Abstract
Human T-cell lymphotropic virus type I (HTLV-I) transforms T cells in vitro, and the viral transactivator Tax functionally impairs the tumor suppressor p53 protein, which is also stabilized in HTLV-I–infected T cells. Thus, the functional impairment of p53 is essential to maintain the viral-induced proliferation of CD4+ mature T cells. However, in the CD4+ leukemic cells of patients with adult T-cell leukemia/lymphoma (ATLL), the viral transactivator does not appear to be expressed, and p53 mutations have been found only in a fraction of patients. We sought to investigate whether p53 function is impaired, in ex vivo samples from patients with ATLL, in the absence of genetic mutations. Here we demonstrate that the p53 protein is stabilized also in ex vivo ATLL samples (10 of 10 studied) and that at least in 2 patients p53 stabilization was not associated with genetic mutation. Furthermore, the assessment of p53 function after ionizing radiation of ATLL cells indicated an abnormal induction of the p53-responsive genes GADD45 and p21WAF1 in 7 of 7 patients. In 2 of 2 patients, p53 regulation of cell-cycle progression appeared to be impaired as well. Because p53 is part of a regulatory loop that also involves MDM2 and p14ARF, the status of the latter proteins was also assessed in cultured or fresh ATLL cells. The p97 MDM2 protein was not detected by Western blot analysis in established HTLV-I–infected T-cell lines or ex vivo ATLL cell lysates. However, the MDM2 protein could be easily detected after treatment of cells with the specific proteasome inhibitor lactacystin, suggesting a normal regulation of the p53–MDM2 regulating loop. Similarly, p14ARF did not appear to be aberrantly expressed in ex vivo ATLL cells nor in any of the established HTLV-I–infected T-cell lines studied. Thus, p53 stabilization in HTLV-I infection occurs in the absence of genetic mutation and alteration of the physiologic degradation pathway of p53.
Human T-lymphotropic virus type I (HTLV-I) infection is endemic in some areas of Africa, Asia, the Caribbean, and Central America.1 Twenty to forty years after infection, HTLV-I induces adult T-cell leukemia/lymphoma (ATLL) in only a fraction (1% to 3%) of persons infected with HTLV-I.2 This long latency suggests that multiple genetic events may occur that culminate in the development of ATLL, and the full spectrum of genes involved remains uncertain.
HTLV-I immortalizes human T cells in vitro3-6 within months of infection, and this model is useful for the identification of genes whose function must be altered to maintain the proliferation of mature CD4+ T cells. p53 stabilization is an early event after in vitro infection of human primary peripheral blood mononuclear cells (PBMC) by HTLV-I.7 It is found in all cells chronically infected with HTLV-I in the absence of genetic mutation,8,9 and p53 stabilization correlates with its functional impairment.8In transient transfection assays, ectopically expressed Tax, the viral transactivator, trans-suppresses p53 transcriptional activity10-12 and reverses p53-induced G1 arrest and apoptosis after DNA damage.10 In addition, T cells immortalized by HTLV-I Tax also have stabilized p53.13Endogenous p53, which is stabilized in HTLV-I–infected cells, seldom carries mutations8,9 but is nevertheless functionally impaired, as suggested by its inability to transactivate exogenously transfected p53-responsive promoters.10,11 It has been proposed that the first 52 amino acids of p53 are involved in Tax-induced stabilization and transcriptional inactivation of p53 and that phosphorylation of amino acid residues within amino-terminus amino acid 15 and between amino acids 387 and 392 is important for this effect.11
Under physiologic conditions, p53 activity is abrogated by the MDM2 oncoprotein that binds the p53 transcriptional-activation domain and blocks its ability to transactivate.14,15 In addition, MDM2 has a ubiquitin–Ligase activity for p53 protein, and the p53–MDM2 complex is targeted for proteasome-mediated degradation through the MDM2 carboxy terminus.16,17 This effect appears to be cell-type specific.18 Interestingly, the conservedBoxI region of p53 located at amino acid 13-19 appears to be essential for binding MDM2, and the lack of binding to MDM2 does not interfere with the ability of the p53ΔI mutant to induce cell-cycle arrest and apoptosis.18 The cleavage of MDM2 by caspase 3 generates a protein of 361 amino acids after the removal of its carboxy-terminus region. The main MDM2 protein product (human, p97; mouse, p90) has been demonstrated to be cleaved in apoptotic cells to a protein of 57 to 60 kd (p60),19 which retains its ability to bind p53. Recent evidence suggests that p60 can also be expressed at high levels in the absence of apoptosis.20 The complexity of the p53–MDM2 regulatory loop is further increased by the recent finding of another protein from the CDKN2A locus on chromosome 9q21 that binds to the p53–MDM2 complex. The CDKN2A locus, which encodes the p16INKα transcript, also generates an alternative transcript (β transcript)—designated as alternative reading frame (ARF)—which encodes a protein of 14 kd (p14ARF).21-27 The human p14ARF causes arrest in the cell-cycle progression with an accumulation of cells in both G1 and G2/M, and this event is associated with an increase in p53, p21CIP,1and MDM2 protein levels. The increase in p53 is thought to result from p14ARF binding to MDM2, which interferes with p53–MDM2 complex formation and proteasome degradation.16,17 Binding of p53 to MDM2 appears to be essential because the p53ΔI mutant, which does not bind MDM2, does not coprecipitate with p14ARF. Interestingly, p14ARF expression inhibited MDM2-induced turnover of p53 and stabilized both proteins independently of proteasome degradation.16 17
Our study addresses, for the first time, the functional status of p53 and the expression of regulatory MDM2 and p14ARF in uncultured ATLL cells, and it demonstrates that p53, in the absence of genetic mutations, is stabilized and transcriptionally impaired. In addition, the p53 regulation of cell-cycle progression appeared to be impaired in ex vivo ATLL cells.
In cultured HTLV-I–infected cells, in which p53 is also stabilized and functionally impaired, the MDM2 protein is undetectable, but MDM2 and p53 stability are increased by proteasome inhibitors, suggesting that p53 is in fact degraded in a physiologic manner. The expression on the same cells of p14ARF, though at a low level, may further contribute to p53 stabilization.
Patients, materials, and methods
Transfection, cell culture, irradiation, and lactacystin treatment
Normal PBMCs, obtained from healthy blood donors, were used as such or were stimulated with phytohemagglutinin (PHA; 4 μg/mL) and IL-2 (Boehringer Mannheim, Indianapolis, IN) (20 U/mL) after macrophage removal by elutriation and were maintained in RPMI containing fetal bovine serum (15%), l-glutamine (0.3 ng/mL), penicillin (100 U/mL), and streptomycin (100 μg/mL).
Exponentially grown cultures of normal PHA-activated PBMC and HTLV-I–infected T-cell lines were γ-irradiated with 10 Gy. After 3 hours, the cells were harvested, washed twice in phosphate-buffered saline (PBS), and lysed in the following lysis buffer: 10 mmol/L Tris-HCl, pH 7.4; 150 mmol/L NaCl; 0.5% NP-40; 1% Triton X-100; 2 mmol/L phenylmethylsulfonyl fluoride; 1 mmol/L dithiothreitol; 1 mmol/L aprotinin; 1 mmol/L leupeptin; and 1 mmol/L sodium orthovanadate. When appropriate, a proteasome inhibitor, lactacystin (synthetic; Calbiochem–Novabiochem, La Jolla, CA) was added to cell culture medium at 10 μmol/L for 8 hours.
293 T cells were transiently transfected with MDM2 cDNA (pMDM2)16 construct using Effectene Transfection Reagent as recommended by the manufacturer (Qiagen, Valencia, CA). For 24 hours before transfection, 5 × 104 cells were plated per well in a 6-well plate with 2 mL Dulbecco's minimum essential medium, 10% fetal bovine serum, 2 mmol/Ll-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. The amount of transfected DNA was normalized to 0.45 μg with pME18S. The protein lysates were used for Western blot analysis.
Detection of p53, MDM2, and p14ARF
For immunoblotting, 30 μg protein was loaded onto 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), subjected to electrophoresis, and electrotransferred to nitrocellulose membrane. For immunoprecipitation, the lysates (1 mg protein) were incubated with antibodies (Ab) at 4°C overnight. The immune complexes were precipitated with 40 μL 50% protein A–agarose for 1 hour at 4°C, washed 3 times in lysis buffer, separated on 10% SDS–PAGE, and electrotransferred to nitrocellulose membranes. For immunoprecipitation, 10 μg conformation-specific anti-p53 Ab 1620 (for wild-type conformation) and Ab 240 (for mutant conformation) (Ab5 and Ab3, CalBiochem, respectively) were used.
Antibody 240 binds to an epitope that is normally folded inside the p53 molecule where it cannot be accessed by the antibody. This antibody does not bind to wild-type p53 in its native conformation, and, because most p53 mutations cause a change in conformation of the p53 molecule that exposes this epitope, antibody 240 binds to mutant p53. Thus, the assay will detect any mutation in p53 that causes conformational change. However, this antibody will not detect some mutations that alter the epitope sequence (amino acid residues 212-217 of human p53 and 206-211 of murine p53). Antibody 240 detects both mutant and wild-type p53 if it is denatured, as in Western blot analysis, because this exposes the epitope and allows the antibody to bind. Antibody 1620 and antibody 7, which recognizes wild-type p53, were also used.
To detect p53 protein, the membranes were blocked with 3% bovine serum albumin (BSA) in PBST (PBS supplemented by 0.05% of Tween-20) for 1 hour at room temperature. Then the membranes were incubated with the first antibody, sheep anti-p53 antibody 7 (CalBiochem), diluted in 1% BSA–PBST at a ratio of 1:1000 for 1 hour at room temperature, and washed 3 times in PBST. The second antibody, antisheep–peroxidase conjugate (Jackson ImmunoResearch, West Grove, PA), diluted in 1% BSA–PBST at ratio of 1:8000, was added, and incubation was performed for 1 hour. In some experiments, 50 μg protein was loaded on an 8% SDS–PAGE, and the p53 antibody (FL-393; Santa Cruz Biotechnology, Santa Cruz, CA) and anti-rabbit second antibody were used.
To detect MDM2, 50 μg protein lysates were separated by 8% SDS–PAGE, transferred to a nitrocellulose membrane, and reacted with antibody against MDM2, which was obtained from Dr Jamil Momand (Beckman Research Institute, National Medical Center); the N-20 antibody directed to the MDM2 amino terminus was purchased from Santa Cruz Biotechnology.
For MDM2 immunoprecipitation, 450 μg protein was reacted overnight at 4°C with MDM2 antibody (N-20; Santa Cruz Biotechnology). Immunocomplexes, bound with protein A–agarose beads, were extensively washed with cold 1 × RIPA buffer and boiled in 1 × SDS–Laemmli buffer (Novex, San Diego, CA). Proteins were analyzed on an 8% SDS–PAGE gel, transferred to nitrocellulose membrane, and detected in Western blot analysis.
To detect p14ARF, 50 μg protein lysate was separated on by 16% SDS–PAGE, and the membrane was blotted with p14ARFantibody (C-20; Santa Cruz Biotechnology). For further confirmation, the antibody preparation was incubated with the appropriate peptide before blotting. Briefly, a 5-fold excess of the blocking peptide was combined with p14ARF antibody and incubated overnight at 4°C, and the mixture applied to the Western blot filter. After several washes in PBST, the filters were incubated in echochemiluminescence detection solutions (Amersham, Arlington Heights, IL) and exposed.
p53 DNA sequences
The DNA of patients 9 and 10, both of whom had ATLL, was studied for abnormalities in the p53 gene sequence. Fragments D, E, F, and G—comprehensive of exons 5, 6, 7, 8, and 9 of the p53 gene—were amplified using primers and polymerase chain reaction (PCR) conditions described by Murakami et al.25 PCR products were excised from 1% low-melting agarose, purified by Wizard PCR Prep DNA Purification Kit (Promega, Madison, WI), and directly sequenced using Big Dye terminator cycle sequencing kit by a DNA sequencer (ABI Prism 310-PE; Applied Biosystem, Branchburg, NJ). Sequencing reactions were performed with the same sense and antisense primers used in PCR amplifications.
Sequencing analysis of p53 exons 5 to 9, considered hot spots for point mutations,26 was performed on the DNA of patients 9 and 10. No point mutations or deletions were observed.
Patients
All patient samples were obtained after informed consent. The clinical diagnosis comprised HTLV-I serologic positivity in Western blot for HTLV-I antigens and the presence of clonal integration of HTLV-I in PBMC. Except for an HTLV-I carrier whose cells were collected from ascites, mononuclear cells were obtained from the blood of patients with ATLL and were separated on Ficoll and maintained in culture for the time necessary to assess the effect of ionizing radiation in RPMI with 10% fetal calf serum.
Results
Detection of low level of wild-type p53 induction in cultured HTLV-I–infected cells
Three IL-2–independent T-cell lines were used that were generated by the coculture of normal human PBMC with cells from patients with ATLL that expressed wild-type p53 (MJ, C91/PL, and C8166) and an IL-2–dependent line that carried a mutation in the fourth exon of p53 (E55/PL) that resulted in a change of codon 47 from a proline to a serine.8 Western blot analysis of total cellular protein lysates revealed higher levels of p53 in HTLV-I–infected cells than normal PBMC (Figure 1A; compare lane 1 to lanes 3, 5, 7, and 9), consistent with the previous observation that p53 is stabilized in HTLV-I–infected T lymphocytes.8,9After ionizing radiation, p53 expression was highly induced in PHA–PBMC but only to a low degree in HTLV-I–infected cells, regardless of their IL-2 requirement for growth (Figure 1A, lanes 2, 4, 6, 8, and 10). Although PHA–PBMC underwent apoptosis, HTLV-I–infected T cells, treated under the same conditions, were arrested in G2 as previously demonstrated8 (data not shown). To assess whether a conformationally altered p53 rather than a wild-type p53 was induced, we used an antibody with the ability to recognize wild-type p53 (antibody 1620) or conformationally altered p53 (antibody 240). Immunoprecipitation with antibody 1620 after ionizing radiation indicated that wild-type p53 expression is partly inducible, though at lower levels in HTLV-I–infected T cells than in normal PBMC (Figure1B). Surprisingly, p53 immunoprecipitation followed by immunoblot revealed apparently high levels of p53 in PBMC in the absence of DNA damage. It is possible that the variation of titers and the affinity of the antibody could explain this finding. The fact that a lower level of p53 was recognized by antibody 240 in normal PBMC and in the HTLV-I–infected cell lines does not indicate that p53 is mutated but rather that our denaturing conditions might have exposed the epitope on p53 recognized by this antibody. Thus, these results confirm and extend previous observations by others and us8 9 that p53 maintains its wild-type genotype in HTLV-I–infected cells.
p53 induction after ionizing radiation treatment of HTLV-I–infected cells.
(A) Western blot analyses of total protein lysate with the anti-p53 antibody 7. (B) Immunoprecipitation was performed with the antibody 1620 (for wild-type conformation) or 240 (for mutant conformation), followed by immunoblotting with anti-p53 antibody 7. + and − refer to the presence and absence, respectively, of ionization–radiation treatment.
p53 induction after ionizing radiation treatment of HTLV-I–infected cells.
(A) Western blot analyses of total protein lysate with the anti-p53 antibody 7. (B) Immunoprecipitation was performed with the antibody 1620 (for wild-type conformation) or 240 (for mutant conformation), followed by immunoblotting with anti-p53 antibody 7. + and − refer to the presence and absence, respectively, of ionization–radiation treatment.
p53 is stabilized in vivo
In ex vivo ATLL samples, the genetic mutation of p53 has been described in one fourth of the patients tested,28,29 but the status and function of p53 in primary ATLL cells have not been previously studied. Cell lysates from the Ficoll-separated PBMC from 5 patients with ATLL and 1 carrier of HTLV-I were obtained (Table1), and p53 expression was analyzed. In the 10 patients studied (an additional 5 patients are presented in Figure4), p53 appears to be stabilized, as demonstrated by the fact that, in normal PBMC, p53 is not detectable under these conditions (Figure2A, top). In the HTLV-I carrier cells analyzed (HC5), a degree of p53 stabilization was also observed, consistent with our in vitro data7 that p53 stabilization is an early event in HTLV-I infection and with the fact that in healthy carriers, the infected cells usually do exceed a few percentage points of the total PBMC.
Clinical features of patients with adult T-cell leukemia/lymphoma (ATLL)
| Patient . | Diagnosis . | White blood cell count (mm3) . |
|---|---|---|
| 1 | Acute ATLL | 77,000 |
| 2 | Acute ATLL | 62,000 |
| 3 | Acute ATLL | n/a |
| 4 | Acute ATLL | 7600 (97%) |
| 5 | HTLV-I carrier | n/a |
| 6 | Acute ATLL | n/a |
| 7 | Acute ATLL | 34,700 (93%) |
| 8 | Chronic ATLL | 36,900 (90%) |
| 9 | Acute ATLL | n/a |
| 10 | Acute ATLL | n/a |
| 30 | Acute ATLL | n/a |
| 31 | Acute ATLL | 11,000 |
| 32 | Acute ATLL | 56,100 |
| 33 | Acute ATLL | 30,600 |
| Patient . | Diagnosis . | White blood cell count (mm3) . |
|---|---|---|
| 1 | Acute ATLL | 77,000 |
| 2 | Acute ATLL | 62,000 |
| 3 | Acute ATLL | n/a |
| 4 | Acute ATLL | 7600 (97%) |
| 5 | HTLV-I carrier | n/a |
| 6 | Acute ATLL | n/a |
| 7 | Acute ATLL | 34,700 (93%) |
| 8 | Chronic ATLL | 36,900 (90%) |
| 9 | Acute ATLL | n/a |
| 10 | Acute ATLL | n/a |
| 30 | Acute ATLL | n/a |
| 31 | Acute ATLL | 11,000 |
| 32 | Acute ATLL | 56,100 |
| 33 | Acute ATLL | 30,600 |
n/a, not available; %, percentage of leukemic cells.
p53 stabilization and transcriptional impairment in ex vivo ATLL cells.
(A) Top panel: Western blot of total cellular lysates from ATLL cells from patients 1 to 4 and 6 or from HTLV-I carrier (HC 5) or normal human PBMC. Bottom panel: Northern blot analysis of the GADD45 and p21WAF1 expression after ionization–radiation treatment (+) of ATLL cells from patients 1, 2, and 4 and from HC 5.5In patients 7 to 10, protein lysates for p53 analyses were unavailable. The numbers designate the same patients in both top and bottom panels of the figure. (B, C) Quantitative summaries of the data presented in the bottom part of A as fold induction of GADD45 mRNA (B) or p21WAF1 mRNA after ionization–radiation. Results were obtained from a single experiment in each patient because of the intrinsic limitation of cells available.
p53 stabilization and transcriptional impairment in ex vivo ATLL cells.
(A) Top panel: Western blot of total cellular lysates from ATLL cells from patients 1 to 4 and 6 or from HTLV-I carrier (HC 5) or normal human PBMC. Bottom panel: Northern blot analysis of the GADD45 and p21WAF1 expression after ionization–radiation treatment (+) of ATLL cells from patients 1, 2, and 4 and from HC 5.5In patients 7 to 10, protein lysates for p53 analyses were unavailable. The numbers designate the same patients in both top and bottom panels of the figure. (B, C) Quantitative summaries of the data presented in the bottom part of A as fold induction of GADD45 mRNA (B) or p21WAF1 mRNA after ionization–radiation. Results were obtained from a single experiment in each patient because of the intrinsic limitation of cells available.
To demonstrate that p53 stabilization in ATLL cells correlated with its transcriptional impairment, normal PBMC and ex vivo ATLL cells were treated with ionizing radiation, and the expression of p53 and the subsequent induction of p53-responsive genes, such as p21WAF1 and GADD45, were quantitated. In normal PHA–PBMC and in resting PBMC, the mRNA encoding GADD45 and the p21WAF1 were induced 10-fold to 25-fold on DNA damage (Figures 2A-C). After DNA damage, Northern blot analysis indicated that the induction of the GADD45 and p21WAF1 mRNA was consistently lower in ATLL cells than was observed in control or resting PHA–PBMC (Figures 2A,B), which suggests p53 transcriptional impairment. In all cases, the induction of GADD45 and p21WAF1 expression was reduced by at least 50% (Figure 2B, C). Parallel experiments were performed to demonstrate equal loading of RNA using the glyceraldehyde-3-phosphate dehydrogenase probe, as previously described8 (data not shown). Altogether, these data demonstrated that in vivo p53 was stabilized in ex vivo ATLL cells and that, on DNA damage, the induction of the expression of p53-responsive genes was significantly less than in normal PBMC.
p53 regulation of cell-cycle progression is impaired in ATLL cells in the absence of genetic mutations
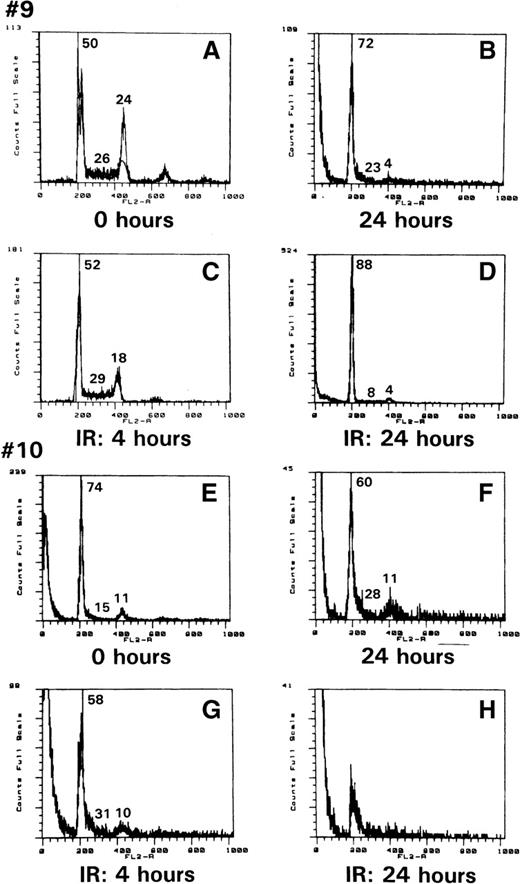
As demonstrated above, p53 was stabilized in ATLL cells and was impaired in its transcriptional activity, as indicated by the defective inductions of GADD45 and p21WAF1 expression. To assess whether any of the other functions of p53, such as G1-arrest induction or apoptosis in response to genotoxic stress, were altered in ATLL cells, PBMC from patients 9 and 10 were exposed to 10 Gy ionizing radiation, and their DNA content was analyzed by propidium iodide staining. As demonstrated in Figure 3, at the time of collection, the cells from patient 9 were cycling (Figure3A) and were mainly arrested in G1 at 24 hours in culture (Figure 3B). After ionizing radiation exposure, however, though cell death was observed (Figure 3C), apoptosis was not readily evident, and the remaining cells were arrested in G1 (Figure 3D). Similar events were also observed in the cells from patient 10. However, in this patient, necrosis was evident in most cells 24 hours after irradiation. Therefore, the effect of ionic radiation on ATLL cells does not parallel normal human PHA–PBMC, in which a clear pre-G1 apoptotic peak can be observed after ionic irradiation.8
ATLL cell resistance to ionization–radiation-induced apoptosis.
Propidium iodide staining of cells from patients 9 (A) and 10 (E) after Ficoll purification; after 24 hours in culture (B, F), or 4 or 24 hours after ionizing treatment (C and D for patient 9, G and H for patient 10). The number refers to the percentage of cells in G1, S, or G2/M.
ATLL cell resistance to ionization–radiation-induced apoptosis.
Propidium iodide staining of cells from patients 9 (A) and 10 (E) after Ficoll purification; after 24 hours in culture (B, F), or 4 or 24 hours after ionizing treatment (C and D for patient 9, G and H for patient 10). The number refers to the percentage of cells in G1, S, or G2/M.
Because we observed p53 stabilization in all patients with ATLL analyzed and the work of others has shown the presence of genetic mutation of p53 in one fourth of patients with ATLL,28 29we assessed whether p53 stabilization could occur in the absence of genetic mutation. Therefore, we obtained DNA from patients 9 and 10 in whose ATLL cells we had demonstrated p53 stabilization and functional impairment (Figures 2 and 3), and we analyzed the nucleotide sequence of exons 5, 6, 7, 8, and 9. In both circumstances, no mutations in the p53 exons were observed (data not shown), indicating that different mechanisms may underlie the p53 functional impairment in ATLL cells.
MDM2 and p14ARF expression in fresh and cultured ATLL cells
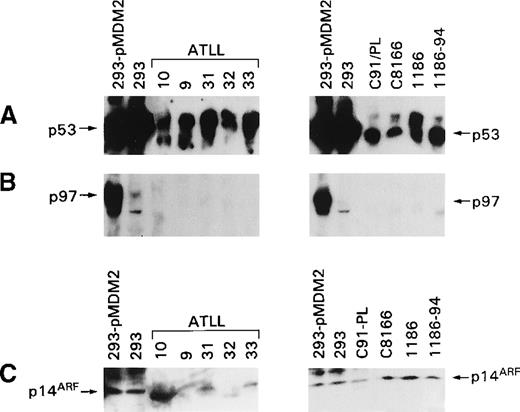
p53 Degradation is mediated by a complex regulatory loop that involves at least 2 other genes, MDM2 and p14ARF. Dysregulation of these 2 proteins' expressions could account for the stabilization of p53 observed in ex vivo ATLL cells and in HTLV-I–infected T-cell lines. Therefore, the expression of both proteins was studied in uncultured and cultured T cells using antibodies able to recognize the amino terminus of MDM2 and the carboxy terminus of p14ARF. In the ex vivo ATLL lysates and in the HTLV-I–infected T-cell lysates, the full-length p97 MDM2 protein was not detectable (Figure 4B, left panel), whereas p53 was readily detectable (Figure 4A). Analysis of p14ARF expression revealed the presence of low levels of p14ARF in some, but not all, ATLL cells, and in 2 patients (patients 10 and 32), the protein detected appeared to migrate faster than p14ARF (Figure 4C). In the cultured HTLV-I–infected cells, variable levels of p14ARF were also observed. The specificity of p14ARF detection was demonstrated by the decreased intensity of the band in the presence of specific blocking peptides (data not shown).
Expression of MDM2, p14ARF, and p53 in ex vivo ATLL cells and HTLV-I–infected cells.
(A) Detection of p53 with the antibody FL-393. (B) Detection of MDM2 with the N-20 antibody. (C) Detection of p14ARF with antibody (C20) that recognizes the p14ARF carboxy terminus.
Expression of MDM2, p14ARF, and p53 in ex vivo ATLL cells and HTLV-I–infected cells.
(A) Detection of p53 with the antibody FL-393. (B) Detection of MDM2 with the N-20 antibody. (C) Detection of p14ARF with antibody (C20) that recognizes the p14ARF carboxy terminus.
Effect of proteasome inhibitors on the p53–MDM2 complex in HTLV-I–infected cells
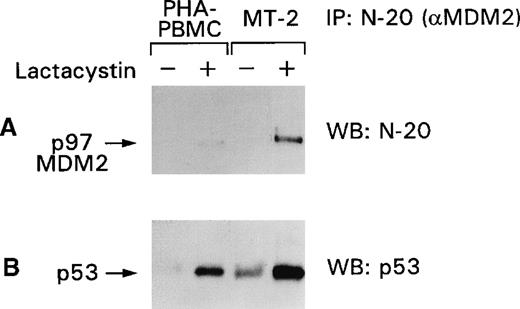
Among other genes, p53 induces the expression of MDM2, which in turn regulates p53 after translation by binding and targeting p53 to the proteasome. Because we were unable to detect the p97–MDM2 proteins in HTLV-I–infected cells (Figure 4), we investigated the status of the MDM2 protein by assessing the responsiveness of the p53–MDM2 complex to proteasome inhibitors. As demonstrated in Figure5A, p97–MDM2 became detectable by the N-20 antibody in PHA–PBMC and more so in the MT-2 cell-line lysates after lactacystin treatment. Immunoblot analysis of the same membranes with the anti-p53 antibody revealed that the amount of p53 bound to p97–MDM2 was also augmented (Figure 5B) and was higher in MT-2 than in PBMC, indicating that p53 is, in part, degraded in the proteasome. Similar results were obtained when the lysates of the C91/PL and C8166 cell lines were used in the same experimental conditions. These data suggest that though most of the p53 is stabilized in HTLV-I–infected cells, a portion of p53 is regulated by the MDM2 targeting of the complex to the proteasome.
p53 stabilization is increased by lactacystin treatment of PHA–PBMC and MT-2 cells.
Normal PHA–PBMC and MT-2 cells were treated with lactacystin, and the cell lysates were immunoprecipitated with the N-20 antibody. The blots were immunoblotted first with the N-20 antibody (A) and, after stripping, with the FL-393 antibody (B). + and − refer to the presence or absence of the drugs, respectively.
p53 stabilization is increased by lactacystin treatment of PHA–PBMC and MT-2 cells.
Normal PHA–PBMC and MT-2 cells were treated with lactacystin, and the cell lysates were immunoprecipitated with the N-20 antibody. The blots were immunoblotted first with the N-20 antibody (A) and, after stripping, with the FL-393 antibody (B). + and − refer to the presence or absence of the drugs, respectively.
Discussion
The immortalization and transformation mechanisms of CD4+ T cells by HTLV-I3-6 include multiple events that affect cell-cycle regulation and T-cell growth.7,30-34 In the last few years, numerous studies reported the alteration of a new class of tumor suppressor genes, the CDKI (p21WAF1, p27KIP1, p16INK4A, p15INK4B, p18INK4C, p19INK4D) in ATLL cells.31
In HTLV-I–infected T cells in vitro, the viral transactivator Tax plays a key role in maintaining a high expression of p21WAF1, 8,10-13,30,32-36 and it may be responsible for functional inactivation of the p53 protein,10-12 inactivation of the p16INK4Aprotein,30,33,34 and interference with the G2 mitotic point by binding to MAD-1.32 These multiple functions of Tax are thought to be at the basis of the unregulated growth of HTLV-I–infected cells in vitro. The impairment of p53 function appears to be essential as well because in HTLV-I infection, in vitro p53 appears to be stabilized very early.7,8,13 However, in uncultured ATLL cells, in which Tax is expressed at a very low level, if at all, mutations in the p53 gene have been found in a fraction of patients, whereas deletion of the p16INK4A gene appears to be frequent.37 Here we investigated the p53 functional status in vivo using ex vivo cells from 12 patients with ATLL and 1 carrier of HTLV-I.
We found that p53 was stabilized in uncultured ATLL cells, and in 2 patients we demonstrated that stabilization occurs in the absence of genetic mutation. Furthermore, stabilization correlated with functional impairment, as indicated by a defective induction of the p53-induced GADD45 and p21WAF1 mRNA after ionization–radiation exposure of leukemic cells. Although these data underscore the importance of p53 inactivation in vivo, the mechanism of p53 inactivation in ATLL cells remains unclear because mutation in the p53 gene has been reported in only one fourth of the patients. In this study we demonstrated that p53 stabilization can occur in the absence of genetic mutation. Because Tax does not appear to be expressed in ATLL cells, other mechanisms, including the mutation or alteration of other genes, may affect p53 function. The data presented here suggest that alteration of the p97–MDM2-p14ARF loop is not a likely mechanism of p53 inactivation of in vitro infected T cells because both genes are expressed within normal levels. However, because treatment with specific proteasome inhibitors was not performed on ATLL cells, the importance of this regulatory loop could not be directly assessed in vivo.
The results of our study suggest that, though in vitro Tax suppresses the activity of p53, the in vivo expansion of leukemic T-cell clones may require the silencing of viral expression to evade the host immune system and that, in the absence of Tax expression, pressure for selecting T cells with alteration in the p53 regulatory pathways may be high. Further studies on patients with ATLL will help to identify defects on other regulatory pathways and perhaps will provide tools to assess prognostic markers to evaluate the progression to ATLL in persons infected with HTLV-I.
Acknowledgments
We thank Steven Snodgrass for editorial assistance. We also thank Masao Matsuoka, Cathryn Lee, and Jeffrey White for help with the patient samples.
Supported in part by l'Istituto Superiore di Sanita', Rome, Italy (R.T.).
Reprints:Genoveffa Franchini, 41/0804 Basic Research Laboratory, Division of Basic Sciences, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal