To the editor:
FANCA protein binds FANCC and FANCG proteins in an intracellular complex
Dr Alan D'Andrea's group, having created antisera with specificity for the 3 well-characterized Fanconi anemia gene products FANCA, FANCC, and FANCG, has described an interesting model by which these 3 gene products interact. Using a variety of immunoprecipitation/immunoblot (“pull-down”) methods, they have reported that: (1) the FANCC and FANCA proteins interact1and they interact strongly,2 (2) the FA protein complexes are found in both the cytoplasm and nucleus of normal cells,3 and (3) the proteins bearing inactivating mutations do not interact. The findings have been controversial. Kruyt and Youssoufian, using reagents sent to them by D'Andrea, published work that concluded that the FANCC and FANCA proteins did not associate.4
An increasingly common feature of the Fanconi genes is that they encode proteins with no revealing domains. Any clues to their function in somatic cells are helpful to all investigators in the Fanconi research community. Consequently, the capacity of these proteins to associate, and D'Andrea's observations that the A and C proteins do not associate in most mutant cells—even those from afflicted children with complementation groups other than A or C—is an important model to confirm. Having focused on the hematopoietic function of the FANCC gene product for the past 6 years and recognizing the potential importance of the D'Andrea model, we have sought to test it directly.
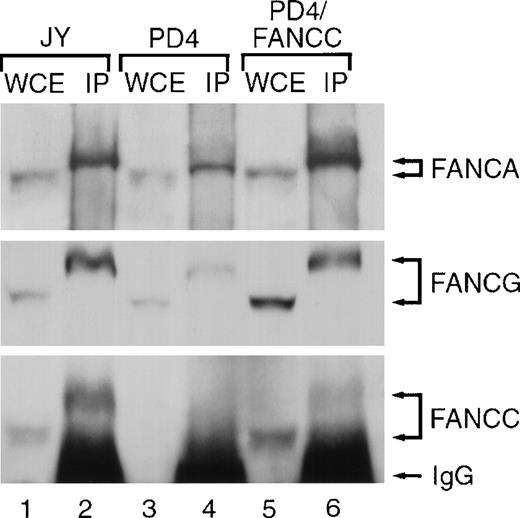
Using affinity-purified antisera from our own laboratory and antisera kindly provided to us by Dr D'Andrea, we found that immunoprecipitation with FANCA antisera resulted in a slower migration of all three FA proteins on the gel than the migration rate of these proteins electrophoresed as whole cell extracts (without an immunoprecipitation step). The differential rate of migration of FANCA, FANCG, and FANCC proteins is shown in Figure1. The apparent shift in molecular weight of the FANCC protein on our denaturing gel after immunoprecipitation was unexpected. However, it is our policy to include lysates of the cell prior to the immunoprecipitation step as one control lane in co-immunoprecipitation experiments. On each occasion we detected FANCC clearly at 58 kd, but after co-immunoprecipitation we detected FANCC at 64 kd. That the slowly migrating signal represented the FA proteins was supported by the observation that FANCC expressed in retrovirally complemented cells (PD4/FANCC) exhibited the identical migration differences (lower panel, lanes 5 and 6). Findings were identical whether we used our own FANCC antiserum or the FANCC antisera from Dr D'Andrea or whether we used lymphoblast cell lines or HeLa cells. Although this “shift” from 58 kd to 64 kd was not reported in the published work from D'Andrea, having shown our co-immunoprecipitation blots to the D'Andrea team, they stated that this phenomenon has been universal in their experiments.
The differential rate of migration of FANCA, FANCG, and FANCC proteins.
FANCA antiserum immunoprecipitates FANCA and co-precipitates FANCC and FANCG proteins. Whole cell extracts (WCEs) were prepared from lymphoblast lines including: JY (normal), PD4 (Fanconi group C mutant cells expressing no detectable FANCC protein) and PD4/FANCC (isogenic PD4 cells complemented with normal FANCC cDNA). Extracts (100 μg) were electrophoresed either without prior manipulation (lanes 1, 3, and 5) or following immunoprecipitation (IP) (3 mg for each sample) using affinity purified anti-FANCA anti-serum (lanes 2, 4, and 6). Fanconi anemia proteins in each lane were detected on immunoblots with affinity purified anti-FANCA, anti-FANCC, and anti-FANCG serum as indicated.
The differential rate of migration of FANCA, FANCG, and FANCC proteins.
FANCA antiserum immunoprecipitates FANCA and co-precipitates FANCC and FANCG proteins. Whole cell extracts (WCEs) were prepared from lymphoblast lines including: JY (normal), PD4 (Fanconi group C mutant cells expressing no detectable FANCC protein) and PD4/FANCC (isogenic PD4 cells complemented with normal FANCC cDNA). Extracts (100 μg) were electrophoresed either without prior manipulation (lanes 1, 3, and 5) or following immunoprecipitation (IP) (3 mg for each sample) using affinity purified anti-FANCA anti-serum (lanes 2, 4, and 6). Fanconi anemia proteins in each lane were detected on immunoblots with affinity purified anti-FANCA, anti-FANCC, and anti-FANCG serum as indicated.
Using conditions that exactly reproduced D'Andrea's, we have repeatedly confirmed quite unambiguously that the FANCC and FANCA proteins interact, that they are readily detected in whole-cell extracts, and that the FANCA and FANCG proteins interact. We hope that our confirmation of the D'Andrea model will help put this debate, one that has been associated with some degree of public acrimony, to rest. The model is instructive and compels us now to focus on the hundreds of questions that need to be answered about the function of these proteins on their own, in complex with each other, and in complex with other informative gene products with more well-understood functions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal