Abstract
Ferrochelatase, the last enzyme in the heme pathway, chelates protoporphyrin IX and iron to form heme and is mutated in protoporphyria. The ferrochelatase gene is expressed in all tissues at low levels to provide heme for essential heme-containing proteins and is up-regulated during erythropoiesis for the synthesis of hemoglobin. The human ferrochelatase promoter contains 2 Sp1 cis-elements and GATA and NF–E2 sites, all of which bind their cognatetrans-acting factors in vitro. To investigate the role of these elements during erythropoiesis, we introduced expression of the green fluorescent protein (EGFP) transgenes driven by various ferrochelatase promoter fragments into a single locus in mouse embryonic stem cells. EGFP expression was monitored during hematopoietic differentiation in vitro using flow cytometry. We show that a promoter fragment containing the Sp1 sites, the NF–E2 and GATA elements, was sufficient to confer developmental-specific expression of the EGFP transgene, with an expression profile identical to that of the endogenous gene. In this system the −0.275 kb NF–E2 cis-element is required for erythroid-enhanced expression, the GATA cis-element functions as a stage-specific repressor and enhancer, and elements located between −0.375kb and −1.1kb are necessary for optimal levels of expression. Ferrochelatase mRNA increased before the primitive erythroid-cell stage without a concomitant increase in ferrochelatase protein, suggesting the presence of a translational control mechanism. Because of the sensitivity of this system, we were able to assess the effect of an A-to-G polymorphism identified in the promoters of patients with protoporphyria. There was no effect of the G haplotype on transcriptional activity of the −1.1 kb transgene.
Heme is required at low levels in all cells for respiratory cytochromes and other essential heme-containing proteins, and it is required at high levels during erythropoiesis to provide heme for hemoglobin. Ferrochelatase, the last enzyme in the heme biosynthetic pathway, chelates ferrous iron and protoporphyrin IX to form heme. The human ferrochelatase gene is nuclear transcribed and, after translation, is transported to its active site in the inner mitochondrial membrane.1-4 The ferrochelatase protein has a molecular weight of 40 to 42 kd in sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis. Radiation inactivation studies suggest that the ferrochelatase protein acts in situ as an 82- to 84-kd dimer.5 Recently, crystallization of human ferrochelatase confirmed a homodimer structure for the enzyme.6
Investigating ferrochelatase gene regulation has primarily involved assessing enzyme activities in human or mouse erythroleukemic cell lines, identifying ferrochelatase gene transcripts in erythroid differentiated embryonic stem cells using reverse transcription–polymerase chain reaction,7 and analyzingcis-element function using transient transfection of reporter genes into nonerythroid and erythroleukemic cells lines.8-15 The ferrochelatase promoter contains 2 Sp1cis-elements and upstream GATA and NF–E2cis-elements,13,16 and each binds its cognatetrans-acting factors in vitro.13 Transient transfection assays in nonerythroid and erythroid cells demonstrated that the minimal promoter (−0.150 kb), containing Sp1 binding sites, is sufficient to confer erythroid-preferential expression, whereas the −0.375 kb GATA and NF–E2 elements in the extended promoter failed to confer any additional increase in erythroid-preferential expression.13
We hypothesized that appropriate use of these erythroidcis-elements may require organized chromatin structure. Two transgenic mouse lines, one containing the minimal promoter (−0.150 kb) and another containing the extended promoter (−4.0 kb), were generated. Unlike the transient transfection studies, the transgenic mice studies demonstrated thatcis-elements 5′ of the −0.150 kb Sp1 sites are required for maximal erythroid-specific expression of the ferrochelatase gene and furthermore implied that thesecis-elements require a structured chromatin environment for appropriate tissue-specific expression.17 The −4.0 kb ferrochelatase promoter fragment contains the −0.375 kb GATA and NF–E2 cis-elements, but it also contains 3.8 kb of a sequence that may contain additional functional cis-elements. Because the studies in transgenic mice used only the minimal promoter (−0.150 kb) or the extended promoter (−4.0 kb), we were unable to precisely identify the cis-elements required for erythroid-preferential up-regulation of the ferrochelatase gene.
Protoporphyria is associated with a partial deficiency in ferrochelatase18,19 and is typically transmitted as an autosomal dominant disease with variable penetrance.20,21Patients with protoporphyria consistently exhibit ferrochelatase activities that are 15% to 30% of normal.21,22 They also exhibit severe photosensitivity and sometimes hepatobiliary disease, which may necessitate liver transplantation. Ferrochelatase mutations identified in patients with protoporphyria are heterogeneous23 and include missense and nonsense mutations24-28 and, most commonly, mutations in splice donor or acceptor sites that lead to aberrant splicing and exon skipping.29-35
It has been proposed that the less than 50% ferrochelatase activities described for protoporphyria arise from the combination of a null allele and a “low-expressing” allele.36 Probable promoter defects that could result in a low-expressing allele include mutations in the ferrochelatase promoter and aberrant methylation of the CpG island in the promoter. In this regard, an A-to-G polymorphism has been identified in the ferrochelatase promoter (−251 bp from the start codon) that segregates with the protoporphyric phenotype.36 37 The effect of the G haplotype on ferrochelatase gene expression is unknown.
Based on our current understanding of transcription factor binding to the NF–E2 and GATA sites at −0.375 kb of the ferrochelatase promoter, we predicted that use of the NF–E2 and GATAcis-elements would be required during erythropoietic up-regulation of the ferrochelatase gene, and because the A-to-G polymorphism lies in a transcriptionally active region between the −0.150 kb Sp1 sites and the −0.375 kb GATA site, we thought the G haplotype might play a role in decreasing expression of the ferrochelatase gene.
To test these hypotheses, we used a gene-targeting method designed to analyze single-copy transgenes integrated into a single locus, 5′ of the HPRT gene.38 We modified an HPRT gene-targeting vector to contain ferrochelatase promoter fragments driving expression of the green fluorescent protein (EGFP) and assessed transgene expression in live cells using fluorescence-activated flow cytometry. This method has advantages over traditional stable cell lines containing luciferase or β-galactosidase reporter genes in that EGFP transgene expression can be analyzed in individual, living cell populations devoid of multiple transgene copy numbers and integration sites.
Materials and methods
Reporter plasmid and targeting vector construction
The expression of green fluorescent protein (EGFP) reporter gene vector, pEGFP, (Clontech, Palo Alto, CA) was modified by adding a poly-cloning linker 5′ of the EGFP cDNA that containedSalI and SacI restriction endonuclease sites. The EGFP vector contains an enhanced version of the Aequorea victoriaprotein39 with the poly A signal from SV40 and the neomycin resistance gene driven by the SV40 promoter. Various regions of the human ferrochelatase promoter, originally cloned into a luciferase reporter gene construct,13 17 were digested withSacI and SalI restriction endonucleases, and theSalI/SacI fragments were subcloned into the modified pEGFP vector.
NF–E2 and GATA mutations were introduced into the ferrochelatase promoter using mutant oligonucleotides; the NF–E2 mutations are as described.13 Mutations abolishing the GATA consensus site were introduced into an oligonucleotide (mutations are underlined: TCCTGGAAAGGAAA). For the GATA/NF–E2 double mutant, the same GATA mutations combined with the NF–E2 mutations were introduced into an oligonucleotide (NF–E2 mutations are underlined: TTTGCATCGTCA). Each mutant oligo was annealed to its complementary sequence. Overhangs of the double-stranded oligos yielded the restriction endonuclease sites HindIII and BamHI. All mutant double-stranded oligonucleotides were ligated into theHindIII and BamHI sites of the −0.375 kb EGFP.
To generate a reporter gene construct that contained the A-to-G polymorphism, genomic DNA was isolated from a patient with protoporphyria who had only 1 allele of the ferrochelatase gene40 with the G haplotype. A 1.1-kb fragment of the ferrochelatase promoter was amplified using polymerase chain reaction (PCR). The presence of the G haplotype (−251) in this patient was demonstrated by di-deoxy sequencing of the 1.1-kb PCR fragment. The PCR product was digested with XbaI and SacII and then cloned into the XbaI and SacII site of pBluescript SK+. A clone that contained the G haplotype was digested with SalI and SacI and cloned into the SalI and SacI pEGFP reporter gene construct. The resultant pEGFP construct was digested with EcoRI and SacI, and the promoter fragment was ligated into the EcoRI and SacI sites of the HPRT/EGFP construct.
The HPRT targeting vector pMP8SKB38 (kindly provided by Drs S. Bronson and O. Smithies) contains a polycloning site flanked 5′ by DNA from the HPRT locus located upstream of the HPRT promoter and flanked 3′ by the promoter and exons 1, 2, and 3 of the HPRT gene. To facilitate cloning, we modified the cloning site by adding additional restriction endonuclease sites and the cDNA for EGFP by standard cloning methods. Additionally, the NotI andSacI sites in the HPRT sequence were eliminated by digestion with the appropriate restriction endonuclease, filling in of the 5′ overhang with Klenow, and performance of a standard ligation reaction. The mutant and wild-type human ferrochelatase promoter regions from the pEGFP vectors (described earlier) were isolated usingEcoRI and SacI restriction endonuclease digests and subcloned into the EcoRI/SacI site of the HPRT targeting vector.
Cell culture/microscopy
The mouse embryonic stem (ES) cell line BK4, a subclone of E14TG2a, was maintained in an undifferentiated state on embryonic fibroblast feeder cells in Dulbecco's modified essential medium (DMEM) high glucose, supplemented with 15% fetal bovine serum (ES qualified; Gibco/BRL, Gaithersburg, MD), 0.1 mmol/L β-mercaptoethanol, 2 mmol/L l-glutamine, 100 U/mL penicillin, and 1 mg/mL streptomycin. ES cells were grown at 37°C in a humidified atmosphere containing 5% CO2. Before primary differentiation, the ES cells were weaned from the feeder cells by passaging 2 to 3 times in DMEM-H, 15% fetal calf serum (FCS; ES qualified; Gibco/BRL), 2 mmol/L glutamine, 100 U/mL penicillin, 1 mg/mL streptomycin, 2U/mL LIF, and 125 μmol/L β-mercaptoethanol on tissue culture-treated plates coated with 0.1% gelatin in phosphate-buffered saline (PBS). For primary differentiation,41 42 approximately 1.5 × 104 cells were plated in 6-cm Petri dishes containing differentiation media (Iscove's–MDM with 10% FCS (Hyclone, Logan, UT), 4.5 × 10−4 mol/L monothioglycerol (MTG), 25 μg/mL ascorbic acid, 200 μg/mL iron-saturated transferrin, 2 mmol/L glutamine). Cells were grown in a humidified atmosphere containing 5% CO2 at 37°C for 3 to 4 days. After 3 to 4 days in primary differentiation conditions, embryoid bodies were washed once in PBS and disrupted by trypsinization for 1 minute (using 0.5× concentration; 0.025% trypsin, 0.26 mmol/L EDTA). Approximately 1.0 to 3.0 × 105 cells were plated in a secondary methylcellulose culture with IMDM containing the above concentrations of glutamine, penicillin/streptomycin, ascorbic acid and MTG, 10% plasma-derived serum (Anatech, Tyler, TX), 5% protein-free hybridoma medium (PFHM II; Gibco/BRL), 1% methylcellulose, 2 U/mL erythropoietin (R&D Systems, Minneapolis, MN), and 100 ng/mL stem cell factor (R&D Systems). Cells were grown in a humidified atmosphere containing 5% CO2 at 37°C for an additional 4 to 8 days.
To determine the half-life of EGFP, 5 to 8 × 105pluripotent cells (from 2 different EGFP-expressing ES clones) were plated into 6-well plates. Twenty-four hours later, 100 μg/mL cyclohexamide43 was added at 2-hour intervals for 6 hours. EGFP levels were then quantified by flow cytometry as later described.
An Olympus IX70 (Olympus, Melville, NY) was used for bright-field and fluorescence microscopy. EGFP was detected using a filter that allowed excitation at 488 nm, and porphyrins were detected using a filter that allowed excitation at 385 nm. Images were captured using 200 ASA Kodak Elite slide film (Kodak, Rochester, NY). The film image was then digitized using a transparency scanner, and image overlays were generated using Adobe Photoshop software (Adobe Systems, San Jose, CA). For bright-field, porphyrin, and EGFP overlays, levels of red fluorescence above background levels were digitally selected and overlaid onto the bright-field image.
Stable integration of reporter gene constructs
Undifferentiated ES cells were isolated by trypsinization, and approximately 2 × 107 cells were electroporated with 15 μg of an EGFP/HPRT targeting vector linearized withPvuI. The cells were suspended in 0.5 mL PBS with the DNA and pulsed in a 4-mm gap electroporation cuvette with 350 V, 50 μF for 1 second (Bio-Rad, Hercules, CA). Twenty-four hours later, the growth medium was replaced with HAT (final concentration of 0.1 mmol/L hypoxanthine, 0.4 μmol/L aminopterin, 16 μmol/L thymidine) selection medium. HAT-resistant clones were isolated and expanded on day 10 after initiation of selection. Individual clonal populations were used in all experiments unless otherwise indicated.
Flow cytometry
A Becton Dickinson FACScan (Franklin Lakes, NJ) was used to analyze ES cells for fluorescence intensities. Pluripotent ES cells, grown in the absence of feeders, were trypsinized, and washed once in growth medium. Then 1 × 106 cells were suspended in 1.0 mL PBS for analysis. For differentiated ES cells, embryoid bodies (EB) were isolated by centrifugation and washed once with PBS. After that, EBs were disrupted by brief trypsinization (using 0.25× trypsin solution for 1 minute), washed once in growth medium, and finally suspended in 1.0 mL PBS for analysis. Each sample was collected at 24-hour intervals. All cells were analyzed for EGFP expression within 30 minutes of trypsinization.
Forward and side scatter plots were used to gate on the live population of cells. The fluorescence of 50,000 to 80,000 cells was measured using the FL1 (or fluorescein isothiocyanate) channel of the FACScan. EGFP intensities were calculated from maximal fluorescence peak heights using Cyclops software (Fort Collins, CO).
Northern blot and immunoblot analyses
Total RNA was isolated at 24-hour intervals during ES cell differentiation using Trizol (Gibco/BRL, Gaithersburg, MD), and 15 μg was separated on a 1.5% agarose/formaldehyde gel, transferred to nylon membrane by capillary action in 20× SSC (3.0 mol/L NaCl, 0.3 mol/L sodium citrate), UV cross-linked in a Stratalinker (Stratagene, San Diego, CA) on automatic setting, and prehybridized in Rapid-hyb buffer (Amersham, Arlington Heights, IL). A random primed32P-labeled mouse ferrochelatase, human EKLF, mouse ALAS-E, or mouse β-globin cDNA probe was applied to the membrane at 1 × 106 cpm/mL and incubated at 60°C for 4 hours. The membrane was washed once in 2× SSC/0.1% SDS at 21°C and twice at 55°C for 15 minutes. Then the wash buffer was adjusted to 0.1× SSC/0.1% SDS and applied to membrane twice at 60° for 15 minutes. The membrane was then exposed to X-OMAT AR film (Kodak) with intensifying screens for 14 hours (ferrochelatase) or analyzed on a Storm 840 Phosphor Imager (Molecular Dynamics, Sunnyvale, CA) for 8 hours (EKLF) or for 24 hours (ALAS-E and β-globin).
To detect ferrochelatase protein, whole protein extracts were isolated at 24-hour intervals in RIPA buffer (0.15 mol/L NaCl, 50 mmol/L Tris-Cl, pH 7.2, 1% deoxycholic acid, 1% Triton-X 100, 0.1% SDS) containing 10 μg/mL phenylmethylsulfonyl fluoride, and 15 μg whole cell lysate was separated on a 12% SDS–polyacrylamide gel and electrophoretically transferred to nitrocellulose. Equal loading of protein was assessed by Ponceau S staining. The membrane was then blocked with BLOTTO (5% wt/vol nonfat dried milk in water) for 15 minutes at room temperature. A 1:500 dilution (in BLOTTO) of anti-recombinant human ferrochelatase polyclonal antibody44was applied to the blot and allowed to incubate at room temperature for 4 hours. The blot was washed once with BLOTTO and twice in Tris-buffered saline containing 0.1% TWEEN-20 (TBS–TWEEN). A goat anti-rabbit alkaline phosphatase-conjugated secondary antibody (Santa Cruz Biotechnologies, Santa Cruz, CA) was applied at a 1:1000 dilution in BLOTTO and allowed to incubate for 1 hour at room temperature. The membrane was washed once in TBS-TWEEN, once in BLOTTO, and twice in TBS-TWEEN for 10 minutes, each time at room temperature. Western Blue substrate (Promega, Madison, WI) was added, and the immunoblot was allowed to develop for approximately 20 minutes.
Results
Single-copy, single-site integration of the ferrochelatase promoter transgene
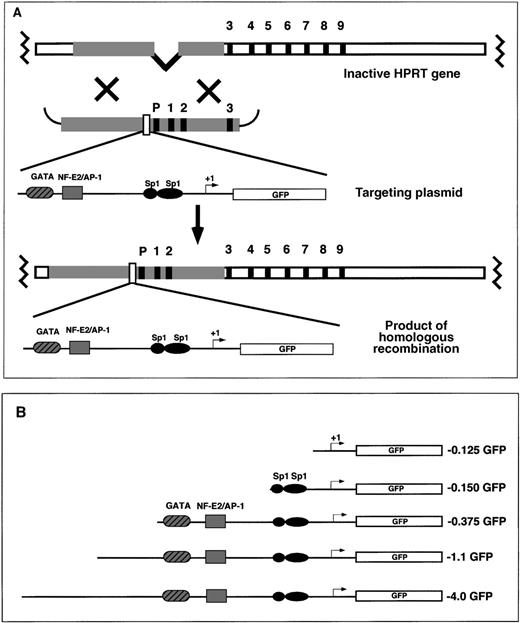
EGFP reporter gene-targeting constructs were introduced into the mouse HPRT locus by electroporation to the mouse ES cell line, BK4. This ES cell line is male-derived and is functionally deficient in the X-linked HPRT gene resulting from a deletion of the HPRT promoter and exons 1 and 2.45 On homologous recombination between the endogenous HPRT gene and the exogenous HPRT targeting vector, the HPRT mutation is corrected, and successfully targeted cells are able to grow in HAT selection media. The HAT-resistant clones contain 1 copy of the reporter gene located 5′ of the HPRT gene (Figure1A). Random integration of the targeting vector does not reconstitute HPRT expression and the ability to grow in HAT selective media.38 45 Various fragments of the human ferrochelatase promoter were cloned into the HPRT targeting vector for site-specific integration experiments (Figure 1B). The ferrochelatase promoter fragments driving reporter gene expression contained −0.125 kb sequence with no functional elements; −0.150 kb sequence containing 2 Sp1 elements; −0.375 kb sequence containing the 2 erythroid elements, GATA and NF–E2 (with an overlapping AP-1 site); or 2 extended promoter fragments containing −1.1 kb sequence or −4.0 kb sequence (Figure 1B).
Targeting the mouse HPRT locus with the human ferrochelatase promoter transgenes.
(A) To introduce ferrochelatase promoter genes 5′ of the HPRT locus, we designed HPRT targeting vectors that contained human ferrochelatase promoter fragments driving expression of the EGFP cDNA flanked 5′ and 3′, with HPRT sequences containing the HPRT promoter, and exons 1, 2, and 3. After electroporation, a homologous recombination event between the HPRT gene and the targeting vector reconstitutes the HPRT gene function, which allows for selection in HAT media, and introduces a single copy of the ferrochelatase promoter transgene into the genomic DNA. (B) EGFP reporter genes contain various fragments of the human ferrochelatase promoter.
Targeting the mouse HPRT locus with the human ferrochelatase promoter transgenes.
(A) To introduce ferrochelatase promoter genes 5′ of the HPRT locus, we designed HPRT targeting vectors that contained human ferrochelatase promoter fragments driving expression of the EGFP cDNA flanked 5′ and 3′, with HPRT sequences containing the HPRT promoter, and exons 1, 2, and 3. After electroporation, a homologous recombination event between the HPRT gene and the targeting vector reconstitutes the HPRT gene function, which allows for selection in HAT media, and introduces a single copy of the ferrochelatase promoter transgene into the genomic DNA. (B) EGFP reporter genes contain various fragments of the human ferrochelatase promoter.
Up-regulated expression of the −4.0 EGFP reporter gene in erythroid colonies
To determine whether expression of the extended promoter transgene (−4.0 kb) in the HPRT locus was responsive to erythroid cell lineage trans-activation, we differentiated pluripotent ES cells in methylcellulose cultures to favor primitive and definitive erythroid cell formation.41 46 After 4 to 5 days of secondary differentiation in methylcellulose, globinized erythroid colonies became visible and continued to proliferate and differentiate until day 8 (Figure 2A). Although differentiation conditions favored hematopoietic development, nonerythroid cells proliferated and surrounded the erythroid colonies from days 7 to 8 of methylcellulose differentiation (Figure2A).
Transgene expression in erythroid colonies.
ES cells containing the targeted −4.0 kb transgene were differentiated in primary culture for 5 days. Then, embryoid bodies (EB) were disrupted to single cells and plated in a secondary methylcellulose culture. Between 4 and 8 days of secondary culture, colonies of erythroid cells were visible as foci of red cells (globinized areas indicated with red arrows) with nonglobinized cells (black arrows). The erythroid colonies were surrounded by nonerythroid cells of unknown lineage. Original magnification, × 200. (A) Erythroid colonies were observed using either visible, (B) porphyrin-specific, or (C) EGFP-specific wavelengths. (Yellow areas indicate emission filter bleed-through of porphyrin fluorescence combined with green fluorescence) (D) To observe areas of fluorescence in the erythroid colony, the porphyrin fluorescence image was overlaid with the visible image, or (E) the EGFP fluorescence image was overlaid with the visible image. (F) To identify areas of both porphyrin and EGFP expression in the erythroid, the images from porphyrin fluorescence and EGFP fluorescence were overlaid.
Transgene expression in erythroid colonies.
ES cells containing the targeted −4.0 kb transgene were differentiated in primary culture for 5 days. Then, embryoid bodies (EB) were disrupted to single cells and plated in a secondary methylcellulose culture. Between 4 and 8 days of secondary culture, colonies of erythroid cells were visible as foci of red cells (globinized areas indicated with red arrows) with nonglobinized cells (black arrows). The erythroid colonies were surrounded by nonerythroid cells of unknown lineage. Original magnification, × 200. (A) Erythroid colonies were observed using either visible, (B) porphyrin-specific, or (C) EGFP-specific wavelengths. (Yellow areas indicate emission filter bleed-through of porphyrin fluorescence combined with green fluorescence) (D) To observe areas of fluorescence in the erythroid colony, the porphyrin fluorescence image was overlaid with the visible image, or (E) the EGFP fluorescence image was overlaid with the visible image. (F) To identify areas of both porphyrin and EGFP expression in the erythroid, the images from porphyrin fluorescence and EGFP fluorescence were overlaid.
To confirm early heme-pathway gene expression, erythroid colonies were observed using fluorescence microscopy. ∂-Aminolevulinic acid synthase–erythroid form (ALAS-E) is the first and rate-limiting enzyme in the heme biosynthetic pathway. Its product, ∂-ALA, is quickly converted to protoporphyrin. When erythroid colonies were observed using wavelengths that excite porphyrins, red fluorescence emanated exclusively from cells of the erythroid colony, indicating heme pathway gene up-regulation (Figure 2B).
To confirm that porphyrins were producing the red fluorescence in the erythroid colonies, we isolated approximately 50 red-fluorescing erythroid colonies on day 4 to day 5 of secondary differentiation, extracted the pigments, and identified the porphyrins using reverse-phase high-performance liquid chromatography (HPLC).47 48 In the erythroid colonies there were high levels of uroporphyrin, coproporphyrin, and protoporphyrin, as indicated by an HPLC retention time and spectrofluorometric signatures unique for each porphyrin (not shown). Levels of porphyrins in nonerythroid cell extracts were below the level of detection using this method.
To determine whether the ferrochelatase promoter EGFP-reporter gene was expressed in cells of the erythroid colony, we viewed cells containing the −4.0 kb promoter transgene with EGFP-specific wavelengths. High levels of green fluorescence were observed in cells of the erythroid colony, whereas no green fluorescence was found in nonerythroid cells surrounding the erythroid colony (Figure 2C), indicating that erythroid-preferential expression is controlled by elements located within 4.0 kb of the ferrochelatase promoter.
Interestingly, there was a different temporal expression for early heme pathway genes (indirectly monitored by porphyrin fluorescence) than for the −4.0 kb ferrochelatase promoter transgene. Using fluorescence microscopy, the fluorescence of individual erythroid colonies was followed over a 5-day period. High levels of EGFP fluorescence were observed from day 4 of primary differentiation through day 4 of secondary differentiation. Porphyrin fluorescence was only observed beginning on day 3 to 4 of secondary differentiation, which was followed by globinization on day 5 to 6 of secondary differentiation. Bright-field and fluorescence images were overlaid to demonstrate expression patterns of globins, early heme pathway genes, and the ferrochelatase promoter transgene. Globinized primitive erythroid cells (Figure 2A, red arrows) were observed in the erythroid colony as were nonglobinized erythroid precursor cells (Figure 2A, black arrows). To identify areas of early heme pathway gene expression in cells of the erythroid colony, the porphyrin fluorescence image (Figure 2B) was overlaid with the bright-field image (Figure 2D). To identify areas of −4.0 kb transgene expression, the EGFP fluorescence image (Figure 2C) was overlaid with the bright-field image (Figure 2E). To identify areas of early heme pathway expression and −4.0 kb EGFP expression, the porphyrin fluorescent image was overlaid with the EGFP fluorescent image (Figure 2F). Overlaid images demonstrated different temporal expression of globins, early heme pathway genes, and the ferrochelatase reporter gene (Figures 2D,2E, and 2F).
Endogenous ferrochelatase gene and transgene expression are up-regulated before the primitive erythroid cell stage
In primary cultures of −4.0 EGFP ES cells that contained early hematopoietic lineages, there was a high level of green fluorescence that appeared to peak on day 3 to 4 of differentiation (Figures3A, 3B). After day 4, the green fluorescence was markedly decreased, and by day 6 to 7 it was below the level of visual detection. Immunoblots for EGFP also indicated a decrease in EGFP protein after day 4 of differentiation, corroborating the fluorescence data (not shown). Flow cytometric analysis of live cells confirmed the increase in transgene expression beginning on day 2, peaking on day 4, and decreasing by day 5 of primary differentiation (Figure 3C). Single-peak histograms were observed for the gated population of live cells, indicating equivalent expression of the transgene in each cell. The examination of early heme pathway gene expression using fluorescence microscopy and HPLC analysis of porphyrins indicated no detectable porphyrin production at any stage of primary differentiation (not shown).
Expression of the ferrochelatase promoter transgene during primary in vitro differentiation.
ES cells containing the −4.0 kb transgene were differentiated in a liquid culture (primary differentiation) for 5 days. Original magnification, × 100. (A) Bright-field images and (B) green fluorescent images were taken from day 0 (undifferentiated) through day 5 using identical exposure times. A transgene-negative, 4-day differentiated EB was included as a negative control (TG-D4, A and B). Arrows point to fluorescing cell clusters. (C) Quantitation of green fluorescence was determined using flow cytometry. Each histogram represents the fluorescent intensities of approximately 50 000 cells determined at each 24-hour time point. Relative EGFP intensities are indicated above each histogram.
Expression of the ferrochelatase promoter transgene during primary in vitro differentiation.
ES cells containing the −4.0 kb transgene were differentiated in a liquid culture (primary differentiation) for 5 days. Original magnification, × 100. (A) Bright-field images and (B) green fluorescent images were taken from day 0 (undifferentiated) through day 5 using identical exposure times. A transgene-negative, 4-day differentiated EB was included as a negative control (TG-D4, A and B). Arrows point to fluorescing cell clusters. (C) Quantitation of green fluorescence was determined using flow cytometry. Each histogram represents the fluorescent intensities of approximately 50 000 cells determined at each 24-hour time point. Relative EGFP intensities are indicated above each histogram.
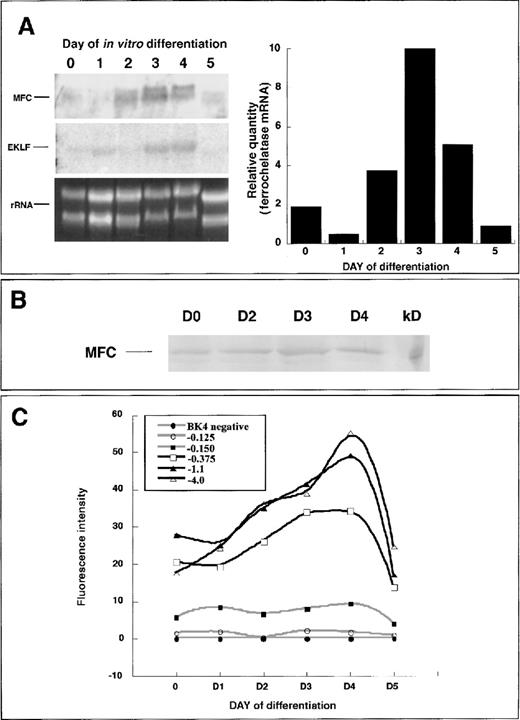
Fluorescence microscopy and flow cytometry data indicated that the ferrochelatase promoter transgene was up-regulated early in hematopoietic development, before the primitive erythroid-cell stage. Therefore, we used primary differentiated embryoid bodies for conducting expression analysis of the endogenous ferrochelatase gene and the ferrochelatase promoter transgenes. Endogenous ferrochelatase mRNA expression was up-regulated beginning on day 2, peaking on day 4, and decreasing by day 5 (Figure 4A). Erythroid Krüppel-like factor (EKLF), an erythroid-specific transcription factor, was temporally expressed in a parallel fashion to the ferrochelatase gene, indicating the erythropoietic potential of cells in the embryoid bodies (Figure 4A).
Endogenous ferrochelatase during primary in vitro differentiation.
(A) (left) Total RNA was isolated from primary differentiated EBs over a 5-day period. Northern blot analysis was conducted using 15 μg total RNA and probed with a random-primed mouse ferrochelatase cDNA (MFC). The membrane was stripped of the ferrochelatase probe and reprobed with a random-primed human EKLF cDNA (EKLF). Amounts of RNA loaded for each time point were assessed using ethidium bromide staining of 18S and 28S ribosomal RNA (rRNA). (right) Graphical representation of ferrochelatase mRNA levels depicted in Northern blot (left). Values were normalized to rRNA levels using scanning densitometry. (B) Endogenous ferrochelatase protein is demonstrated by immunoblotting. (C) Ferrochelatase promoter transgene expression during primary in vitro differentiation. Transgene expression was determined at each 24-hour time point by disrupting the EBs with brief trypsinization and quantifying EGFP expression in live cells using flow cytometry. Black lines represent transgenes that have −0.150 kb Sp1 elements and the −0.375 kb NF–E2 and GATA sites. Gray lines represent the −0.150 kb transgene that contains only the Sp1 elements, the −0.125 kb transgene that contains no known functional elements, and the transgene-negative cell line BK4.
Endogenous ferrochelatase during primary in vitro differentiation.
(A) (left) Total RNA was isolated from primary differentiated EBs over a 5-day period. Northern blot analysis was conducted using 15 μg total RNA and probed with a random-primed mouse ferrochelatase cDNA (MFC). The membrane was stripped of the ferrochelatase probe and reprobed with a random-primed human EKLF cDNA (EKLF). Amounts of RNA loaded for each time point were assessed using ethidium bromide staining of 18S and 28S ribosomal RNA (rRNA). (right) Graphical representation of ferrochelatase mRNA levels depicted in Northern blot (left). Values were normalized to rRNA levels using scanning densitometry. (B) Endogenous ferrochelatase protein is demonstrated by immunoblotting. (C) Ferrochelatase promoter transgene expression during primary in vitro differentiation. Transgene expression was determined at each 24-hour time point by disrupting the EBs with brief trypsinization and quantifying EGFP expression in live cells using flow cytometry. Black lines represent transgenes that have −0.150 kb Sp1 elements and the −0.375 kb NF–E2 and GATA sites. Gray lines represent the −0.150 kb transgene that contains only the Sp1 elements, the −0.125 kb transgene that contains no known functional elements, and the transgene-negative cell line BK4.
To analyze ferrochelatase protein expression during hematopoietic development, we isolated whole cell extracts from undifferentiated and 2-, 3-, and 4-day differentiated embryoid bodies and assessed ferrochelatase protein levels by immunoblotting. There were equivalent levels of ferrochelatase protein in undifferentiated and 2-, 3-, and 4-day differentiated cells (Figure 4B).
cis-Elements located between −0.150 kb and −0.375 kb are required for erythroid preferential expression, but maximal erythroid enhancement requires elements between −0.375 kb and −1.1 kb of the ferrochelatase promoter
To identify regions within the ferrochelatase promoter that contributed to regulating ferrochelatase gene expression, we generated HPRT-targeted clonal cell lines containing a single copy of various regions of the ferrochelatase promoter-driving expression of EGFP (Figure 1B). Transgene expression was assessed over a 5-day period of primary culture differentiation using flow cytometry. In pluripotent cells there were higher expression levels from the −0.375 kb, −1.1 kb, and −4.0 kb ferrochelatase promoter transgenes compared to the −0.125 kb or −0.150 kb ferrochelatase promoter transgenes (Figure 4C). Ferrochelatase promoter transgenes containing the GATA and NF–E2 sites showed enhanced expression beginning on day 2 of differentiation, peaking maximally on day 4, and dropping to levels similar to those of pluripotent cells by day 5 (Figure 4C). Between day 0 and day 4 (maximal induction), there was a 1.9 ± 0.07-fold induction for the −1.1 kb transgene, a 2.4 ± 0.1-fold induction for the −4.0 kb transgene, and a 1.45 ± 0.35-fold induction for the −0.375 kb transgene. Cells containing the −0.125 or −0.150 kb ferrochelatase promoter transgenes showed no enhanced expression at any point of differentiation. Maximal expression of ferrochelatase promoter transgenes required additional sequences upstream of the GATA site (from −0.375 kb to −1.1 kb).
The NF–E2 cis-element is required for erythroid-enhanced expression, and the GATA cis-element functions as a stage-specific repressor and enhancer
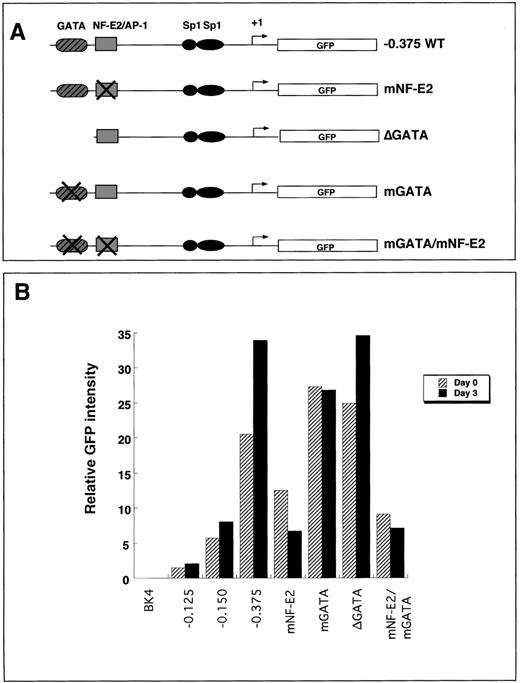
To elucidate the roles of the −0.375 kb NF–E2 and GATAcis-elements, we mutated either the NF–E2 site, the GATA site, or both and assessed expression of the wild-type and mutant transgenes integrated into the HPRT locus. Mutations within the NF–E2 site abolished binding activity of a protein corresponding to the migration pattern of NF–E2 but did not abolish binding of AP-1, as determined by mobility shift assays.13 GATA binding was abolished by either deleting the site (ΔGATA) or by introducing point mutations (mGATA) (Figure 5A).
Expression of transgenes that contain mutant GATA sites, NF–E2 sites, or both in undifferentiated and 3-day differentiated embryoid bodies.
(A) Summary of the constructs that contained mutations in either GATA, NF–E2, or both (mGATA, mNF–E2, or mGATA/mNF–E2). The mutant constructs contain point mutations that abolish the consensus site for its transcription factor. δGATA contains a complete deletion of the GATA cis-element. All mutations were introduced into the −0.375 kb promoter fragment. (B) Either undifferentiated (D0, black bars) or 3-day differentiated (D3, hatched bars) ES cells were disrupted to single cells by brief trypsinization and analyzed for EGFP transgene expression using flow cytometry. The graph is representative of 3 independent experiments.
Expression of transgenes that contain mutant GATA sites, NF–E2 sites, or both in undifferentiated and 3-day differentiated embryoid bodies.
(A) Summary of the constructs that contained mutations in either GATA, NF–E2, or both (mGATA, mNF–E2, or mGATA/mNF–E2). The mutant constructs contain point mutations that abolish the consensus site for its transcription factor. δGATA contains a complete deletion of the GATA cis-element. All mutations were introduced into the −0.375 kb promoter fragment. (B) Either undifferentiated (D0, black bars) or 3-day differentiated (D3, hatched bars) ES cells were disrupted to single cells by brief trypsinization and analyzed for EGFP transgene expression using flow cytometry. The graph is representative of 3 independent experiments.
Transgene expression was determined using flow cytometry in pluripotent or 3-day differentiated clonal populations of ES cell lines containing either the NF–E2 mutation (mNF–E2), the GATA mutations (mGATA or ΔGATA), or both mutations (mNF–E2/mGATA). In pluripotent ES cells, there was a 40% to 50% reduction in transgene expression in cell lines containing the NF–E2 mutation compared to cells expressing the −0.375 kb wild-type promoter transgene (Figure 5B). In 3-day differentiated cells, there was an 80% decrease in transgene expression in the mutant NF–E2 transgenic cell line compared to expression in the −0.375 kb wild-type promoter transgene cell lines (Figure 5B).
In pluripotent ES cells containing either a deletion of the −0.375 kb GATA site or a mutated GATA site, there was a 20% to 25% increase in transgene expression compared to cells containing the −0.375 kb wild-type promoter (Figure 5B). In 3-day differentiated cells, there was an 18% decrease in transgene expression in the mutant GATA cell line (mGATA) and an equivalent level of expression in the cells containing the GATA deletion (ΔGATA) compared to cells containing the −0.375 kb wild-type promoter (Figure 5B).
When both the NF–E2 and the GATA sites were mutated, the transgene expression was similar to the NF–E2 mutation alone. In the pluripotent mGATA/mNF–E2 cell lines, transgene activity was decreased by 55% compared to the −0.375 kb wild-type promoter transgene expression. In 3-day differentiated cells that contained the double NF–E2/GATA mutation, there was an 80% decrease in transgene expression compared to the −0.375 kb wild-type promoter transgene. Furthermore, in pluripotent ES cells, the NF–E2 mutation abrogated the increased expression resulting from the GATA mutation.
Transgene expression was determined using flow cytometry in pluripotent or 3-day differentiated clonal populations of ES cell lines containing either the NF–E2 mutation (mNF–E2), the GATA mutations (mGATA or δGATA), or both mutations (mNF–E2/mGATA). In pluripotent ES cells, there was a 40% to 50% reduction in transgene expression in cell lines that contained the NF–E2 mutation compared to cells expressing the −0.375 kb wild-type promoter transgene (Figure5B). In 3-day differentiated cells, there was an 80% decrease in transgene expression in the mutant NF–E2 transgenic cell line compared to expression in the −0.375 kb wild-type promoter transgene cell lines (Figure 5B).
In pluripotent ES cells containing either a deletion of the −0.375 kb GATA site or a mutated GATA site, there was a 20% to 25% increase in transgene expression compared to cells containing the −0.375 kb wild-type promoter (Figure 5B). In 3-day differentiated cells there was no increase in transgene expression in the mutant GATA cell line (mGATA), but there was a 30% increase in the cells containing the GATA deletion (δGATA) compared to cells containing the −0.375 kb wild-type promoter (Figure 5B). When both the NF–E2 and the GATA sites were mutated, the transgene expression was similar to the NF–E2 mutation alone. In the pluripotent mGATA/mNF–E2 cell lines, transgene activity was decreased by 55% compared to the −0.375 kb wild-type promoter transgene expression. In 3-day differentiated cells that contained the double NF–E2/GATA mutation, there was an 80% decrease in transgene expression compared to the −0.375 kb wild-type promoter transgene. Furthermore, in pluripotent ES cells, the NF–E2 mutation abrogated the increased expression resulting from the GATA mutation.
A-to-G polymorphism identified in the promoter of patients with protoporphyria has no effect on transcription of the ferrochelatase promoter transgene
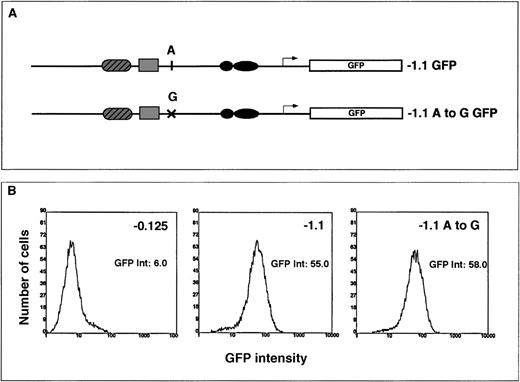
It has been proposed that the “wild-type” allele of the ferrochelatase gene is low expressing36 and contributes to the less than 50% ferrochelatase protein levels and activities observed in patients with protoporphyria. To determine whether an A-to-G polymorphism identified in the promoter of patients with protoporphyria36 contributed to a low-expressing allele, we constructed an HPRT-targeting vector that contained the A haplotype or the G haplotype in the context of the −1.1 kb ferrochelatase promoter transgene (Figure 6A).
The transcriptional effect of an A-to-G polymorphism identified in the ferrochelatase promoter of patients with protoporphyria.
(A) Summary of the HPRT targeting constructs used to assess the effect of A or G polymorphism located at −251 bp from the start codon. The G haplotype was introduced into the −1.1 kb reporter gene construct by PCR amplification of this region from genomic DNA from a patient with protoporphyria known to have the G polymorphism. The −1.1 kb fragment was then cloned into the EGFP reporter gene vector (−1.1 A to G). (B) Cells containing either the A polymorphism or the G polymorphism were differentiated for 3 days in primary culture. The embryoid bodies were disrupted by brief trypsinization, and EGFP expression was determined in live cells using flow cytometry. The EGFP intensities for the −0.125 (control transgene), the −1.1 kb or the −1.1 A to G transgenes were determined using maximal peak heights and are noted to the right of each histogram.
The transcriptional effect of an A-to-G polymorphism identified in the ferrochelatase promoter of patients with protoporphyria.
(A) Summary of the HPRT targeting constructs used to assess the effect of A or G polymorphism located at −251 bp from the start codon. The G haplotype was introduced into the −1.1 kb reporter gene construct by PCR amplification of this region from genomic DNA from a patient with protoporphyria known to have the G polymorphism. The −1.1 kb fragment was then cloned into the EGFP reporter gene vector (−1.1 A to G). (B) Cells containing either the A polymorphism or the G polymorphism were differentiated for 3 days in primary culture. The embryoid bodies were disrupted by brief trypsinization, and EGFP expression was determined in live cells using flow cytometry. The EGFP intensities for the −0.125 (control transgene), the −1.1 kb or the −1.1 A to G transgenes were determined using maximal peak heights and are noted to the right of each histogram.
Using flow cytometry, we measured reporter gene expression in pluripotent and 3-day differentiated cells containing a single copy of the transgene integrated into the HPRT locus. Based on the ferrochelatase activities observed in protoporphyria, we predicted that transcription from a low-expressing promoter would range from 30% to 60% of a normal promoter. However, there was no significant difference in transcriptional activity from a promoter transgene that contained the G haplotype and a promoter transgene that contained the A haplotype in either pluripotent (not shown) or 3-day differentiated cells (Figure6B).
Discussion
The ability to use conventional stable cell lines to analyze and compare functional elements in promoters that require organized chromatin for proper tissue-specific expression is confounded by the inability to control chromosomal integration site and copy number of transgenic reporter constructs. We have eliminated variable copy number and integration site by using a gene-targeting method to introduce a single copy of an EGFP reporter gene in a single locus at the 5′ end of the mouse HPRT gene. We have validated this method by showing that ferrochelatase promoter transgene expression in the HPRT locus is regulated in a tissue-specific manner and a time-course similar to the endogenous gene. This technique has enabled us to identify that the proximal NF–E2 element is required for enhanced expression during hematopoiesis and that the GATA element functions as a stage-specific repressor in pluripotent cells and an enhancer during early erythroid development. Furthermore, we have demonstrated that undefined elements between −0.375 kb and −1.1 kb are required for maximal enhancement of ferrochelatase gene transcription during erythroid cell development.
EGFP has been shown to be very stable in Chinese hamster ovary (CHO) cells.43 The implication of a highly stable EGFP reporter protein is that transgene induction rates may indicate an accumulation of EFGP protein and not actual induction rates. We addressed this concern by determining the half-life of EGFP in ES cells. Contrary to the high stability of EGFP in CHO cells, our findings indicated that in ES cells, the half-life of EGFP is between 3 and 4 hours (not shown; see “Materials and methods”). Therefore, the induction rates of the ferrochelatase reporter genes in this study most probably reflect actual induction rates.
Our data indicate that transcription of the human ferrochelatase promoter transgene is an early event during hematopoietic differentiation of ES cells in culture that precedes both ALAS-E expression and globin expression. In primary differentiation conditions, the ferrochelatase promoter transgene expression (−4.0 kb) increases on day 2, peaks on day 4, and decreases to low basal levels by day 5 (Figures 3 and 4). Using Northern blot analysis, we confirmed that the endogenous ferrochelatase gene followed an expression pattern and a time course similar to that of the transgene. No ALAS-E mRNA or β-globin mRNA was observed in Northern blot experiments. Fluorescence microscopy and HPLC analysis of porphyrins demonstrated no porphyrin expression during any stage of primary differentiation (not shown). Porphyrin expression was observed just before the primitive erythroid cell stage (ie, globinization), indicating late-stage transcription, translation, or both of ALAS-E mRNA.
Although ferrochelatase mRNA levels increased 5-fold on day 3 of primary differentiation, ferrochelatase protein levels remained equivalent to levels in undifferentiated cells (Figures 4A, 4B). A hallmark of erythroid cell differentiation is selective expression and stabilization of mRNA required by the cell after enucleation. Stabilization of α- and β-globin mRNA requires cis-acting elements in the 3′-UTR of both transcripts,49,50 and expression of ALAS-E mRNA is controlled by a 5′-UTR stem loop structure.51 The absence of increased ferrochelatase protein expression during transcriptional induction may indicate a translational control mechanism involving repressive stabilization of ferrochelatase mRNA in noncommitted precursors with later expression in committed erythroid cells.
We show that the NF–E2 cis-element is necessary for both the basal expression in pluripotent cells and the up-regulated expression of the ferrochelatase gene during erythroid cell development. It is noteworthy that AP-1 is still able to bind to the mutant NF–E2cis-element,13 suggesting that AP-1 fails to complement for the function of NF–E2 in this system. The failure of AP-1 to compensate for NF–E2 function has also been observed in the PBGD promoter.52 Transcription factors capable of binding the NF–E2 cis-element (besides Jun family factors) include p45/Maf (NF–E2)53-55 and Bach/Maf dimers.56,57The transcriptional enhancing mechanism of NF–E2 involves chromatin remodeling in the human β-globin and ε-globin locus.58Bach/Maf is highly expressed in fetal hematopoietic tissues before NF–E257 and is also thought to enhance transcription through chromatin rearrangement.57 Because ferrochelatase up-regulation is an early hematopoietic event, we speculate that enhanced transcription of the ferrochelatase gene may be initially regulated through a Bach/Maf complex.
GATA-1 is known to be a positive regulator of erythroid gene expression,59-61 and it is essential for erythroid differentiation.62-64 Murine GATA-1 binds the −0.375 kb ferrochelatase GATA element in vitro13; therefore, we predicted that the GATA site would be required for increased expression during erythroid cell development. The results show that in pluripotent ES cells, the GATA element functions as a transcriptional repressor, and in 3-day differentiated cells the GATA element functions as a transcriptional enhancer. The functional difference between the mGATA transgene and the δGATA transgene probably reflects the presence of 5′-flanking sequence in the mGATA construct, which is absent in the δGATA construct. One explanation for repression through the GATAcis-element in pluripotent ES cells is that another GATA-binding factor mediates this role. GATA-2 has previously been shown to be a repressor.65 It is expressed initially in the erythroid precursors, and later it is down-regulated as GATA-1 transcription increases.66,67 In addition, recent evidence shows that GATA-1, GATA-2, and GATA-4 are able to mediate transcriptional repression through a ternary complex with FOG factors (Friend-of-GATA) and CtBP2.68 69
Although −0.375 kb of the ferrochelatase promoter is sufficient for erythroid-enhanced expression, maximal expression during erythroid cell development requires sequences between −0.375 kb and −1.1 kb. This upstream region has not been formally studied for transcription factor binding; however, a computer databasecis-element search has identified several putative transcription factor-binding sites, GATA-1, CAC-binding protein,70 and CACCC (EKLF),71 that may contribute to up-regulation of the transgene in early erythroid cells (Figure 7). We are currently investigating transcription factor binding to these cis-elements in vitro.
Summary of putative cis-elements located between −0.375 kb and −1.1 kb of the human ferrochelatase promoter.
The TESS computer database was used to search for putative binding sites for erythroid-specific or muscle-specific trans-acting factors. The locations of these sites are indicated by circles (TESS URL, http://www.cbil.upenn.edu/tess/).
Summary of putative cis-elements located between −0.375 kb and −1.1 kb of the human ferrochelatase promoter.
The TESS computer database was used to search for putative binding sites for erythroid-specific or muscle-specific trans-acting factors. The locations of these sites are indicated by circles (TESS URL, http://www.cbil.upenn.edu/tess/).
Phenotypic expression of “dominantly” transmitted protoporphyria is thought to require inheritance of a null ferrochelatase allele and a “low-expressed” ferrochelatase allele.36 The G haplotype segregates with the protoporphyric phenotype and, in all but 1 reported case,37 segregates with a low-expressing allele. We tested the effect of the G haplotype on expression of the −1.1 kb ferrochelatase promoter transgene using the gene-targeting method. There was no difference in transcriptional activity between the transgene containing the G haplotype and the transgene containing the A haplotype (Figure 6).
This result suggests there is another mutation located in the promoter or elsewhere that contributes to a low-expressing allele. Hyper-methylation of CpG islands in some promoters causes a decrease in gene expression and disease.72,73 It is conceivable that aberrant methylation of the ferrochelatase promoter could contribute to decreased transcriptional activity from the imprinted allele. An alternative possibility is that the presence of intron mutations, conferring cryptic splice sites, could result in mildly defective splicing, an unstable ferrochelatase mRNA, and lower levels of normal ferrochelatase mRNA. Such mutations have been implicated in bipolar disorder74 and Rabson–Mendenhall syndrome.75
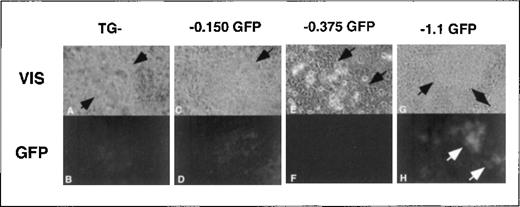
Although we predicted high EGFP expression levels in erythroid colonies, we unexpectedly observed equally intense EGFP expression in rhythmically contracting foci of cells containing the −4.0 kb ferrochelatase promoter transgene (Figure8). When ES cells were differentiated under conditions that favored cardiomyocyte formation,76,77enhanced transgene expression correlated with the presence of −0.375 kb to −1.1 kb of the ferrochelatase promoter (Figure8). Up-regulation of the ferrochelatase promoter transgene in rhythmically contracting foci may indicate an increased demand for heme in cardiomyocytes to produce myoglobin. Using the TESS computer database search, we identified the putative cis-elements, myoD, myogenin, and MEF2 in the region from −0.375 kb to −1.1 kb of the ferrochelatase promoter that may contribute to increased expression of the promoter transgene in cardiomyocytes (Figure 7). MyoD,78 myogenin, and MEF-279 are muscle tissue-specific transcription factors belonging to the basic helix–loop–helix family, and are all involved in developmental up-regulation of muscle-specific genes.80 81
Transgene expression in cardiomyocyte foci.
Areas of early cardiac cell lineages were visually identified at approximately day 10 of secondary culture in methylcellulose in foci of rhythmically contracting cells (A, C, E, G, arrows indicate areas of contracting cells). Cardiomyocyte foci with either no transgene (TG−) or the −0.150 kb, −0.375 kb, or −1.1 kb transgenes were assessed for EGFP expression using fluorescence microscopy (B, D, F, H). Note that the fluorescence images from the TG negative and −0.150 kb cells (B, D) have 3 times the exposure length as the −0.375 kb and −1.1 kb cells (F, H). Original magnification, × 400.
Transgene expression in cardiomyocyte foci.
Areas of early cardiac cell lineages were visually identified at approximately day 10 of secondary culture in methylcellulose in foci of rhythmically contracting cells (A, C, E, G, arrows indicate areas of contracting cells). Cardiomyocyte foci with either no transgene (TG−) or the −0.150 kb, −0.375 kb, or −1.1 kb transgenes were assessed for EGFP expression using fluorescence microscopy (B, D, F, H). Note that the fluorescence images from the TG negative and −0.150 kb cells (B, D) have 3 times the exposure length as the −0.375 kb and −1.1 kb cells (F, H). Original magnification, × 400.
Gene targeting using a single-copy, single-integration site of an EGFP reporter gene proves to be a sensitive and powerful method for the study of cis-element use during differentiation of ES cells. The pluripotency of ES cells enables the transgene to be studied in various cell lineages in vitro. In vivo studies are possible by using the targeted ES cells to generate chimeric mice or mice that are heterozygous or homozygous for the transgene.38 This method can also be used to assess the transcriptional role of polymorphic regulatory regions in genes of patients with inherited diseases.
Acknowledgments
We thank Oliver Smithies and Sarah Bronson for the HPRT targeting vector and Nobuyo Maeda for consultation and cell culture support. We also thank the staff at the Center for Gastrointestinal and Biological Diseases for computer support (DK34987) and at the University of North Carolina for use of the flow cytometry core facility.
Supported by grants DK47361 and DK34987 (D.A.B.). A.T. was supported by a fellowship from the Fundacion Dr Manuel Morales (Tazacorte, Isla de La Palma, Canary Islands, Spain).
Reprints:David A. Brenner, Department of Medicine, CB#7038, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7038; e-mail: dab@med.unc.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal