Abstract
We have developed a gene trap approach to select specific cytokine receptor/ligand responsive genes in the cell line TF-1. This cell line exhibits a dependency on granulocyte-macrophage colony-stimulating factor (GM-CSF) or interleukin-3 (IL-3) and responds to interleukin-5 (IL-5). In an attempt to detect genes modulated by one of these factors, cells were infected with the Rosaβgeo retrovirus in the presence of GM-CSF, IL-3, or IL-5 and clones were selected for retroviral integration on the basis of G418 resistance. Housekeeping and cytokine-regulated trapped genes were then differentiated on the basis of G418 resistance versus sensitivity in the presence of the different cytokines. To determine the reliability of this screen, DNA sequences upstream of the proviral integration site were identified by 5′ rapid amplification of DNA ends polymerase chain reaction (RACE PCR) from selected GM-CSF–treated and –infected clones. Comparison of the sequences with those in the Genbank database revealed that 2 sequences correspond to known genes: NACA and RBM3. NACAwas recently defined as a coactivator of c-jun–mediated transcription factors in osteoblasts, and RBM3 as a protein from the heterogeneous nuclear ribonucleoprotein family. Data from transcriptional analysis of these 2 genes in TF-1 cells showed a specific up-regulation by GM-CSF. Both transcripts were also found to be up-regulated in purified CD34+ cells, suggesting their involvement in proliferative processes during hematopoiesis. Interestingly, down-regulation was observed during monocytic differentiation of TF-1 cells, suggesting their extinction could contribute to monocytic lineage development. This study demonstrates that this gene trap approach is a useful method for identifying novel, specific cytokine-responsive genes that are involved in the regulation of hematopoiesis.
The role of cytokines in survival, proliferation, and differentiation of hematopoietic cells has been extensively studied (reviewed in Metcalf1 and Ogawa2). However, the molecular mechanisms that underlie a specific pattern of biologic effects for 1 given cytokine remain unclear. It is likely that these mechanisms involve sets of unidentified genes. One illustration is the lack of correlation between intracellular events in known and shared pathways after the binding of granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), or interleukin-5 (IL-5) to their respective receptors and the divergent spectrum of their biologic functions.3 Sequentially or together, these cytokines are required for a variety of hematopoietic functions such as proliferation of hematopoietic progenitors4 or stimulation of formation of multipotent colonies that contain granulocytes, erythroid cells, monocytes, and megakaryocytes (reviewed in Clark and Kamen5and Sato and Miajima6). In addition, complete maturation within various lineages (ie, erythropoiesis, granulopoiesis, or basophil/mast cell development) requires IL-3. IL-5, a more restrictive factor,7 mainly controls eosinophil and basophil lineages, but also acts on overlapping compartments. Identification of a common βc chain,8 shared by these cytokine receptors, which activates a combination of signaling molecules such as Jak/Stat9-12 and Ras-Raf-MAPK,3,13-17 offers only a partial explanation for the apparent common functions. Evidence for a role in the signaling of the respective cytokine receptor alpha chain18-22 enlightens divergent signaling pathways and indicates that cytokines may simultaneously recruit different pathways.
In an attempt to isolate cytokine-responsive genes, we developed an in vitro gene trap strategy. Similar strategies used to study different systems such as embryogenesis23-30 or cell growth and differentiation in other tissues31-35 have allowed identification of novel genes. We took advantage of the well-characterized gene trap retroviral vector Rosaβgeo,23 24 which places the geo reporter gene (a chimera between lacZ and neo) under the control of the regulatory elements of genes disrupted by retroviral insertion. The presence of a splice acceptor sequence in the vector allows the production of a chimeric transcript containing at least the ATG of geo, from which the Geo protein expression can be monitored by G418 resistance or sensitivity of the cells.
Because hematopoietic progenitor cells are difficult to manipulate in vitro, we first evaluated the feasibility of the method in the human premyeloid TF-1 cell line.36 TF-1 cells require GM-CSF or IL-336 for growth and respond to IL-5,36,37thus making this cell line a suitable in vitro model for the study of specific versusoverlapping genes involved in the proliferative activity of these cytokines. Here, we describe the specific transcriptional modulation by GM-CSF of 2 trapped genes (NACAand RBM3),38 39 not yet reported to be cytokine regulated, but seemingly involved in the molecular mechanism of differentiation in other tissues. Our study indicates the feasibility of identifying new genes specifically involved in regulation of hematopoiesis downstream of individual cytokines, despite overlapping signal transduction machinery. The importance of yet unknown mechanisms involved in the cytokine regulation of hematopoiesis is discussed.
Materials and methods
Retrovirus production and gene trap vector
The GP+EnvAm12 packaging cells40 were transfected with lipofectin (Life-Technologies SARL, Cergy Pontoise, France) according to the manufacturer's instructions with the retroviral vector pRosaβgeo.24 Briefly, this vector has a deletion of viral enhancers and a splice acceptor in front of a promoterless georeporter gene (a fusion gene between lacZ and neo), allowing the generation of fusion transcripts under the control of the regulatory elements of the trapped gene. The reporter gene carries its own ATG, enabling the detection of integration into untranslated sequences.
Cells and culture
The GP+EnvAm12 packaging cells were maintained in DMEM medium containing 10% fetal calf serum (FCS), l-glutamine (2 mmol/L), penicillin (100 U/mL), and streptomycin (100 mg/mL). Retrovirus-producing cells were selected with G418 (0.5 mg/mL active powder (Life-Technologies SARL) for 2 weeks, and the virus titer was determined by the number of G418-forming units per milliliter in infected rat-2 fibroblast cells.
The GM-CSF–dependent TF-1 cell line used for retroviral infection was provided by Dr T. Kitamura36 and maintained in RPMI medium supplemented with 10% FCS, l-glutamine (2 mmol/L), penicillin (100 U/mL), streptomycin (100 mg/mL), and recombinant GM-CSF (5 ng/mL, Sandoz, Rueil-Malmaison, France). Monocytic differentiation was induced with 10 nmol/L phorbol 12-myristate acetate (PMA) (Sigma, St Louis, MO).
TF-1 cells used for transcriptional analysis (provided by Dr H. Gascan, INSERM U298, France) were a subclone of the original cells that responded to a wider range of cytokines.
Actinomycin D experiments were conducted with the treatment of TF-1 cells with 10 μg/mL of actinomycin D (Sigma) for different periods with or without GM-CSF (5 ng/mL).
Enrichment of peripheral blood CD34+ cells
Apheresis samples were taken after informed consent was obtained from 2 adult patients with non-Hodgkin lymphoma, mobilized with chemotherapy and rhG-CSF. CD34+ cells were obtained by a combination of density separation, adherence, depletion, and immunoselection using the Magnetic Activating Cell Sorting device (MACS, Miltenyi Biotech GmbH, Bergish Gladbach, Germany), according to the manufacturer's recommendations.
Retroviral infection and selection of G418-resistant TF-1 clones
One day before infection, a confluent 100-mm plate of GP+EnvAm12 Rosaβgeo vector-producing cells was maintained in RPMI medium. The next day the culture supernatant containing the retroviruses was used to infect the human erythroleukemic cell line TF-1 (2 × 105 cells) maintained in GM-CSF, IL-3, or IL-5, pretreated for 30 minutes with polybrene (5 μg/mL) (Sigma). After 16 hours, TF-1 cells were washed in phosphate buffered saline (PBS) and seeded in 96-well plates (102 cells per well), in the presence of G418 (1 mg/mL active powder) and GM-CSF (5 ng/mL), IL-3 (1 ng/mL, R&D Systems, Oxon, UK), or rhIL-5 (0.5 ng/mL, R&D Systems). After 3 weeks, surviving clones (1 of 103 cells) of an average size of 1000 cells were expanded and submitted to a cytokine-dependent proliferative assay.
Proliferative assay
1 × 105 cells per milliliter uninfected or infected TF-1 cells were washed and starved overnight in RPMI medium supplemented with 10% FCS, l-glutamine (2 mmol/L), penicillin (100 U/mL), and streptomycin (100 mg/mL). Cells were seeded in a 96-well plate (104 cells per well) and cultured in the presence or absence of G418 (1 mg/mL active powder) with either rhGM-CSF (5 ng/mL, Sandoz), rhIL-3 (1 ng/mL, R&D Systems), or rhIL-5 (0.5 ng/mL, R&D Systems) for 68 hours or 5 days. Cells were then pulsed with 37 Bq (1.78 Tbq/mmol [3H]-thymidine, Amersham Life Science, UK) for 4 hours, harvested on fiber paper, and the labeled cellular DNA was measured using a β-counter Rackbeta Compact 1212-411 (LKB, Uppsala, Sweden). Triplicate samples were assayed for each set of conditions.
Isolation of 5′ flanking sequences
Complementary DNA (cDNA), corresponding to gene trapped-geo fusion transcripts, as isolated using a 5′ RACE (rapid amplification of cDNA ends) kit (Life-Technologies SARL), according to the manufacturer's instructions. Reverse transcription using SuperScript II RTase (Promega France, Charbonnières, France) was performed using the oligonucleotide primer 5′ATGCGCTCAGGTCAAATTCAGACGG complementary to the lacZ sequence. After RNAse H treatment and cDNA purification, an anchor sequence was added to the 3′ end of the cDNA using dCTP and TdT. Polymerase chain reaction (PCR)-amplification was performed on dCTP-tailed cDNA using a 3′ oligonucleotide primer 5′CCGTGCATCTGCCAGTTTGAGGGGA complementary to the lacZ sequence and the described AP (anchor primer from Life-Technologies SARL). After 30 cycles (94°C/1 min; 55°C/1 min; 72°C/1 min), PCR products were agarose gel-sized (above 400 nucleotide [nt]) and excised. Agarose gel was melted and DNA was diluted 1:100; and reamplified under the same conditions with a different nested oligonucleotide primer 5′TACCGTCGATCCCCACTGGAAA complementary to the lacZ sequence and with the described 5′ UAP (universal anchor primer from Life-Technologies SARL). PCR products were purified using the GlassMAX DNA Isolation Spin Cartridge System (Life-Technologies), cloned in the SmaI site of pUC18 and sequenced with the T7 polymerase Sequencing kit (Pharmacia, Uppsala, Sweden).
GenBank database searches
Probes
The 5′RACE PCR isolated sequences (198 base pairs [bp] from clone 50, 47 bp from clone 134, and 44 bp from clone C) were extended by PCR amplification using oligonucleotide primers complementary to the EST sequences downstream from the 5′RACE PCR sequence, to generate products about 300 nt length. Amplification products were cloned in a pUC-18 plasmid (Pharmacia) for clones 50 and C, and into the pGEM-T vector using the pGEM-T vector systems (Promega France) for clone 134. Miniprep plasmid DNA was sequenced with the T7 polymerase Sequencing kit (Pharmacia).
The lacZ sequence used as a probe was obtained by digestion of pRosaβgeo with ClaI and subcloned into the ClaI site of the pBluescript plasmid (StrataGene, La Jolla, CA).
Probes were labeled by the random oligonucleotide priming method.42 A 28S oligoprobe (5′-TGAATCCTCCGGGCGGACT) was γ32P-labeled with the T4 polynucleotide kinase (New England Biolabs, Ozyme, Montigny, France).
Southern and Northern blot analyses
Total RNA and genomic DNA were prepared by the guanidium/CsCl method.43 Southern and Northern blots were conducted as described.43 Fifteen micrograms genomic DNA was digested with EcoRI for Southern blot analysis and 20 μg total RNA of a subclone from the original TF-1 cell line was used for Northern blot studies. Membranes were scanned and analyzed with a PhosphorAnalyst (Phosphorimager, Biorad SA, Ivry-sur-Seine, France).
Expression of transcripts 50 and 134 by reverse transcriptase-polymerase chain reaction in enriched CD34+ cells
Messenger RNA (mRNA) from enriched CD34+ cells was extracted using the Quick prep Micro mRNA purification kit (Pharmacia Biotech Products, Saclay, France), was treated with DNAse I (Boehringer Mannheim, Roche Diagnostics, Meylan, France), and was reverse transcribed (RT) with the Superscript preamplification system (Life-Technologies SARL) according to the manufacturer's instructions, using a random primer. For PCR, 2 μL of cDNA was taken from each RT reaction volume and samples were submitted to either 22 and 26 (product of RBM3) or 26 and 28 (product of NACA) cycles of PCR in a Perkin-Elmer thermal cycler (Perkin-Elmer, Norwalk, CT) using 1 minute of denaturation at 94°C, 1 minute of annealing at either 55°C for RBM3 or 60°C for NACA, and 1 minute of polymerization at 72°C. A final polymerization step was performed for 5 minutes at 72°C. The primers used were 5′CCAGGACTTGAACTGCCATG (sense) and 5′CAGACTTCCTGCCATGATCC (antisense), amplifying a product of 445 bp in the RBM3 gene; and 5′GCTACAGAGCAGGAGTTGCCA (sense) and 5′TAACCACCCTGGTTTCTGCC (antisense), amplifying a product of 567 bp in the NACA gene. To monitor the amount of RNA per tube and the efficiency of the reactions, β2 microglobulin transcript was assessed using the sense primer: 5′CCAGCAGAGAATGGAAAGTC (contained in the first exon of the gene) and the antisense primer: 5′GATGCTGCTTACATGTCTCG (contained in the second exon of the gene). The size of the amplified product was 268 bp.
The products were electrophoresed on 2% agarose gel, blotted, and hybridized with γ32P-labeled oligonucleotides chosen within the amplified PCR region. Control samples not RT were used to monitor for possible genomic DNA contamination.
Results
Selection for cytokine-regulated integrations
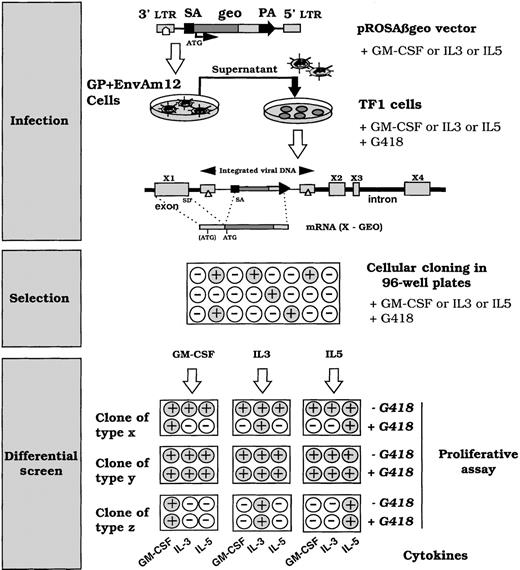
In an attempt to identify genes differentially regulated by the GM-CSF/IL-3/IL-5 group of cytokines, TF-1 cells were infected with the Rosaβgeo retroviral vector24 (Figure1) in the presence of GM-CSF, IL-3, or IL-5 to favor integration events respectively into GM-CSF, IL-3, or IL-5 transcriptionally active genes. This retrovirus contains a splice acceptor 5′ to a promoterless reporter gene, encoding a fusion protein of β-galactosidase and neomycin phosphotransferase. Clones with retroviral integrations into expressed genes will therefore produce an active Geo protein, conferring G418 resistance. After 3 weeks of selection with G418 and GM-CSF, 173 clones grew out of 2 × 105 infected cells. The growth of clones from IL-3– or IL-5–cultured and infected cells was delayed by 10 days. In addition, the number of clones from IL-5–cultured and infected cells was roughly a third of those recovered from IL-3– or GM-CSF–cultured and infected cells. These phenomena reflect differences in the long-term proliferation efficiency of IL-3, IL-5, and GM-CSF with the clones of the TF-1 cells used in our study. G418 resistance can be obtained by expressed transcripts representing both cytokine-responsive genes and noncytokine-regulated genes. To screen for integrations into differentially cytokine-expressed genes, clones were submitted to a proliferative assay in the presence of G418 and a panel of different cytokines (GM-CSF, IL-3, or IL-5) as shown in Figure 1. Acquisition of G418 resistance in the presence of a specific cytokine and G418 sensitivity to a different cytokine suggested an integration event into a cytokine-regulated gene. This phenotype reflects the fact that the copy number of the transcript varies with growth in a cytokine-dependent manner. Figure 2 shows a typical pattern of differential cytokine proliferation monitored by [3H]-thymidine incorporation after at least 3 days of cell culture. Results are represented as the relative ratio of [3H]-thymidine incorporation for each cytokine-grown condition compared with incorporation after exposure to the same cytokine and G418 during culture. A threshold of significant difference was set at 0.35, representing a 65% inhibition of G418 resistance for a given cytokine. The residual resistance probably reflects differences in cell cycle synchronization of the cells or leaky expression of the vector product geo, or both. Figure 2 summarizes the outcome of the screen. Uninfected TF-1 cells displayed complete G418 sensitivity after 3 days of growth, and the pattern of clone C is representative of unregulated integration. From GM-CSF–cultured and infected cells, 7 of 80 clones showed a clear differential G418 behavior (resistance and sensitivity within the same clone) in the presence of IL-5 and G418. In addition, 2 of these clones showed a differential response with IL-3 and IL-5 when the proliferative assay was conducted after 5 days of growth. In IL-3– or IL-5–cultured and infected cells, differential responses were observed after 3 days of exposure of the cells to the cytokines before [3H]thymidine incorporation assay (Figure 2B). Five days of growth did not modify the G418 responses for clones showing either incomplete (+/−), or complete G418 resistance to a cytokine other than the one used at the time of infection. This suggests that modulation of the insertion site by several cytokines occurred. Considering these results, we delineated cytokine-modulated genes either by more than a single cytokine or by all 3 cytokines tested. In addition, we obtained clones with single cytokine-modulated loci.
Schematic representation of the screening used to identify integration in cytokine-selective inducible loci.
The Rosaβgeo retroviral gene trap contains a splice acceptor (SA) 5′ to a promotorless fusion reporter gene (geo), which when integrated, generates a fusion transcript between the reporter gene and the trapped gene. The Geo protein will be produced from the ATG of the trapped gene or at least from the ATG in the GEO sequence. TF-1 cells were infected with the pRosaβgeo and diluted in 96-well plates under G418 selection for 3 weeks. Surviving clones were expanded, plated in duplicate and submitted to a proliferative assay with different cytokines: GM-CSF, IL-3, or IL-5, in the absence or presence of G418 (1 mg/mL). ○ cells that do not proliferate • growing cells. Clones of type X from GM-CSF–, IL-3–, or IL-5–cultured and infected cells and selected for survival in the presence of G418, and the cytokine present at the time of infection, exhibit a differential cytokine response, ie, if cultured and selected in GM-CSF, it remains sensitive to G418 in the presence of IL-5 and IL-3, whereas it is G418 resistant in the presence of GM-CSF. This indicates that neither IL-3 nor IL-5 induces transcription of a chimeric transcript, whereas GM-CSF does. Clones of type Y do not present a differential cytokine response based on the acquisition of G418 resistance. It acquires G418 resistance in the presence of all the tested cytokines. It corresponds to integration into either housekeeping genes or nonspecifically cytokine-modulated genes in this assay. Clones of type Z correspond to cells that do not proliferate in the presence of the tested cytokine, reflecting a cytokine-response heterogeneity of the cell line. These clones were not studied further.
Schematic representation of the screening used to identify integration in cytokine-selective inducible loci.
The Rosaβgeo retroviral gene trap contains a splice acceptor (SA) 5′ to a promotorless fusion reporter gene (geo), which when integrated, generates a fusion transcript between the reporter gene and the trapped gene. The Geo protein will be produced from the ATG of the trapped gene or at least from the ATG in the GEO sequence. TF-1 cells were infected with the pRosaβgeo and diluted in 96-well plates under G418 selection for 3 weeks. Surviving clones were expanded, plated in duplicate and submitted to a proliferative assay with different cytokines: GM-CSF, IL-3, or IL-5, in the absence or presence of G418 (1 mg/mL). ○ cells that do not proliferate • growing cells. Clones of type X from GM-CSF–, IL-3–, or IL-5–cultured and infected cells and selected for survival in the presence of G418, and the cytokine present at the time of infection, exhibit a differential cytokine response, ie, if cultured and selected in GM-CSF, it remains sensitive to G418 in the presence of IL-5 and IL-3, whereas it is G418 resistant in the presence of GM-CSF. This indicates that neither IL-3 nor IL-5 induces transcription of a chimeric transcript, whereas GM-CSF does. Clones of type Y do not present a differential cytokine response based on the acquisition of G418 resistance. It acquires G418 resistance in the presence of all the tested cytokines. It corresponds to integration into either housekeeping genes or nonspecifically cytokine-modulated genes in this assay. Clones of type Z correspond to cells that do not proliferate in the presence of the tested cytokine, reflecting a cytokine-response heterogeneity of the cell line. These clones were not studied further.
Summary of the cytokine-dependent proliferative responses.
G418 resistant TF-1 clones dependent on the cytokine present at the time of infection (ie, GM-CSF, IL-3, or IL-5) were tested for their resistance in a proliferative assay (as described in “Materials and methods”). The rates of proliferation were compared in the absence (▭) or presence ( after 3 days; •• after 5 days) of G418 for each cytokine. Results are represented as the relative ratio of [3H]-thymidine incorporation (cpm) obtained without G418 to that with G418. The G418 resistance was evaluated after 3 or 5 days of culture by the amount of [3H]-thymidine uptake. After 72 hours with G418, TF-1 cells did not incorporate [3H]-thymidine. (A) Representative profiles of clones derived from cells infected in the presence of GM-CSF (GM-50, GM-C), or IL-3 (IL-3-4) or IL-5 (IL-5-16). GM-C represents a clone in which no specific cytokine modulations occurred. (B) Schematic representation of the results of [3H]-thymidine incorporation for the selected clones from cells infected in the presence of GM-CSF (upper panel), IL-3 (left lower panel), or IL-5 (right lower panel) infected cells. −: no proliferation, +/−: difference in proliferation when compared with and without G418, +: similar proliferation with and without G418.
after 3 days; •• after 5 days) of G418 for each cytokine. Results are represented as the relative ratio of [3H]-thymidine incorporation (cpm) obtained without G418 to that with G418. The G418 resistance was evaluated after 3 or 5 days of culture by the amount of [3H]-thymidine uptake. After 72 hours with G418, TF-1 cells did not incorporate [3H]-thymidine. (A) Representative profiles of clones derived from cells infected in the presence of GM-CSF (GM-50, GM-C), or IL-3 (IL-3-4) or IL-5 (IL-5-16). GM-C represents a clone in which no specific cytokine modulations occurred. (B) Schematic representation of the results of [3H]-thymidine incorporation for the selected clones from cells infected in the presence of GM-CSF (upper panel), IL-3 (left lower panel), or IL-5 (right lower panel) infected cells. −: no proliferation, +/−: difference in proliferation when compared with and without G418, +: similar proliferation with and without G418.
Summary of the cytokine-dependent proliferative responses.
G418 resistant TF-1 clones dependent on the cytokine present at the time of infection (ie, GM-CSF, IL-3, or IL-5) were tested for their resistance in a proliferative assay (as described in “Materials and methods”). The rates of proliferation were compared in the absence (▭) or presence ( after 3 days; •• after 5 days) of G418 for each cytokine. Results are represented as the relative ratio of [3H]-thymidine incorporation (cpm) obtained without G418 to that with G418. The G418 resistance was evaluated after 3 or 5 days of culture by the amount of [3H]-thymidine uptake. After 72 hours with G418, TF-1 cells did not incorporate [3H]-thymidine. (A) Representative profiles of clones derived from cells infected in the presence of GM-CSF (GM-50, GM-C), or IL-3 (IL-3-4) or IL-5 (IL-5-16). GM-C represents a clone in which no specific cytokine modulations occurred. (B) Schematic representation of the results of [3H]-thymidine incorporation for the selected clones from cells infected in the presence of GM-CSF (upper panel), IL-3 (left lower panel), or IL-5 (right lower panel) infected cells. −: no proliferation, +/−: difference in proliferation when compared with and without G418, +: similar proliferation with and without G418.
after 3 days; •• after 5 days) of G418 for each cytokine. Results are represented as the relative ratio of [3H]-thymidine incorporation (cpm) obtained without G418 to that with G418. The G418 resistance was evaluated after 3 or 5 days of culture by the amount of [3H]-thymidine uptake. After 72 hours with G418, TF-1 cells did not incorporate [3H]-thymidine. (A) Representative profiles of clones derived from cells infected in the presence of GM-CSF (GM-50, GM-C), or IL-3 (IL-3-4) or IL-5 (IL-5-16). GM-C represents a clone in which no specific cytokine modulations occurred. (B) Schematic representation of the results of [3H]-thymidine incorporation for the selected clones from cells infected in the presence of GM-CSF (upper panel), IL-3 (left lower panel), or IL-5 (right lower panel) infected cells. −: no proliferation, +/−: difference in proliferation when compared with and without G418, +: similar proliferation with and without G418.
Analysis of the retroviral integrations
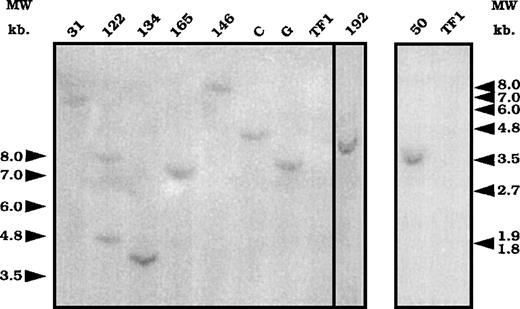
To associate the differential cytokine response with either 1 or several exon-trapped gene(s), we determined the number of integration events in 9 clones from GM-CSF–cultured and infected cells. Clones 122, 134, 165, 146, 192, and 50 presented differentially cytokine-regulated trapped genes, whereas clones 31, C, and G were defined by our screen as not being specifically modulated by the tested cytokines. After EcoRI digestion, which cut the retroviral vector DNA once, genomic DNA from 9 clones was analyzed by Southern blot with a lacZ probe. Eight clones had a single retroviral integration, as only 1 band was revealed (Figure3), whereas 2 fragments were detected for clone 122. Different sized fragments were obtained, suggesting that 1 individual retroviral integration occurred per clone (except for clone 122).
Southern blot analysis of the number of integrations.
Genomic DNA prepared from the different clones was digested withEcoR1 and submitted to a Southern blot analysis using a lacZ probe. DNA from uninfected cells was used as a control for hybridization (lane TF1). Numbers and letters at the top represent the different clones. Arrows represent the molecular weight markers in kilobase (kb).
Southern blot analysis of the number of integrations.
Genomic DNA prepared from the different clones was digested withEcoR1 and submitted to a Southern blot analysis using a lacZ probe. DNA from uninfected cells was used as a control for hybridization (lane TF1). Numbers and letters at the top represent the different clones. Arrows represent the molecular weight markers in kilobase (kb).
We then used 5′RACE-PCR on purified total RNA from these clones to isolate the coding sequences flanking the proviral integration site. After cloning the PCR products, different sequences ranging from 44 to 353 bp were identified (Table 1). Only 1 sequence was obtained from clone 122, suggesting that either only 1 trapped locus is transcribed or that, of the 2 putative amplification products, only 1 was detected. All the sequences were different from each other, except sequence 192 and sequence 165. These results suggest that the described integration events in the 9 clones took place at 8 different loci in cellular chromosomal DNA. Alternatively, some integrations may have occurred in identical loci at different integration sites, resulting in different and nonoverlapping sequences. This phenomenon was detected for at least 1 locus (165/192) and was identified from the overlapping sequences. Because this locus is “trapped” twice, it might represent a site of high chromatin accessibility for retroviral integration.
Result of analysis of the 5′RACE PCR trapped sequence in the GenBank database
| Clone number . | Gene description . | 5′RACE flanking fragment size (bp) . | Overlap length (bp) . | % Identity in the overlap length . | GenBank accession number . |
|---|---|---|---|---|---|
| 50 | hnRNPL (RBM3) | 198 | 177 | 100 | (h) U28686 |
| 134 | NACA | 47 | 47 | 100 | (h) X80909 |
| 18 | hnRNPF | 86 | 28 | 97 | (h) AA310292 |
| 30 | Unknown | 111 | 62 | 82 | (m) D77412 |
| 165/(192) | Unknown | 170/33 | 127 | 100 | (h) AA38369 |
| 11 | Unknown | 175 | 50 | 94 | (h) AA77426 |
| 122 | Unknown | 114 | 106 | 97 | (h) AA326035 |
| G | Unknown | 79 | None | ||
| 31 | Jumonji | 353 | 202 | 99.5 | (m) D311967 |
| C | L18a | 44 | 44 | 100 | (r) X14181 |
| Clone number . | Gene description . | 5′RACE flanking fragment size (bp) . | Overlap length (bp) . | % Identity in the overlap length . | GenBank accession number . |
|---|---|---|---|---|---|
| 50 | hnRNPL (RBM3) | 198 | 177 | 100 | (h) U28686 |
| 134 | NACA | 47 | 47 | 100 | (h) X80909 |
| 18 | hnRNPF | 86 | 28 | 97 | (h) AA310292 |
| 30 | Unknown | 111 | 62 | 82 | (m) D77412 |
| 165/(192) | Unknown | 170/33 | 127 | 100 | (h) AA38369 |
| 11 | Unknown | 175 | 50 | 94 | (h) AA77426 |
| 122 | Unknown | 114 | 106 | 97 | (h) AA326035 |
| G | Unknown | 79 | None | ||
| 31 | Jumonji | 353 | 202 | 99.5 | (m) D311967 |
| C | L18a | 44 | 44 | 100 | (r) X14181 |
Overlap length is represented as the number of nucleotides present in the 5′RACE PCR sequence matching a GenBank sequence over the total length of the 5′RACE PCR sequence. The sequence of the known gene is present in the 5′RACE PCR sequence for clones 50, 31, 134, and C. For clone 18, the 5′RACE PCR sequence is contained in an EST sequence (AA310292) from which the 3′end overlaps (208/1902) with the 5′end sequence encoding an hnRNPF(L28010). % Identity is calculated with the number of nucleotides with a complete match in the overlapping GenBank sequence. (h) human; (m) mouse; (r) rat.
Comparison of the 5′ RACE PCR sequences to the GenBank database sequences indicated that sequences from clones 165/192, 11, and G correspond to EST sequences or do not match any known sequences, whereas clones 134, C, 50, 31, and 18 correspond to known genes (Table1 and Figure 4).
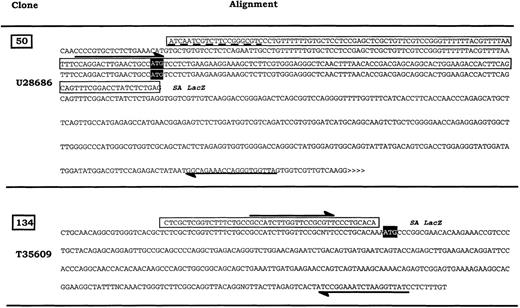
Generation of probes for expression analysis of 2 different cellular transcripts.
The selected cellular DNA sequences flanking retroviral integrations were compared with gene sequences in the GenBank database. The sequence “50” is similar to the human putative RNA-binding protein RNPL (U28686). The sequence “134” overlaps with the 5′ end EST sequence T35609 similar to human alpha NAC mRNA (X80909). Respective fragments were used as probes in subsequent Northern blot analyses. ⇌ Oligonucleotides used to amplify fragments from TF-1 total RNA ▭ 5′RACE sequence; >>>>> EST sequence continue; ---- Mismatches between alignment, SA: Splice acceptor, open reading frame for the production of the related proteins.
open reading frame for the production of the related proteins.
Generation of probes for expression analysis of 2 different cellular transcripts.
The selected cellular DNA sequences flanking retroviral integrations were compared with gene sequences in the GenBank database. The sequence “50” is similar to the human putative RNA-binding protein RNPL (U28686). The sequence “134” overlaps with the 5′ end EST sequence T35609 similar to human alpha NAC mRNA (X80909). Respective fragments were used as probes in subsequent Northern blot analyses. ⇌ Oligonucleotides used to amplify fragments from TF-1 total RNA ▭ 5′RACE sequence; >>>>> EST sequence continue; ---- Mismatches between alignment, SA: Splice acceptor, open reading frame for the production of the related proteins.
open reading frame for the production of the related proteins.
Transcriptional studies
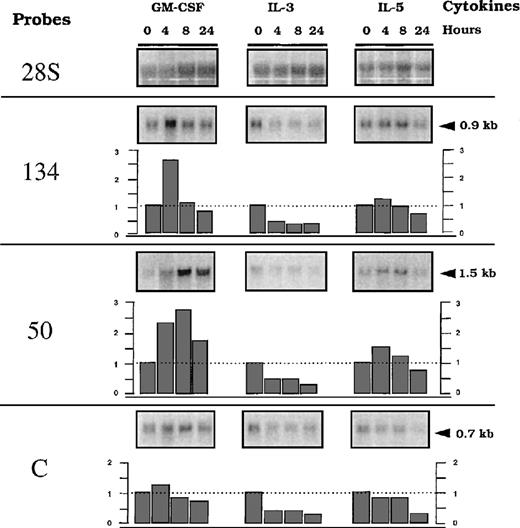
We further analyzed transcriptional modulation of 2 clones showing differential cytokine responses when tested in the presence of G418; ie, NACA, the alpha chain of the polypeptide-associated complex, recently identified as a coactivator of the transcription factor jun44 and RBM3, a protein from the hnRNP family. We included analysis of the expression of the transcript from clone C, with the presumed unregulated insertion. We used PCR amplification with 2 oligonucleotide primers derived from an identified sequence of the 5′ RACE PCR fragment and a sequence from the cDNAs encoding the known genes (Figure 4). Further Northern blots using these fragments as probes were performed with total RNA, including a time-course analysis (up to 24 hours) after stimulation with GM-CSF, IL-3, or IL-5. Figure 5 shows representative data. A basal level of expression was detected for each transcript, indicating that the integration events occurred in active units with low levels of constitutive transcription, because similar levels were found after long-term culture of the cells in the presence of different cytokines (results not shown). Half of the basal level remained detected with actinomycin D treatment for up to 8 hours, suggesting that it represented stable RNA (Figure6). From this basal level, specific GM-CSF modulation, estimated using the ratio of the [32P] signals associated with a specific band over the 28S, was observed forRBM3 and NACA. Maximum expression was reached at different times for the 2 loci: 4 hours for NACA versus 8 hours for RBM3. IL-3 and IL-5 did not significantly modulate the expression (ie, relative index less than 1 ± 0.5) of these loci. These results indicate that up-regulation of transcripts encodingNACA and RBM3 is associated with a GM-CSF–dependent proliferative response of the TF-1 cells. In addition, this modulation was not observed in the presence of actinomycin D and GM-CSF, indicating that GM-CSF–dependent up-regulation was transcriptionally controlled (Figure 6). To test the differentiation effect on the expression of the transcripts, we then investigated their behavior during monocytic/granulocyte differentiation of the TF-1 cells (24 hours exposure to PMA).36 45 The results showed that both transcripts were down-modulated in cells treated with PMA (Figure7).
Time-course analysis of the trapped genes RNA (50, 134, and C) after exposure of parental TF-1 cells to different cytokines.
RNA was obtained from TF-1 cells starved for 4 hours and stimulated with GM-CSF, IL-3, or IL-5, according to the indicated time course. Northern blot membranes were hybridized sequentially with the32P-labeled probes prepared for detection of 50, 134, (described in Figure 4) or C (described in “Materials and methods”). A probe of 28S RNA was used as a control to quantify the amount of RNA. Indices of modulation were calculated as a ratio of signals obtained with the specific probes versus the 28S probes, using a BIO-RAD Phosphor Analyst/PC program. Arrows represent the molecular weight markers in kilobases (kb).
Time-course analysis of the trapped genes RNA (50, 134, and C) after exposure of parental TF-1 cells to different cytokines.
RNA was obtained from TF-1 cells starved for 4 hours and stimulated with GM-CSF, IL-3, or IL-5, according to the indicated time course. Northern blot membranes were hybridized sequentially with the32P-labeled probes prepared for detection of 50, 134, (described in Figure 4) or C (described in “Materials and methods”). A probe of 28S RNA was used as a control to quantify the amount of RNA. Indices of modulation were calculated as a ratio of signals obtained with the specific probes versus the 28S probes, using a BIO-RAD Phosphor Analyst/PC program. Arrows represent the molecular weight markers in kilobases (kb).
Effect of actinomycin D on the expression of transcripts 50 and 134.
TF-1 cells were starved for 4 hours (lane 1) and exposed for 4 hours (transcript 134) or 8 hours (transcript 50) to either GM-CSF alone (lane 2), or actinomycin D alone (lane 3) or actinomycin D and GM-CSF. Total RNA was submitted to Northern blot analysis using specific probes for detection of transcripts 50, 134, or 28S as described in Figure5.
Effect of actinomycin D on the expression of transcripts 50 and 134.
TF-1 cells were starved for 4 hours (lane 1) and exposed for 4 hours (transcript 134) or 8 hours (transcript 50) to either GM-CSF alone (lane 2), or actinomycin D alone (lane 3) or actinomycin D and GM-CSF. Total RNA was submitted to Northern blot analysis using specific probes for detection of transcripts 50, 134, or 28S as described in Figure5.
Effect of PMA on the expression of 50 and 134 transcripts.
RNA from TF-1 cells exposed to either GM-CSF alone (−), or GM-CSF + PMA (+) were submitted to Northern blot analysis using specific probes for detection of transcripts 50, 134, and 28S, respectively. Relative indices of modulation were calculated as described in Figure 5.
Effect of PMA on the expression of 50 and 134 transcripts.
RNA from TF-1 cells exposed to either GM-CSF alone (−), or GM-CSF + PMA (+) were submitted to Northern blot analysis using specific probes for detection of transcripts 50, 134, and 28S, respectively. Relative indices of modulation were calculated as described in Figure 5.
To establish whether the GM-CSF up-regulation of the transcript expression uncovered using TF-1 cells was observed in primary cells, we used highly enriched CD34+ cells from peripheral blood, unstimulated or stimulated for 4 or 8 hours with either GM-CSF or IL-3. The transcripts were up-regulated (at 4 hours for NACA and 8 hours for RBM3) after GM-CSF stimulation (Figure 8).
RT-PCR analysis of the transcripts 50 and 134 from CD34+ cells.
mRNA from enriched CD34+ cells either nonstimulated (0) or stimulated (4 or 8 hours) with GM-CSF was RT. cDNA from each RT reaction was PCR-amplified using specific primers for the detection of a 445 bp in the RBM3 transcript or a 567 bp in the NACAtranscript sequences, together with the amplification of the β2 microglobulin transcript (268 bp). The products were electrophoresed on a 2% agarose gel, blotted, and hybridized with oligonucleotides within the RBM3, NACA, and β2 microglobulin (β2 m) respective amplified PCR region.
RT-PCR analysis of the transcripts 50 and 134 from CD34+ cells.
mRNA from enriched CD34+ cells either nonstimulated (0) or stimulated (4 or 8 hours) with GM-CSF was RT. cDNA from each RT reaction was PCR-amplified using specific primers for the detection of a 445 bp in the RBM3 transcript or a 567 bp in the NACAtranscript sequences, together with the amplification of the β2 microglobulin transcript (268 bp). The products were electrophoresed on a 2% agarose gel, blotted, and hybridized with oligonucleotides within the RBM3, NACA, and β2 microglobulin (β2 m) respective amplified PCR region.
Discussion
Rapid and transient expression of many genes is seen after stimulation with either the same or subfamilies of cytokines, suggesting that a specific intracellular response occurs further downstream and involves modulation of a variety of other genes. To identify such genes, we used a gene trap approach in the TF-1 premyeloid cell line and screened for genes regulated during proliferation in GM-CSF, IL-3, or IL-5, a group of cytokines that exhibit overlapping signal transduction intermediates.16
Comparison of the proliferative responses with different cytokines (GM-CSF, IL-3, or IL-5), in the presence of G418, resulted in the selection of differential cytokine-modulated trapped genes. With GM-CSF–cultured and infected cells, 5 days were necessary for selection of IL-3–unregulated genes. This result suggests differences in the proliferative behavior of GM-CSF grown cells when they are pre-exposed to IL-3 for 3 days, whereas this effect is no longer apparent between 3 and 5 days of the assay. It is likely that G418 resistance and sensitivity correlate with the mitogenic effect of the cytokine. Consequently, cells containing non-IL3–regulated retroviral insertions became sensitive to G418 at day 5 of the bioassay. In addition, it is possible that a residual activity of the Geo protein produced in GM-CSF–cultured cells maintains G418 resistance until a proliferation rate similar to that given by GM-CSF is reached. As 3 days were enough to select for IL-5–unregulated genes, IL-5 and GM-CSF act similarly in proliferation on GM-CSF–grown cells.
From IL-3– or IL-5–grown cells before the infection, the 10-day delay for selection of G418-resistant clones in the respective cytokine indicates a difference in the long-term proliferation of the TF-1 cells in IL-3 or IL-5. When the clones grew, they proliferated rapidly enough in the cytokine tested (ie, GM-CSF and IL-5 for the IL-3–cultured cells and GM-CSF and IL-3 for the IL-5–cultured cells) to display G418 sensitivity, when the expression of trapped genes was not sustained by the cytokines tested.
These results indicate that culturing the cells in a specific cytokine and maintaining the cytokine present at the time of infection favored the “turning on” of cytokine-dependent genes. This study shows that it is possible to select for specific-cytokine–modulated insertion site products among the other trapped genes. Identification of the trapped exon indicates that the insertion occurred in the 5′ sequence of the gene, which may contain 5′-specific sequences involved in the regulation of hematopoeisis. Most of the trapped genes were not previously known to be cytokine-regulated and may be important in the regulation of hematopoiesis. They are likely to be a target for earlier genes, participating in the specificity of 1 given cytokine response, because we selected for retroviral integration into GM-CSF or IL-3 or IL-5 active loci, using G418 sensitivity, and therefore favored selection of rather “late” cytokine-responsive genes.
Transcriptional analysis of 2 cloned cDNA sequence clones identical to known genes (NACA and RBM3) revealed that insertions occurred at loci transcribed constitutively at a Northern blot detectable level, from which a transcriptional specific GM-CSF–dependent up-modulation occurred in TF-1 and primary CD34+ cells. The genes trapped (NACA andRBM3) may represent a downstream response involving GM-CSF–specific signaling. Because part of the cytokine specificity resides in the composition of their receptors consisting of specific (α) and common (βc) chains,46 it is possible thatNACA and RBM3 are activated through either the alpha chain or the specific α and βc association chains of the GM-CSF receptor.
The importance of the GM-CSF modulation/PMA extinction of the described trapped proteins identical to RBM339 andNACA38 has not yet been explored in hematopoiesis. Recent data demonstrate that in osteoblasts, NACA47functions as a transcriptional coactivator, leading to sustained interaction between jun and its AP-1 DNA binding site. During monocyte-macrophage differentiation, the arrangement of the members of the AP-1 complexes determines the differentiation of a specific subset of hematopoietic precursor (reviewed by Liebermann et al48). The observed transcriptional GM-CSF/PMA regulation of NACA in proliferating or differentiated TF-1 cells may regulate the AP-1 partner complexes.
The other GM-CSF–dependent modulated molecule, RBM3, belongs to the hnRNP family. Several studies demonstrate that these proteins are multifunctional and participate at different steps of gene expression, including transcriptional and posttranscriptional events (reviewed by Dreyfuss et al,49 Weighardt et al,50 and Ladomery51). Different mechanisms have been associated with described hnRNPs, such as a shuttle between the cytoplasm and the nucleus,52 transcriptional transactivation,53 or a transcription factor.54Whether these processes are involved in the observed GM-CSF–dependent overexpression of hnRNP remains to be explored. Interestingly, a 3-fold cAMP-inducible hnRNP-like molecule was identified in a human B-cell line.55 This study reported that the control of the expression of the complement receptor 2 (CR2) gene involves up-regulation of this hnRNP and implies the recognition of a novel DNA motif.
Such described genes (NACA and RBM3) may be important in hematopoiesis because processes that govern cell proliferation/differentiation are undoubtedly complex and probably involve a combination of cell-specific (transiently) and widely expressed proteins. These genes were not previously known to be involved in hematopoiesis and may be important for cytokine specificity in maintaining GM-CSF–dependent myeloid precursor cells in a proliferative stage by preventing their differentiation.
Our results suggest that it is possible to trap genes differentially modulated by cytokines in the TF-1 cell line. The use of a panel of cytokines as modulators and transcriptional studies of the selected clones from the infected cells revealed specific modulation of the cytokine-responsive trapped genes. The modulations reported in the cell line were reproduced in primary cells, indicating that the regulation of the studied genes may be physiologically relevant. The mechanism involving NACA and RBM3 in maintaining GM-CSF–dependent proliferation or PMA-dependent differentiation remains to be established.
Acknowledgments
We thank Dr Soriano for the gift of the vector pRosaβgeo; Dr Chabannon and Thierry Alario for providing the CD34+ cells; Dr P. Pontarotti for help with computer analysis of databases; and E. Lecocq for technical assistance. We also thank laboratory members for helpful discussion and comments.
Supported in part by funds from INSERM (Institut National de la Santé et de la Recherche Médicale), FEGEFLUC (Féderation Nationale des Groupements des Entreprises Françaises dans la Lutte contre le Cancer), and ARC (Association de la Recherche contre le Cancer).
S.B. is a recipient of a predoctoral fellowship from the Ministère de la Recherche et de l'Enseignement.
Reprints:Sophie Gomez, Laboratoire de Cancérologie expérimentale, U119 INSERM, 27 Bld Leï Roure, 13009 Marseille, France; e-mail: gomez@marseille.inserm.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 2. Summary of the cytokine-dependent proliferative responses. / G418 resistant TF-1 clones dependent on the cytokine present at the time of infection (ie, GM-CSF, IL-3, or IL-5) were tested for their resistance in a proliferative assay (as described in “Materials and methods”). The rates of proliferation were compared in the absence (▭) or presence ( after 3 days; •• after 5 days) of G418 for each cytokine. Results are represented as the relative ratio of [3H]-thymidine incorporation (cpm) obtained without G418 to that with G418. The G418 resistance was evaluated after 3 or 5 days of culture by the amount of [3H]-thymidine uptake. After 72 hours with G418, TF-1 cells did not incorporate [3H]-thymidine. (A) Representative profiles of clones derived from cells infected in the presence of GM-CSF (GM-50, GM-C), or IL-3 (IL-3-4) or IL-5 (IL-5-16). GM-C represents a clone in which no specific cytokine modulations occurred. (B) Schematic representation of the results of [3H]-thymidine incorporation for the selected clones from cells infected in the presence of GM-CSF (upper panel), IL-3 (left lower panel), or IL-5 (right lower panel) infected cells. −: no proliferation, +/−: difference in proliferation when compared with and without G418, +: similar proliferation with and without G418.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/12/10.1182_blood.v95.12.3750/8/m_bloo01224002x.jpeg?Expires=1763631468&Signature=X~3xHy-PMPNJSAcbJdNtdZiVR1XELHZ-RIyCf0ThGc3OI8XKID~d7OKxBTpTV5PyKrKs-zSdtTNTmU23CFDu7lkoKwCE5MDdN1QjaddQeHJ3XHokU2Ed--qh7NZ3ptaw6pomtjuWSrs5BFcwV4J7hO73O04IqPwOPZ5EnSevYHaHLMbDqVtEJShObsbcXbu-aBp5HBaYrQpbNKKG1PU5XheN2Jn~~oVqQAXBIPocmt0fVRwFIoStO~VjEGmRDpp2tXFrzlfjTpvzYMAYK8is-9g~OcV2KKzHIep9SOkyS7a0axBVlsBZnFIfrm8LCozTjKEYl1mdKWYgmBVc9LPrYw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal