-5 (control) cells, were selected by incubation in the presence of 1 μg/mL puromycin for 1 week. The puromycin-resistant cells were infected with combinations of ActR-I (QD), ActR-IB (TD), and ActR-II viruses, as indicated. After another week of culture, a portion of the culture was used for benzidine staining and the rest were used for RT-PCR analysis of β-globin gene expression.
Effect of Smad7 on erythroid differentiation induced by activated forms of type I activin receptors.
(A) Promotion of erythroid differentiation of F5-5 cells by expression of constitutively activated ActR-I (QD) or ActR-IB (TD). Cells were infected with various combinations of vector viruses expressing ActR-I (QD), ActR-IB (TD), and ActR-II. Infected cells were supplemented with medium to achieve the initial density of 3 × 104cells/mL and incubated for a week. At the end of the incubation period, a portion of the culture was examined by using benzidine staining for hemoglobin and the rest were collected for RNA extraction. The results shown are the percentages of total cells that contained hemoglobin. Values are the mean and SD from triplicate assays. (B) Total RNA harvested from the above cells was analyzed with RT-PCR for the presence of β-globin mRNA as a marker of erythroid differentiation. G3PDH served as a loading control. (C and D) Smad7 efficiently inhibits erythroid differentiation induced by ActR-I (QD) but only inefficiently inhibits that induced by ActR-IB (TD). F5-5 cells were infected with puromycin-selectable virus expressing Smad7 or with control virus. Stably transduced cells, F5-5 (Smad7) cells and F5-5 (control) cells, were selected by incubation in the presence of 1 μg/mL puromycin for 1 week. The puromycin-resistant cells were infected with combinations of ActR-I (QD), ActR-IB (TD), and ActR-II viruses, as indicated. After another week of culture, a portion of the culture was used for benzidine staining and the rest were used for RT-PCR analysis of β-globin gene expression.
To investigate the effects of Smad7 on the erythroid differentiation induced by stable expression of the constitutively active receptors, we prepared a Smad7 construct using a puromycin-selectable retroviral vector for another series of infection and selection. In the experiments that followed, cells were first selected for transduction of Smad7 (or for control infection) and the surviving cell population was used for infection with the activin receptors. As shown in Figures5C and 5D, prior expression of Smad7 severely suppressed the hemoglobin synthesis induced by ActR-I (QD) with ActR-II, a finding consistent with our observations about the effect of Smad7 on the activin-stimulated differentiation of F5-5 cells. On the other hand, Smad7 only inefficiently attenuated the increase in hemoglobin-containing cells induced by ActR-IB (TD) with or without Act-RII, indicating only partial inhibition of the differentiation. These analyses clearly indicate that Smad7 prevents ActR-I signaling more potently than it does ActR-IB signaling, not only in the early transcriptional response but also in the cell differentiation induced by activin.
Discussion
The growth factors of the TGF-β superfamily influence, either physiologically or pathologically, various cellular regulatory pathways, including growth, proliferation, and differentiation, in a cell-type–specific manner. Specific expression of their receptors and signaling mediators provides particular cells with unique responsiveness to certain ligands.63,64 Conversely, a selective inhibitor inside the cells would be expected to control the ligand effects by attenuating a specific signaling pathway. Smad family proteins are essential molecules for signaling of TGF-βs downstream of their receptors,5,6 and Smad7 has been characterized as one having an inhibitory function.16-20,65 66Our results in this study indicate that Smad7 expression is specifically absent in hematopoietic malignant cells that respond to activin by undergoing erythroid differentiation, whereas various other cells responsive to activin usually express Smad7. Also, ectopic production of Smad7 in the erythroid leukemia cells causes them to be resistant to activin-induced promotion of hemoglobin synthesis. By using the phenotypic expression of these cells as a sensitive marker in a functional assay system, we have provided evidence that Smad7 efficiently blocks ActR-I signaling but does not effectively interfere with ActR-IB signaling. Similarly, this receptor-dependent inhibition by Smad7 was observed for the early transcriptional response to activin in transduced cells. These results strongly suggest that Smad7 acts as a selective blocker of different signaling pathways of activin.
The pleiotropic factor activin is endogenously present in bone marrow, where, in adult mammals, hematopoietic cells undergo continuous propagation followed by serial maturation. In primary cultures, activin can alter the growth and differentiation lineage of colonies derived from various multipotential precursor cells.67-69 In particular, erythroid progenitor cells, which have receptors for activin, produce an increased proportion of colonies containing hemoglobin in response to activin.27,28,70,71 Our results indicate that among cell lines established from hematic-tissue origins, Smad7 expression is very low, specifically in cells that can differentiate along the erythroid lineage. Mouse F5-5 cells were cloned from leukemic cells in a Friend virus-induced splenofocus,72 whereas human K562 cells were derived from a patient with chronic myelogenous leukemia with theBCR/ABL oncogene of the Philadelphia chromosome translocation,73 and HEL erythroleukemia cells were established from human peripheral blood.74 These cells of widely different origins shared the characteristics of responsiveness to activin and a lack of Smad7 expression. The results suggest that the common lack of Smad7 is closely related to the potential of these cells to undergo erythroid differentiation and was not caused by nonspecific inactivation of gene expression during the course of tissue culture.
Indeed, a variety of other cell lines representing different hematopoietic lineages and stages of maturation, including myeloid, lymphoid, and megakaryocyte lineage cells, were found to express Smad7 with varied abundance. Activin was shown to antagonistically affect monocyte differentiation of M1 myeloblast cells and to induce growth arrest of several plasmacytomas, including Sp2 cells,54thereby indicating that these myeloid and lymphoid lineage cells, which express substantial amounts of Smad7, respond to activin. These observations, together with our results, indicate that Smad7 is not a simple inhibitor that blocks all the activin signals, a finding consistent with its selective interference with different pathways of activin signaling. Although it was shown that levels of Smad7 mRNA are transiently increased by laminar-fluid shear stress in vascular endothelial cells,75 the transcriptional control of Smad7 has remained unclear. Clarification of the pattern and regulatory mechanism of Smad7 expression during hematocyte differentiation in embryos and adults will supply valuable information about hematopoietic regulation.
Comparisons of epithelial tumor cells have shown a loss of responsiveness to the regulatory actions of TGF-β as carcinogenesis progresses.76 In such tumor cells, Smad genes are frequently inactivated; Smad4 gene (DPC4) mutation is detected in many pancreatic tumors,12 whereas the Smad2 gene is frequently mutated in colorectal tumors.77 Loss of these Smads appears to play a role in the insensitivity of various types of carcinoma cells to proliferation arrest by TGF-β.78 79 In contrast, our results indicated that inappropriate or unregulated increase of Smad7 expression can lead activin-responsive leukemic cells to escape from activin-induced differentiation, which suggests that the Smad7 level may be related to the degree of malignancy of erythroid leukemia cells.
Two serine kinase receptors, ActR-I and ActR-IB, have been identified as activin type I receptors because of their ability to bind to activin in concert with ActR-II or ActR-IIB.31-36 In K562 cells, increasing the levels of ActR-IB and ActR-II by use of an inducible promoter enhanced the effects of activin on erythroid differentiation, demonstrating that ActR-IB and ActR-II are physiologically important receptors for activin.30 Our findings using activated variants of type I receptors in studies in F5-5 cells provide further insight into signaling by these receptors. Continuous signaling from ActR-IB (TD) alone induced hemoglobin production in cells in the absence of activin. Furthermore, ActR-IB (TD) overwhelmed the blockage by Smad7 of erythroid differentiation. On the other hand, expression of ActR-I (QD) alone was not sufficient for the induction of hemoglobin synthesis in F5-5 cells. These results may indicate a qualitative difference between the signaling pathways of the 2 type I receptors. It was reported that ectopic expression of either of the Xenopusactivin receptors corresponding to ActR-I (QD) and ActR-IB (TD) in frog embryos commonly induced transcripts of Xbra, a general mesodermal marker,80 in ectodermal grafts, although only ActR-IB (TD) was capable of inducing a secondary axis.41 These results indicate that activation of each of the 2 type I receptors can cause partly common and partly distinct responses to activin.81-83 Our findings indicate that, in concert with ActR-II, constitutive activation of ActR-I or ActR-IB ultimately produces the same differentiated phenotype in erythroid leukemic cells, with similar efficiencies. However, the identical results should be achieved uniquely in cells lacking Smad7 because of its effective and selective inhibition of ActR-I signaling.
Despite the high degree of competence of F5-5 cells to differentiate along the erythroid lineage in response to activin (up to 50%), the differentiation that was induced by activation of either the ActR-I or the ActR-IB pathway was relatively inefficient (up to 10%). Previous studies indicated that activin binds to both ActR-I and ActR-IB expressed endogenously in K562 cells.84 Thus, the highly efficient response to activin of the erythroid precursor cells might require simultaneous or sequential activation of signaling by both receptors. The efficient suppression of erythroid differentiation by Smad7, which preferentially inhibits the ActR-I pathway, may be explained by disruption of a synergy of the 2 pathways.
Expression of Smad6 at low levels was shown to specifically inhibit BMP signaling by acting as a Smad4 decoy, yielding an inactive Smad1-Smad6 complex in competition with Smad4.15 Although homo-oligomer formation of Smad7 was detected when the protein was singly produced, we obtained no evidence of a direct interaction of Smad7 with Smad1, Smad2, Smad3, or Smad4 in the presence or absence of activin. Smad2 and Smad3 were shown to be phosphorylated and stimulated downstream of ActR-IB activated by activin, as well as of TGFβR-I activated by TGF-β, and to mediate signals from these ligands.85-89 Thus, the differentiation induced by ActR-IB (TD) in the presence of Smad7 may be mediated by either Smad2 or Smad3, which have been shown to play distinct roles in activin effects.90 On the other hand, the signaling mediator of ActR-I has not been clearly identified.91 92 It is possible that ActR-I signals through a not-yet-identified pathway, which should be sensitive to the inhibitory effect of Smad7.
The present results indicate the existence of a novel activity of Smad7 that inhibits erythroid differentiation of leukemic cells by preventing intracellular signaling of activin. The physiologic importance of this finding is that the level of expression of Smad7 can be a critical determinant of the cell's response and function through selective blockage of the activin signal pathways. Aside from erythroid progenitors, a wide variety of hematopoietic and supporting cells respond to activin with various actions, including secretion of cytokines, retardation of the cell cycle, and progression of differentiation.28,54 67-69 Even in these processes, the distinct responses of cells might be at least partly due to different levels of Smad7, which would affect signal transduction from activin and consequently the cell fate.
Acknowledgments
We thank Yuzuru Eto for recombinant human activin A (EDF); Joan Massagué for p3TP-Lux; George Q. Daley for pGD′; Motoaki Ohtsubo for pMoψ−; Takahiro Kunisada, Masao Matsuoka, Shin-ichi Nishikawa, and Makoto Tsuneoka for cells; Noboru Nakajima for helpful discussions; Elizabeth Nakajima for critical reading of the manuscript; and Yoshiro Shimura for encouragement.
Reprints:Kenji Okazaki, Department of Molecular Biology, Biomolecular Engineering Research Institute, 6-2-3 Furuedai, Suita, Osaka 565-0874, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 1. Specific absence of Smad7 mRNA in particular hematopoietic cells. / (A) Northern blot analysis of mouse Smad7 mRNA. Each lane contained 2 μg of poly(A)+ RNA from the indicated mouse organs. A Smad7 probe (625-base pair [bp] complementary DNA [cDNA] fragment) revealed one major transcript in various tissues. Positions of RNA size markers (kilobase [kb]) are shown on the left. (B) RT-PCR analysis of Smad7 mRNA in different types of cells. The examined cell lines were mink Mv1Lu (lung epithelial), mouse Swiss-3T3 (embryonic fibroblast), F5-5 (erythroid leukemia), M1 (myeloblast), WEHI-3 (myelomonocyte), C7 (macrophage), P815 (mastocytoma), CTLL-2 (T lymphocyte), Sp2 (plasmacytoma), human HEL (erythroleukemia), K562 (chronic myelogenous leukemia), HL-60 (promyelocytic leukemia), and MEG-01 (megakaryoblast). cDNAs to 1 μg of RNA isolated from these cells were synthesized by reverse transcription with random primers. Equal aliquots from each reaction were subjected to PCR to amplify a Smad7 and a control G3PDH cDNA fragment. The PCR primer pairs and conditions were designed for amplification of a 207-bp Smad7 sequence in both rodent and human cells. The data indicate that F5-5, HEL, and K562 cells specifically lack Smad7 expression.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/11/10.1182_blood.v95.11.3371/6/m_bloo01137001w.jpeg?Expires=1767757653&Signature=uLNYrGsNPJJwfdclyC7lAK-IN7RNVFxlU~DCM~rW1E7RAuDJ5wsk-rBGo8lIzVRzXhsRpWt1RliRdbaxSqORtNSw17IlxVWLJy7k9L~zX7p6MBC0aESYz2xMV6n2aT3wvrM2koWbDrjhEH28f3WTNvvadTu8SNpS2~ESouFt482rDLRu9GUlPPJj0s2vap9pl2P9BubnP4ZgwMut0CsPCHbRiSToqDRKxm287dW-4SZ0SMDa8mOwQrH47q6Jf6fTbL1DMSYbHT4cp1a6OXJ0e4FJJtOpKrMJS9OFPRNPGjelyiMXZHYIYR5oCjrt01ry~tDWdQ5x0gKveyhdFsECWQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal