The hallmark of the 8p12 stem cell myeloproliferative disorder (MPD) is the disruption of the FGFR1 gene, which encodes a tyrosine kinase receptor for members of the fibroblast growth factor family.FGFR1 can be fused to at least 3 partner genes at chromosomal regions 6q27, 9q33, or 13q12. We report here the cloning of the t(8;9)(p12;q33) and the detection of a novel fusion betweenFGFR1 and the CEP110 gene, which codes for a novel centrosome-associated protein with a unique cell-cycle distribution. CEP110 is widely expressed at various levels in different tissues and is predicted to encode a 994-amino acid coiled-coil protein with 4 consensus leucine zippers [L-X(6)-L-X(6)-L-X(6)-L]. Both reciprocal fusion transcripts are expressed in the patient's cells. The CEP110-FGFR1 fusion protein encodes an aberrant tyrosine kinase of circa 150-kd, which retains most of CEP110 with the leucine zipper motifs and the catalytic domain of FGFR1. Transient expression studies show that the CEP110-FGFR1 protein has a constitutive kinase activity and is located within the cell cytoplasm.
Two distinct clinical syndromes have been associated with recurrent translocations of the 8p12 chromosomal region: stem-cell myeloproliferative disorder (MPD) and acute myeloid leukemia (AML). The MPD is characterized by B- or T-cell lymphoblastic leukemia/lymphoma, myeloid hyperplasia, and peripheral blood eosinophilia, and it generally progresses to AML (1for review and references therein). The second group of diseases consists of acute myelomonocytic or monocytic leukemia, predominantly associated with erythrophagocytosis [t(8;14)(p11;q11.1);2t(8;16)(p11;p13);3-9 t(8;19)(p11;q13);7t(8;22)(p11;q13)].10 The t(8;16) and t(8;22) translocations fuse the MOZ gene with the genes encoding the transcriptional coactivators CBP11 and p300,12respectively. In MPD, the 8p12 breakpoint, associated with at least 3 different partners—6q27, 9q33, and 13q12—disrupts the FGFR1gene,13 which encodes 1 of the 4 tyrosine kinase receptors for the fibroblast growth factors.14 Two breakpoints associated with FGFR1 have already been characterized.FGFR1 is fused to the FOP gene in t(6;8)(q27;p11)15 and to the FIM/RAMP/ZNF198 gene in t(8;13)(p11;q12).16-20 In both cases, the chimeric proteins with putative oncogenic properties contain potential oligomerization domains encoded by the 6q27 (FOP) and 13q12 (FIM) genes fused to the tyrosine kinase domain of FGFR1. FIM-FGFR1 is constitutively activated16 and predominantly cytoplasmic,20,21 and it has a constitutive dimerization capability mediated by the FIM N-terminus sequences.21
We report here the characterization of the third chromosomal rearrangement event occurring in 8p12 MPD in 1 patient. TheFGFR1 gene is fused to the CEP110 (centrosome protein 110) gene, which encodes a novel protein identified with human autoimmune sera and is associated with the centrosome. One of the 2 fusion products generated by the translocation t(8;9), CEP110-FGFR1 encodes an aberrant tyrosine kinase that is constitutively activated and localized in the cell cytoplasm.
Materials and methods
Patient
A 37-year-old man was examined for fatigue, weight loss, and gingival bleeding. He had hepatomegaly and splenomegaly. The hemogram showed hemoglobin 11 g/L, platelets 63 giga/L, leukocytes 97.8 giga/L with 21% neutrophils, 33% eosinophils, 1% basophils, 13% lymphocytes, 16% monocytes, and 16% immature granulocytes (3% myeloblasts, 7% promyelocytes, 4% myelocytes, 1% metamyelocytes, and 1% eosinophil myelocytes). The leukocyte alkaline phosphatase score was 0. Bone marrow was hypercellular and was characterized by hyperplasia of the granulocytic lineage (86% granulocytes, including 59% eosinophils). Both the peripheral blood and the bone marrow counts were consistent with a diagnosis of eosinophilic chronic leukemia. According to the ISCN nomenclature,22 the karyotype at diagnosis was 46,XY, t(8;9)(p12;q33)[8]/48, idem, +der(9)t(8;9);+21[12].
After 1 month of hydroxyurea (500 mg/d), the myeloproliferative syndrome accelerated to a monocytic blast crisis. The patient did not respond to several courses of chemotherapy induction and died 10 months later. Peripheral blood cells from the patient were obtained after informed consent was given.
Human cell lines and peripheral blood cells
The following cell lines were purchased from the American Type Culture Collection: JY (B-lymphoblastic ALL), U937 (histiocytic lymphoma), KG1 (myeloblastic/promyelocytic AML), Daudi (Burkitt's lymphoma), HSB-2, MO, and MOLT-4 (T-ALL), HEL (erythroleukemia AML), HL60 (promyelocytic AML) cultured in the presence or absence of phorbol esters, MIA PaCa-2 (pancreatic carcinoma), IMR-90 (lung fibroblast), A549 (lung carcinoma), HeLa (epithelioid carcinoma). The IE8 (pre-B/B stage ALL) and SU-DHL-1 (Ki-1 lymphoma cells) cell lines were gifts from T. LeBien (University of Minnesota Medical School, MN) and R. Rimokh (Hôpital E. Herriot, Lyon, France), respectively.
Peripheral blood cells isolated from blood samples obtained from healthy donors (Centre Régional de Transfusion Sanguine, Marseille, France) were fractionated (B and T lymphocytes, granulocytes, and monocytes) as previously described.23
CD34+ cells were purified from mobilized blood cells as described24 (kindly provided by C. Chabannon, Institut Paoli-Calmettes, Marseille, France).
Cloning and sequencing of the t(8;9) fusion cDNA
To isolate 1 of the t(8;9) fusion transcripts, 5′ RACE-PCR was performed using the Marathon cDNA amplification kit (Clontech, Palo Alto, CA). Specifically, the first-strand cDNA was synthesized from 2 μg total RNA from the patient by using primer F5R (Table1). Second-strand cDNA synthesis and cDNA adaptor ligation were performed according to the manufacturer's instructions. The fragment containing the fusion cDNA was amplified by 2 sequentially nested PCRs using F4R (Table 1) and adaptor-specific AP1 primers and then by F3R (Table 1) and adaptor-specific AP2 primers. Polymerase chain reaction (PCR) conditions were as follows: an initial denaturation step at 95°C for 5 minutes, followed by 30 cycles (denaturation, 94°C for 30 seconds; annealing, 60°C for 30 seconds; elongation, 68°C for 4 minutes), and a final extension at 68°C for 10 minutes in a DNA Thermal Cycler 480 (Perkin Elmer Cetus, Montigny-Le-Bretonneux, France). The PCR product was ethanol precipitated and cloned in the pUC18 plasmid by using Sure Clone Ligation Kit (Pharmacia, Uppsala, Sweden). Individual clones were manually sequenced with forward- and reverse-sequencing primers for pUC18 using T7Sequencing Kit (Pharmacia), and products were analyzed on 5% polyacrylamide/urea sequencing gels.
Primers
| Primer Name . | Sequence . |
|---|---|
| F3R | CTTGGAGGCATACTCCACGAT |
| F4R | ACTCTGGTGGGTGTAGATCCG |
| F5R | ATGGACAGGTCCAGGTACTCC |
| F9.2 | CATACTCAGAGACCCCTGCTAGC |
| FA | ATCATCTATTGCACAGGGGCC |
| CEPF | AATGGCAACAATTGAACTGGTAGC |
| CEPR | TCCAAATCTTTTTGGTTTGCC |
| RACE1 | TTAAGTGCTTCCTTCAGGCTGGCC |
| RACE2 | AACTGGTTCCGCAGAAGGCTGA |
| RACE3 | AAGTCTGCCCTCATAGAATCTGCC |
| RACE4 | TCTCCTCTAGCACTGCCACCTG |
| RACE5 | CATCTGCCTTAAAACAACCTGAAGA |
| RACE6 | TCTTCCTCTTTTTCAATTTCCTGC |
| RACE7 | ACGGATGAAACTGACGGACAAATAG |
| RACE8 | AAGAGTTCGGGGTCTAGCAGAAGA |
| β2MF | CCAGCAGAGAATGGAAAGTC |
| β2MR | GATGCTGCTTACATGTCTCG |
| SPINF | AAGCGGGAAGAAAGGTGG |
| SPINR | GGCTGAGGCATTCTTTTCC |
| 21REV3 | CCGCTCATGGTTGTCCTG |
| 21R4 | CTTCTGGAGTCGGGTCTGC |
| 21F4 | GAAGGACATCAGTGAATGGG |
| Primer Name . | Sequence . |
|---|---|
| F3R | CTTGGAGGCATACTCCACGAT |
| F4R | ACTCTGGTGGGTGTAGATCCG |
| F5R | ATGGACAGGTCCAGGTACTCC |
| F9.2 | CATACTCAGAGACCCCTGCTAGC |
| FA | ATCATCTATTGCACAGGGGCC |
| CEPF | AATGGCAACAATTGAACTGGTAGC |
| CEPR | TCCAAATCTTTTTGGTTTGCC |
| RACE1 | TTAAGTGCTTCCTTCAGGCTGGCC |
| RACE2 | AACTGGTTCCGCAGAAGGCTGA |
| RACE3 | AAGTCTGCCCTCATAGAATCTGCC |
| RACE4 | TCTCCTCTAGCACTGCCACCTG |
| RACE5 | CATCTGCCTTAAAACAACCTGAAGA |
| RACE6 | TCTTCCTCTTTTTCAATTTCCTGC |
| RACE7 | ACGGATGAAACTGACGGACAAATAG |
| RACE8 | AAGAGTTCGGGGTCTAGCAGAAGA |
| β2MF | CCAGCAGAGAATGGAAAGTC |
| β2MR | GATGCTGCTTACATGTCTCG |
| SPINF | AAGCGGGAAGAAAGGTGG |
| SPINR | GGCTGAGGCATTCTTTTCC |
| 21REV3 | CCGCTCATGGTTGTCCTG |
| 21R4 | CTTCTGGAGTCGGGTCTGC |
| 21F4 | GAAGGACATCAGTGAATGGG |
The sequence of the primers used in RACE experiments and in PCR assays to amplify either wild-type and chimeric transcripts are listed in a 5′-3′ orientation; the F and R at the end of the names indicate forward (sense) and reverse (antisense), respectively.
CEP110 cDNA cloning and analysis
Two independent approaches were used in the cloning of theCEP110 wild-type gene. (1) Autoimmune serum JK was identified from a serum bank of 25,000 autoimmune sera collected for testing at the Advanced Diagnostic Laboratory (University of Calgary, Canada). This serum was used at a dilution of 1:1000 to immunoscreen a HeLa 5′ stretch λgt11 cDNA expression library (Clontech), as previously described.25 From this screen, 4 reactive cDNA clones were identified. DNA sequencing of these clones revealed a single novel cDNA clone, designated 21.
Additional cDNA clones that overlapped and extended clone 21 were obtained by rescreening the HeLa cDNA library using specific 5′ and 3′ probes by DNA hybridization as described.25Each probe was labeled with [32P]dCTP (random primer labeling kit; Stratagene, La Jolla, CA) and was purified from free nucleotides using microspin G-50 columns (Pharmacia). All cDNA clones were cloned into the pBluescript vector (Stratagene), and the DNA sequence was determined using exonuclease III-generated deletions as described26 and the dye terminator cycle-sequencing ready-reaction kit (Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations. Reactions were analyzed at the University of Calgary DNA sequencing facility. Rescreening of the HeLa cDNA library was continued until the entire coding region of the centrosome protein was identified. The nucleotide and predicted amino acid sequence of CEP110 is available in GenBank under the accession number AF083322. All nucleic acid and protein sequence searches and analyses were conducted on the local network server using the BLAST search program.27
(2) Several rounds of RACE-PCR obtained the full-length cDNA for CEP110 from the t(8;9) patient's RNA. A first round of 5′ RACE-PCR was performed with primers derived from the aa52c11.r1 cDNA clone sequence (accession no. AA491 104). The single-strand cDNA was obtained from 2 μg total placental mRNA using the specific primer RACE1 (Table 1). Second-strand cDNA and adaptor ligation were conducted according to the manufacturer's protocol. Nested PCR was performed using primer pairs RACE2-AP1 and RACE3-AP2 (Table 1). The PCR conditions were the same as described above. Amplified fragments were size-fractionated by agarose gel electrophoresis and cloned in the pUC18 vector. Individual clones were sequenced at Génome Express (Grenoble, France) using an automated sequencer (Applied Biosystems 373; Applied Biosystems). Sequence comparisons with GenBank and dbEST entries were made using BLASTN and TBLASTN.27 These resulted in the retrieval of several cDNA clone sequences. Specific primers RACE4, RACE5, and RACE6 (Table 1) were derived from 1 of the cDNA clones (clone 71) and were used for a second round of 5′ RACE-PCR. A third round of 5′ RACE-PCR was performed using RACE4 primer and nested PCR with RACE7 and RACE8 reverse primers derived from the 5′ untranslated region of CEP110 (see “Results”) and the AP1 and AP2 adaptor primers, respectively. PCR product cloning and sequencing were performed as described above.
Northern blot analyses
The multiple-tissue Northern blots (Clontech; human, no. 7759-1, 7760-1, and 7767-1; mouse, no. 7762-1) were hybridized according to the manufacturer's instructions. Human probes used were a 0.6-kb fragment obtained from the third round 5′ RACE PCR using RACE8 reverse primer corresponding to the human CEP110-5′ untranslated region; a 2.5-kb cDNA fragment obtained from the second round of 5′ RACE-PCR with RACE6 primer corresponding to the insert of cDNA clone 71; and a 1.2-kb insert fragment from cDNA clone mu38gO2 (accession no. AA209914) corresponding to the 3′ part of murineCep110. All probes were labeled with [32P]dCTP in random priming reactions.
Wild-type and fusion gene expression analysis by RT-PCR
Reverse-transcription reactions (RT) were performed using 2 μg total RNA, random hexanucleotides, and SuperScript II reverse transcriptase (Gibco-BRL, Burlington, Ontario, Canada) according to standard procedures. The equivalent of 500 ng reverse-transcribed RNA from either t(8;9) patient's leukemic cells or various cell lines was used for PCR detection of wild-type and fusion gene expression. The primer pairs (Table 1) were as follows: FGFR1 (FA-F9.2, PCR product, 259 bp); CEP110 (CEPF-CEPR, PCR product, 130 bp); FGFR1-CEP110 (FA-CEPR, PCR product, 177 bp); and CEP110-FGFR1 (CEPF-F9.2, PCR product, 212 bp). All PCR amplifications were as follows: initial step of denaturation (95°C for 5 minutes); 30 cycles (denaturation, 95°C for 30 seconds, annealing, 58°C for 30 seconds, extension 72°C for 1 minute); final extension step (72°C for 10 minutes). The amplified fragments were gel-purified, cloned in the pUC18 plasmid, and sequenced as described above. Human β2 microglobulin primer pair (β2MF-β2MR; see Table 1; PCR product, 268 bp) was used as control to estimate reaction efficiencies.
PCR amplification of t(8;9) breakpoints on derivative chromosome 8
Using FA-CEPR primer pair (Table 1), the fusion junctionFGFR1-CEP110 (der(8) chromosome fragment of 272 bp) was amplified from the patient's genomic DNA. PCR products were cloned and sequenced as described above.
Autokinase activity and tyrosine phosphorylation analysis of the CEP110-FGFR1 fusion protein
106 NIH3T3 or Cos-1 cells were transiently transfected using 10 μg reconstructed full-length CEP110-FGFR1cDNA inserted into the pcDNA3 expression vector (Invitrogen) or 10 μg empty vector and 30 μL FuGENE 6 transfection reagent (Roche Diagnostics, Meylan, France) following the manufacturer's recommendations. Twenty-four hours after transfection and 2 hours after serum starvation, cells were cultured in the presence or absence of 10 ng/ml FGF1 plus 10 μg/mL heparin. The FGFR1-expressing cell line NFlg 2628 was used as a positive control. Cell lysates and immunoprecipitation using an antibody directed against an FGFR1 C-terminal peptide (anti-C-FGFR1) (C15; Santa Cruz Biotech, Santa Cruz, CA) were performed as described.29 Half of each immunoprecipitate was challenged for autokinase activity in the presence of γ[32P]ATP and 5 mmol/L MnCl229 and was analyzed by electrophoresis and autoradiography. Phosphorylation on tyrosine and level of expression were analyzed by immunoblotting with an anti-phosphotyrosine antibody (4G10; UBI, Lake Placid, NY) and an anti-C-FGFR1 antibody, respectively.30 31
Recombinant protein production and antibody generation
For antibody Ab72, a 510-bp Ecl136 II/Pvu II digestion fragment of CEP110 (corresponding to amino acids 805-972) was cloned into the Sma I site of the glutathione-S-transferase (GST) expression vector pGEX 3 × (Pharmacia). Antibody 5′2 was generated against amino acids 5-103 of CEP110. Large-scale protein inductions (500 mL) were carried out, and the GST-CEP110 fusion proteins were injected into New Zealand White rabbits for the production of polyclonal antibody as previously described.25
Affinity purification
Purified GST-CEP110 fusion protein and control GST bacterial lysates were dialyzed against 100 mmol/L MOPS (3-[N-Morpholino] propanesulfonic acid) (pH 7) overnight at 4°C. Each protein sample was then covalently coupled to 1 mL Affigel 10 beads (Bio Rad Laboratories, Hercules, CA) according to the manufacturer's instructions in a total volume of 5 mL overnight at 4°C. The protein-bead slurry was then packed into a 10-mL econo-column (Bio Rad Laboratories). Antibacterial and anti-GST antibodies were depleted from 4 mL rabbit serum (Ab72 and Ab5′2) by repeated passage over the protein-bead column. This precleared serum was then repeatedly passed over the GST-CEP110 fusion protein column for 2 to 4 hours at room temperature. The column was washed with 25-bed volumes of tris-buffered saline. Antibody was eluted by passing 15 mL elution buffer (5% acetic acid. 0.15 mol/L NaCl2) over the column and collecting 1-mL fractions into 150 μL 1 mol/L tris-HCl (pH 9). The absorbency of each fraction was then read at OD 280, and all protein-containing fractions were pooled and dialyzed against Dulbecco's phosphate-buffered saline (DPBS) overnight at 4°C. The dialyzed antibody was then concentrated in a Centricon 30 (Amicon, Oakville, Ontario, Canada) according to the manufacturer's instructions.
Indirect immunofluorescence
HeLa cells grown on coverslips were extracted in 0.5% Triton X-100 in DPBS for 2 minutes and fixed in 100% methanol at −20°C for 10 minutes. The coverslips were then blocked with 1:200 dilution of normal goat serum (NGS) in DPBS at room temperature for 30 minutes and then incubated with a 1:50 dilution of affinity-purified anti-CEP110 antibody (Ab72) in NGS-DPBS for 30 minutes at 37°C. After a wash in DPBS, the coverslips were incubated with a 1:10 000 dilution of antipericentrin antibody in NGS-DPBS and further incubated for 30 minutes at 37°C. The coverslips were then washed and incubated with a mixture of Cy3-conjugated antirabbit 1:400 and fluorescein isothiocyanate (FITC)-conjugated antimouse 1:400 secondary antibody (Jackson Laboratories, Oak Grove PA) in NGS-DPBS for 30 minutes at 37°C. After incubation the coverslips were washed in DPBS, counterstained with DAPI (4′,6′-diamidino-2-phenylindole), and mounted in 90% glycerol containing paraphenylenediamine. Cells were observed using a Leica (Heidelberg, Germany) DMRB microscope using a 100× objective. Images were recorded with an RTE/CCD detector (Princeton Instruments, Trenton, NJ) using IPLab Spectrum software (Signal Analytics, Vienna, VA).
Cos-1 cells were grown as monolayers on coverslips the day before transfection (2 × 105 cells/60-mm plates) using 2 μg plasmid DNA and 3 μL FuGENE transfection reagent. Twenty-four hours after transfection, cells were washed once in PBS and fixed in 3.7% paraformaldehyde in PBS for 15 minutes at room temperature. After extensive PBS washes, cells were permeabilized and blocked in 5% fetal calf serum PBS and 0.1% Triton X 100 for 15 minutes. Cells were incubated with either the purified anti-CEP110 antibodies (Ab72 or Ab5′2) or the human immune serum, each used at 1:20 dilution for 1 hour at room temperature, rinsed several times in PBS, and incubated for at least 1 hour with either the Alexa-conjugated antirabbit 1:500 (Molecular Probes, Eugene, OR) or FITC-conjugated F(ab′)2 fragment goat antihuman IgG, FCγ (Immunotech, Marseille, France) secondary antibodies, respectively. Coverslips were then washed several times in PBS and incubated for 1 hour in PBS containing 25 μg/mL 7-AAD (7-aminoactinomycin D; Molecular Probes) used to visualize DNA. After several washes with PBS, coverslips were mounted in Mowiol. Cellular localization of proteins was analyzed by confocal laser system microscopy using a TCS NT Leica apparatus.
Green fluorescent protein transfection constructs and transient transfection
Various portions of the protein-encoding CEP110 cDNA were cloned in-frame into pEGFP-N1 (Clontech), a red-shifted green fluorescent protein (GFP) eukaryotic expression vector (see Figure 7). A 2957-bp fragment of the coding region of CEP110 was amplified by PCR using the primers SPINF and SPINR (Table 1) and was cloned into the Ecl136 II site of pEGFP-N1 to create the construct CEP110-GFP. Fragment A was amplified by PCR using the primers SPINF and 21REV3 (Table 1) and was cloned to the Ecl136 II site of pEGFP-N1 to create expression construct A. Fragment B was amplified by PCR using the primers SPINF and 21R4 (Table 1) and was cloned to the Ecl136 II site of pEGFP-N1 to create construct B. Fragment C was generated by Dra I/Eco RV digestion of the full-length CEP110 cDNA and was cloned to the Eco47III site of pEGFP-N1 to create construct C. Fragment D was generated by digesting the full-length CEP110 cDNA with EcoR V and cloning the C-terminal fragment to the Eco47 III site of pEGFP-N1 to create construct D. Fragment E was generated by digesting full-length CEP110 cDNA with Eco RV and Ecl136II and was cloned to the Eco47 III site of pEGFP-N1 to create construct E. Fragment F was amplified by PCR using the primers 21F4 and SPINR (Table 1) and was cloned to the Ecl136 II site of pEGFP-N1 to create construct F. Fragment G was amplified by PCR using the primers 21F4 and 21REV3 (Table 1) and was cloned to theEcl136 II site of pEGFP-N1 to create expression construct G.
HeLa cell coverslips at 30% to 40% confluence were washed briefly in serum-free medium (Opti-MEM; Gibco-BRL) and were incubated with 2 μg DNA-lipofectin reagent (Gibco-BRL) in Opti-MEM for 3 hours. The transfection medium was then replaced with complete medium (DMEM, 10% fetal calf serum). Twenty-four hours later transfected cells were extracted for 1 minute in 0.5% Triton-X-100 in DPBS and fixed in 4% paraformaldehyde (Sigma, Oakville, Ontario, Canada) in DPBS for 10 minutes. The coverslips were then processed for indirect immunofluorescence with antipericentrin antibody 1:10 000 as described above.
Results
FGFR1 chromosomal translocation
In a previous work we showed that the t(8;9) breakpoint interrupts the FGFR1 locus.13 To determine the fusion partner of FGFR1 in a t(8;9) MPD, RNA was isolated from the patient's leukemic cells and subjected to RACE using nested primers withinFGFR1 exon 14. Two of the retrieved clones were sequenced and had novel sequences upstream of FGFR1 exon 9. To clone more of the 5′ region of the FGFR1 partner gene, 2 rounds of 5′ RACE-PCR were performed, resulting in a 2.8-kb cDNA contig. Database searches with the novel sequence showed 100% identity with several expressed sequence tags (ESTs) and with the CEP110mRNA. Of the retrieved ESTs, EST AA491104, which surrounded the t(8;9) breakpoint, is derived from the cDNA clone aa52c11. This cDNA clone was entirely sequenced and had 100% identity with the 3′ region of CEP110 (GenBank accession no. AF083322), a novel gene that was identified by screening a cDNA expression library with a human autoimmune serum containing autoantibodies to the centrosome. During the cloning of this autoantigen, a set of cDNA clones that showed no significant similarity with any protein of the GenBank database was assembled and spanned 3893 bp. An open-reading frame (ORF) of 2982 bp in this cDNA initiates at nucleotide position 473 and is preceded by several in-frame stop codons. A termination codon for this ORF is found at nucleotide position 3455. Two potential poly(A)+ initiation signals (AATAAA) are found at position 3866-3871 and 3887-3892. During the independent cloning of CEP110 as a novel antigen and as the FGFR1 partner in the t(8;9) multiple CEP110cDNA variants, were identified with different 5′ noncoding and coding regions. Of them, cDNA clones were identified that contained a 141-bp in-frame deletion between nucleotide positions 2755 to 2896 in the ORF. These results indicated that the CEP110cDNA is alternatively spliced.
A search of GeneMap 98 from NCBI (http://www.ncbi.nlm.nih.gov/genemap98/map/loc.cgi?) resulted in the identification of a sequence-tagged site from the Whitehead Institute (Cambridge, MA), WI-11 957, specific toCEP110 and EST AA499 904. As described in the GeneBridge 4 radiation hybrid database, WI-11 957 maps to 9q33, the region of the chromosome 9 breakpoint involved in the t(8;9).
Features of the deduced amino acid sequence of CEP110
The predicted ORF of CEP110 cDNA codes for 994 amino acids. From the deduced amino acid sequence, the molecular weight of CEP110 was calculated as 116 813 d. A GenBank database search with the amino acid sequence revealed no significant similarity with known proteins. However, regions of weak similarity could be observed with a number of coiled-coil proteins, including CENP-E, myosin heavy chain, and kinesin light chain. Analyzing the amino acid sequence with the program COILS32 showed that CEP110 is predicted to have extensive stretches of coiled-coil structure throughout much of its sequence, except between amino acids 65-80, 295-320, and 810-850, and last 114 amino acids of the C-terminus (data not shown).
A protein MOTIF search (http://www.motif.genome.ad.jp/motif-bin/nph-motif2) revealed the possibility of 1 amidation site, 2 N-myristoylation sites, and 1 N-glycosylation site. Furthermore, there are 2 potential cyclic adenosine monophosphate–phosphorylation sites, 19 casein kinase II phosphorylation sites, and 20 protein kinase C phosphorylation sites. In addition, there are 4 predicted leucine zippers (L-X(6)-L-X(6)-L-X(6)-L) at amino acid positions 28-49, 97-118, 496-517, and 689-710 (Figure 1). There are no consensus sequences for microtubule or nucleotide binding. Overall, CEP110 is an acidic protein with a predicted pI of 5.43.
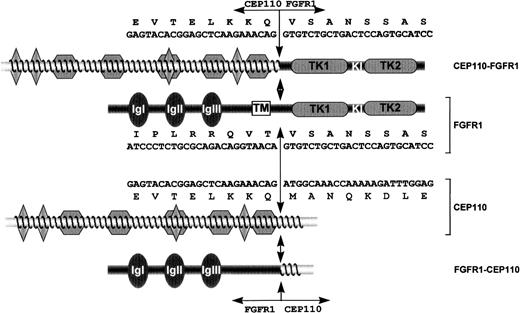
Schematic representation of FGFR1, CEP110, and both CEP110-FGFR1 and FGFR1-CEP110 chimeric proteins.
FGFR1 domains are indicated as follows: IgI, IgII, IgIII, the 3 immunoglobulin-like domains; TM, the transmembrane domain; TK1 and TK2, the tyrosine kinase 1 and 2 subdomains interrupted by a kinase insert (KI). CEP110 regions are indicated as follows: diamonds, leucine zipper motifs; waved lines, predicted coiled-coil region. Double arrows indicate the t(8;9) breakpoint. Nucleotide and amino acids sequences are indicated for CEP110-FGFR1 and both wild-type products.
Schematic representation of FGFR1, CEP110, and both CEP110-FGFR1 and FGFR1-CEP110 chimeric proteins.
FGFR1 domains are indicated as follows: IgI, IgII, IgIII, the 3 immunoglobulin-like domains; TM, the transmembrane domain; TK1 and TK2, the tyrosine kinase 1 and 2 subdomains interrupted by a kinase insert (KI). CEP110 regions are indicated as follows: diamonds, leucine zipper motifs; waved lines, predicted coiled-coil region. Double arrows indicate the t(8;9) breakpoint. Nucleotide and amino acids sequences are indicated for CEP110-FGFR1 and both wild-type products.
Characterization of the fusion junctions in the t(8;9)
We next examined the nucleotide sequence spanning the t(8;9) breakpoint on both parental and derivative chromosomes. RT-PCR assays localized the breakpoint at positions 1259 and 2424 from the start codon in the FGFR1 (accession no. M34 185) and CEP110mRNA, respectively. The translocation leads to the formation of the two reciprocal transcripts. CEP110-FGFR1, transcribed from chromosome derivative 9, encodes a large protein containing all 4 leucine zipper motifs of CEP110 at its N-terminus, and the catalytic domain of FGFR1 at its C-terminus (Figure 1). The junction sequence is described above the representation of the fusion protein in Figure 1.FGFR1-CEP110, transcribed from chromosome derivative 8, encodes a protein containing the FGFR1 N-terminal region with its ligand-binding and transmembrane domains and the CEP110 C-terminal region (Figure 1).
To determine the precise position of the t(8;9) breakpoints, genomic DNA from the patient's leukemic cells was PCR-amplified (FA-CEPR primers; Table 1), sequenced, and compared with the sequence of normal human genomic DNA. In FGFR1, the breakpoint is localized in exon 8, corresponding to position 1259 from ATG, 12 bp upstream of the exon8/intron 8 boundary. In CEP110, the breakpoint occurs at position 1658 of an intron of 1744 bp. The breakpoint intron ofCEP110 contains AluJb and MIR elements (Mask Repeat Sequence, http://ftp.genome.washington.edu/).
Gene expression
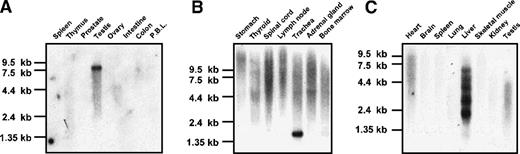
Hybridization of a CEP110 probe to human multiple-tissue Northern blots identified 3 main transcripts of approximately 7.5, 4.5, and 1.5 kb with different rates of expression. A high-level expression of the 7.5- and 1.5-kb transcripts was seen in testis (Figure2A) and trachea (Figure 2B), respectively. The 7.5-kb transcript was weakly expressed in the majority of tissues, including ovary (Figure 2A), trachea, adrenal gland, and bone marrow (Figure 2B). In addition, CEP110 transcripts were barely expressed in thymus and peripheral blood cells (Figure 2A) and heart, brain, and liver (data not shown). To further analyze Cep110 expression, we hybridized a murine Cep110 probe to a murine Northern blot. Six main transcripts were revealed in mouse liver that varied in size from 1.5 kb to 9 kb (Figure 2C).
CEP110 expression.
(A,B) Indicated human poly(A)+ RNA from Clontech Northern blots was hybridized with CEP110 probes either derived from the cDNA insert of CEP110 clone 71 (A) or the 5′ untranslated probe (B). (C) Mouse poly(A)+ Northern blot (Clontech) hybridized with a CEP110 probe from cDNA clone mu38gO2. The marker sizes (in kb) are indicated on the left.
CEP110 expression.
(A,B) Indicated human poly(A)+ RNA from Clontech Northern blots was hybridized with CEP110 probes either derived from the cDNA insert of CEP110 clone 71 (A) or the 5′ untranslated probe (B). (C) Mouse poly(A)+ Northern blot (Clontech) hybridized with a CEP110 probe from cDNA clone mu38gO2. The marker sizes (in kb) are indicated on the left.
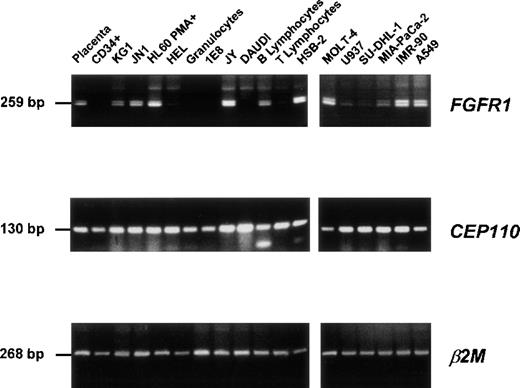
The expression of CEP110 and FGFR1 wild-type genes was assessed by RT-PCR assays from either the t(8;9) patient or various normal and malignant hematopoietic cells (Figure3). Both types of gene were expressed in all tested cells (Figure 3). In contrast, the 2 reciprocal fusion transcripts were only found in the t(8;9) leukemic cells (Figure4). Sequence analysis confirmed that these unique products contained fusions of FGFR1 and CEP110sequences, as expected.
Expression of FGFR1 and CEP110 genes.
RT-PCR products were obtained from a variety of tissues and normal and tumoral hematopoietic cells (listed in “Materials and Methods”) using specific primer pairs of each gene (FGFR1, FA-F9.2; CEP110, CEPF-CEPR; see Table 1). Each panel is a photograph of the ethidium bromide-stained agarose gel in which PCR products were electrophoresed. β2 Microglobulin (β2 mol/L) amplification was used to estimate the efficiency of RT-PCR reactions. Labels of the source of the material and PCR product sizes are indicated at the top of rows and on the left, respectively.
Expression of FGFR1 and CEP110 genes.
RT-PCR products were obtained from a variety of tissues and normal and tumoral hematopoietic cells (listed in “Materials and Methods”) using specific primer pairs of each gene (FGFR1, FA-F9.2; CEP110, CEPF-CEPR; see Table 1). Each panel is a photograph of the ethidium bromide-stained agarose gel in which PCR products were electrophoresed. β2 Microglobulin (β2 mol/L) amplification was used to estimate the efficiency of RT-PCR reactions. Labels of the source of the material and PCR product sizes are indicated at the top of rows and on the left, respectively.
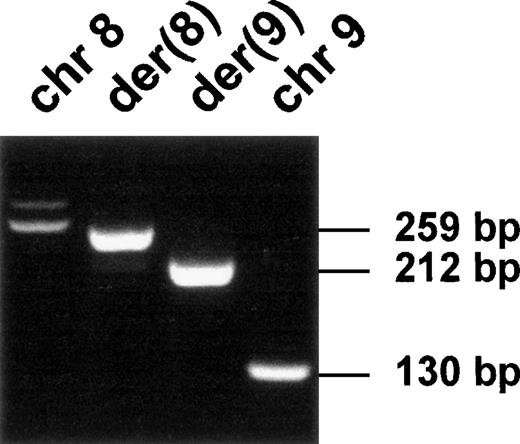
Expression of the fusion transcripts.
RT-PCR was performed using RNA from the t(8;9) patient's malignant cells and specific primers located near the translocation breakpoint (FA-CEPR and CEPF-F9; see Table 1). Chromosomal positions and transcript sizes are indicated at the top and on the right, respectively.
Expression of the fusion transcripts.
RT-PCR was performed using RNA from the t(8;9) patient's malignant cells and specific primers located near the translocation breakpoint (FA-CEPR and CEPF-F9; see Table 1). Chromosomal positions and transcript sizes are indicated at the top and on the right, respectively.
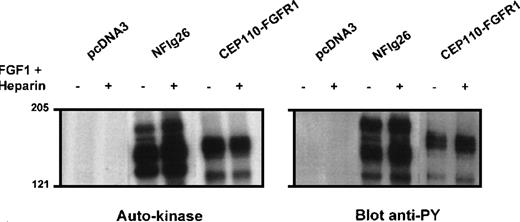
Constitutive kinase activity and tyrosine phosphorylation of CEP110-FGFR1
NIH3T3 or Cos-1 cells were transiently transfected withCEP110-FGFR1 cDNA. Anti-C-FGFR1 immunoprecipitates were immunoblotted with anti-C-FGFR1 antibody to verify the proper expression of the fusion construct (data not shown) and with anti-phosphotyrosine antibody to measure the tyrosine phosphorylation level of the kinase. A portion of the NIH3T3 immunoprecipitate was also assayed for autophosphorylation activity in the presence of γ[32P]ATP. The fusion protein was detectable as a band of approximately 150 kd (Figure 5, right panel, 2 last rows). The fusion protein had autophosphorylation activity and was constitutively tyrosine-phosphorylated regardless of FGF1 stimulation (Figure 5, left and right panels, respectively). In contrast, tyrosine phosphorylation of FGFR1 in NFlg26 cells was stimulated by the addition of FGF1 and heparin (Figure 5, left and right panels). These results indicated that CEP110-FGFR1 is a constitutively activated tyrosine kinase and suggested that this activation may be mediated by dimerization of the CEP110 portion of the fusion protein, which contains the leucine zippers.
Autokinase assay and phosphorylation on tyrosine of the CEP110-FGFR1 fusion protein.
NIH3T3 cells transiently transfected by either CEP110-FGFR1 cDNA (CEP110-FGFR1) or the empty vector (pcDNA3) and FGFR1 overexpressing cells (NFlg26) were cultured in the presence (+) or absence (−) of FGF1 plus heparin. Immunoprecipitates using anti-C-FGFR1 antibody were analyzed for autokinase activity (left panel) (autoradiography of 4 hours for all samples) and phosphorylation on tyrosine after Western blot with anti-phosphotyrosine antibody 4G10 (right panel). The position of molecular mass standards (in kd) is indicated.
Autokinase assay and phosphorylation on tyrosine of the CEP110-FGFR1 fusion protein.
NIH3T3 cells transiently transfected by either CEP110-FGFR1 cDNA (CEP110-FGFR1) or the empty vector (pcDNA3) and FGFR1 overexpressing cells (NFlg26) were cultured in the presence (+) or absence (−) of FGF1 plus heparin. Immunoprecipitates using anti-C-FGFR1 antibody were analyzed for autokinase activity (left panel) (autoradiography of 4 hours for all samples) and phosphorylation on tyrosine after Western blot with anti-phosphotyrosine antibody 4G10 (right panel). The position of molecular mass standards (in kd) is indicated.
Immunolocalization of CEP110 during the cell cycle
To further characterize CEP110, we determined its localization in the cell. Affinity-purified Ab72 antibody was used in indirect immunofluorescence studies in conjunction with a mouse antibody raised against the human centrosome protein pericentrin. Preimmune rabbit sera showed no specific reactivity in these studies (data not shown).
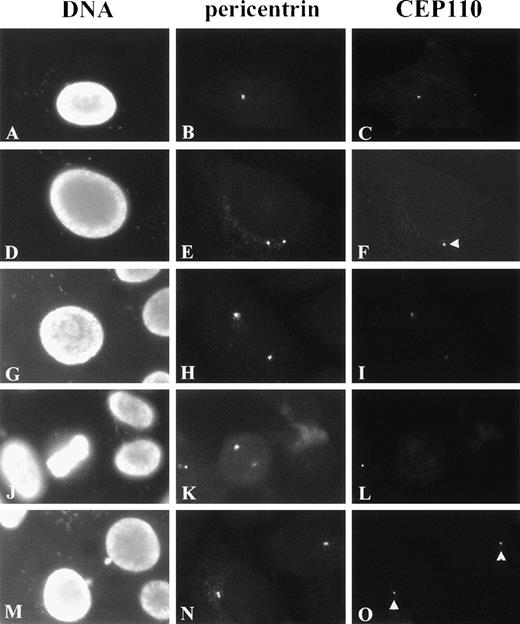
As shown in Figures 6A to 6C, G1 cells revealed CEP110 staining as single, small foci colocalizing with pericentrin reactivity at the centrosome. In all the cells examined, the area of CEP110 reactivity at the centrosome was smaller than that of pericentrin reactivity and was often seen at the center or periphery of pericentrin staining (data not shown), suggesting the association of CEP110 with the centrosome. Before and during centrosome separation, CEP110 was observed in association with only 1 of the 2 centrosome duplexes (Figure 6F, arrowhead). However, before prophase, CEP110 reactivity was observed at both centrosome duplexes (Figures 6G to 6I). At early prophase, as pericentrin reactivity at the centrosome increased, CEP110 reactivity at both centrosomes appeared to decrease and was barely detectable in most cases. Further, from metaphase to anaphase, only weak reactivity to CEP110 could be detected at the spindle poles (Figures 6J to 6L and data not shown). By the end of telophase and the beginning of G1 of the next cell cycle, CEP110 was once again observed as a small focus at the centrosome of each daughter cell (Figures 6N, 6O). CEP110 reactivity was not observed at the intercellular bridge or midbody, where other centrosome components can be found.
Immunolocalization of CEP110 during the cell cycle.
HeLa cells were double-stained for CEP110 (C, F, I, L, O) and pericentrin (B, E, H, K, N) and counterstained for DNA with DAPI (A, D, G, J, M). In early interphase, CEP110 is seen as a small focus at the centrosome (C). In later interphase cells, when duplicated centrosomes begin to separate CEP110 reactivity, is only found at 1 centrosome (F, arrowhead). In late interphase–early prophase cells CEP110 reactivity can be found on both centrosomes (I). Pericentrin reactivity is prominent at the spindle poles during metaphase (K), but CEP110 reactivity is barely detectable (L). At telophase, CEP110 reactivity is detected as small foci at the centrosome in the daughter cells (O, arrowheads). Bar = 10 μm.
Immunolocalization of CEP110 during the cell cycle.
HeLa cells were double-stained for CEP110 (C, F, I, L, O) and pericentrin (B, E, H, K, N) and counterstained for DNA with DAPI (A, D, G, J, M). In early interphase, CEP110 is seen as a small focus at the centrosome (C). In later interphase cells, when duplicated centrosomes begin to separate CEP110 reactivity, is only found at 1 centrosome (F, arrowhead). In late interphase–early prophase cells CEP110 reactivity can be found on both centrosomes (I). Pericentrin reactivity is prominent at the spindle poles during metaphase (K), but CEP110 reactivity is barely detectable (L). At telophase, CEP110 reactivity is detected as small foci at the centrosome in the daughter cells (O, arrowheads). Bar = 10 μm.
Fusion of CEP110 to GFP
The localization of CEP110 to the centrosome is likely mediated through protein-protein interactions. However, several protein-protein interaction motifs are found throughout CEP110, among them 4 leucine zippers, and several coiled-coil regions. Thus, to define which region(s) of CEP110 are necessary for its interaction with the centrosome, a series of CEP110 deletion constructs fused toGFP were created and transfected in HeLa cells. After 24 hours, the transfected cells were fixed and immunostained with antibody to pericentrin to identify the centrosome. Figure7 summarizes the structure of each CEP110-GFP deletion construct and its ability to localize to the centrosome. These results demonstrated that the region necessary for targeting CEP110 to the centrosome was confined to a 170-amino acid fragment of the C-terminus.
Schematic diagram of GFP-tagged CEP110 constructs.
Various regions of CEP110 were tagged with GFP at their C-terminus, transfected, and examined for their ability to target the centrosome. Colocalization of GFP with pericentrin staining was scored asyes, whereas no colocalization was scored as no. The amino acid positions of CEP110 in each construct are shown. Shaded ovals indicate the GFP portion of the fusion protein.
Schematic diagram of GFP-tagged CEP110 constructs.
Various regions of CEP110 were tagged with GFP at their C-terminus, transfected, and examined for their ability to target the centrosome. Colocalization of GFP with pericentrin staining was scored asyes, whereas no colocalization was scored as no. The amino acid positions of CEP110 in each construct are shown. Shaded ovals indicate the GFP portion of the fusion protein.
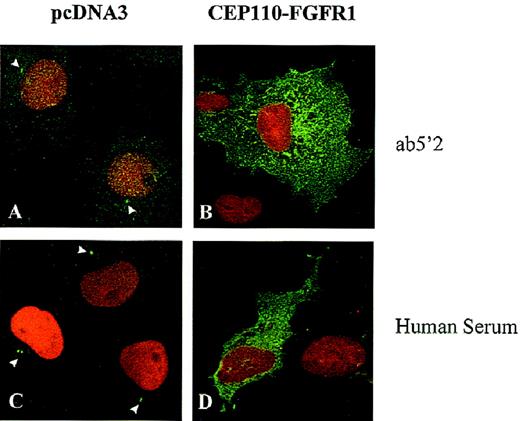
CEP110-FGFR1 is found in the cytoplasm
The localizations of fusion CEP10-FGFR1 and endogenous CEP110 proteins were compared by indirect immunofluorescence in transiently transfected Cos-1 cells. In control cells transfected with the empty vector (pcDNA3) and revealed by the purified antibody directed against the CEP110 N-terminus (Ab5′2), the endogenous CEP110 was found in the same localization, ie, concentrated in the centrosome (Figure8A). Similar patterns were found using the C-terminus CEP110 antibody (Ab72) (data not shown) and the human serum (Figure 8C). In contrast, the fusion protein CEP110-FGFR1 was detected mainly in the cytoplasm (Figures 8B, 8D). The fusion protein was not detected by the CEP110 C-terminus antibody (Ab72), which recognizes an epitope not present in the chimeric protein (data not shown). Altogether, the CEP110-FGFR1 fusion protein resulting from the t(8;9) was translocated to a subcellular compartment different from both CEP110 and FGFR1 wild-type proteins, which are centrosome and plasma membrane-bound, respectively.
Immunolocalization of endogenous CEP110 and CEP110-FGFR1 fusion protein.
Cos-1 cells transfected with the expression vector pcDNA3, either empty (A, C) or containing CEP110-FGFR1 fusion cDNA (B, D), were subjected to double- immunofluorescence staining with anti-CEP110 antibodies (Ab5′2 and human immune serum), revealed by Alexa-conjugated antirabbit (for Ab5′2) or FITC-conjugated antihuman (for human serum) secondary antibodies and 7-AAD to visualize endogenous CEP110 (arrowheads) and CEP110-FGFR1 (both in green) and the DNA (in red), respectively. Magnification, × 1000.
Immunolocalization of endogenous CEP110 and CEP110-FGFR1 fusion protein.
Cos-1 cells transfected with the expression vector pcDNA3, either empty (A, C) or containing CEP110-FGFR1 fusion cDNA (B, D), were subjected to double- immunofluorescence staining with anti-CEP110 antibodies (Ab5′2 and human immune serum), revealed by Alexa-conjugated antirabbit (for Ab5′2) or FITC-conjugated antihuman (for human serum) secondary antibodies and 7-AAD to visualize endogenous CEP110 (arrowheads) and CEP110-FGFR1 (both in green) and the DNA (in red), respectively. Magnification, × 1000.
Discussion
In a previous work,13 we identified FGFR1 as the gene involved in the specific stem cell myeloproliferative disorder associated with the 8p12 region. We report here the molecular characterization of the third event associated with MPD, ie, the t(8;9)(p12;q33). In this case, the FGFR1 gene was fused in-frame to CEP110, a novel gene coding for a centrosome protein that behaved as an autoantigen in a human autoimmune disease.
The t(8;9) breakpoint in the FGFR1 gene is localized in exon 8, 12 bp upstream of the exon 8/intron 8 junction. It is distinct from the breakpoints in the t(6;8)15 and t(8;13)16-20 that are located slightly more 3′ within FGFR1 intron 8, but it preserves the same FGFR1 sequence in the chimeric protein. Such fairly precise rearrangements, clustered either in an intron or with a small deletion of the 3′ end of the upstream exon, have already been observed, for example, in the RET oncogene in thyroid carcinoma.33
Analysis of the deduced amino-acid sequence of CEP110 reveals extensive regions of coiled-coil structure. In this respect, CEP110 is similar to other centrosome proteins that function as antigens, among them pericentrin,34 ninein,35 and CEP250/CNAP1.25 In addition to the coiled-coil structure, 4 leucine zippers are found in CEP110. Leucine zipper motifs are well-characterized motifs involved in protein-protein interaction36,37 and are found in other centrosome proteins including ninein35 and CEP250 (Mack and Rattner, unpublished observations).
In the MPD linked to 8p12, the key oncogene isFGFR1.13 In the t(8;9) patient's leukemic cells, 2 reciprocal fusion genes were identified as a result of the translocation. CEP110-FGFR1 transcript encodes an aberrant tyrosine kinase of approximately 150 kd, which contains the 808 first amino acid residues from CEP110 (81.3% of the entire protein), including the leucine zippers, joined to the FGFR1 intracellular region minus the major part of its juxtamembrane domain. CEP110-FGFR1 is constitutively phosphorylated in transfected cells. This is the case for the 2 other fusion proteins involved in the 8p12 MPD, FIM-FGFR1,16 and FOP-FGFR1 (Guasch et al, January 1999, unpublished data). These activated aberrant tyrosine kinases are likely to promote leukemogenesis through constitutive activation of the FGFR1 kinase mediated by the N-terminus homodimerization motifs of the FGFR1 partners, which contain either zinc fingers for FIM/ZNF198,16-20 leucine-rich region for FOP,15 or leucine zippers for CEP110. In a recent work, 21 we showed that FIM-FGFR1 has constitutive dimerization capability mediated by the FIM N-terminus sequences.
The fusion of 2 proteins resulting from a chromosomal translocation event often creates an aberrantly located protein.38 We show here that CEP110 is a centrosome component. Based on the indirect immunofluorescence pattern, we suggest that CEP110 is a component of the mature centrosome and has a function in centriole maturation. In human cells, the centrosome is a distinct cytoplasmic protein complex that is the primary microtubule-organizing center of the cell (see Brinkley39). To identify the domains of CEP110 that specify its centrosome localization, various regions of the protein were fused to GFP and transfected to HeLa cells. The centrosome-binding domain of CEP110 was found to lie within a 170-amino acid region near the C-terminus (amino acids 617-787). This region includes the fourth leucine zipper motif and is primarily of coiled-coil structure. The presence of a single leucine zipper in the centrosome-binding domain of CEP110 is similar to that found for the centrosome-targeting domain of CEP250 (Mack and Rattner, June 1998, unpublished data). Because the region targeting CEP110 to the centrosome is retained in CEP110-FGFR1, there is the possibility that the constitutively activated fusion protein is located within this organelle, therein inducing disruption of centrosome structure, function, or both. Indeed, transient overexpression of centrosome-associated kinases PLK1 and STK15/BTAK/aurora2 in NIH3T3 cells results in centrosome amplification, aneuploidy, and transformation.40-43 We demonstrate here that CEP110-FGFR1 has an aberrant cellular localization compared to its normal counterparts. Although endogenous FGFR1 is at the membrane and CEP110 has a unique centrosome localization, CEP110-FGFR1 predominantly localizes in the cytoplasm and is not confined to either the membrane or the centrosome (however, we cannot completely eliminate the possibility that a proportion of CEP110-FGFR1 may go to the centrosome and exert a potential effect there but is undetected under the conditions used in this study). FIM-FGFR1, another fusion protein involved in the 8p12 MPD, also localizes within the cytoplasm.21 Thus, in this particular syndrome, fusion proteins may affect the growth of hematopoietic stem cell through continuous kinase stimulus—presumably triggered by its dimerization mediated by the protein–protein interaction motifs of the FGFR1 protein partner—and inappropriate recruitment in the cytoplasm of signaling substrates. The oncogenic activation of protein tyrosine kinases through structural alterations produced by chromosomal rearrangements has been frequently associated with human hematologic malignancies.16,44-47 Another mechanism of activation involves gene dysregulation, in particular alterations of other FGFR family members in multiple myeloma48 or in osteosarcoma.49 The oncogenic kinases are constitutively activated and variably affect the signaling pathways.
The 3 FGFR1 partners are different, unrelated proteins with different cellular localizations, ie, nuclear and nucleolar for FIM,21 cytoplasmic for FOP (Guasch et al, January 1999, unpublished data), and centrosome for CEP110 (as described in the current work). Accumulating data (clustered breakpoints in FGFR1, presence of dimerization motifs in each partner, cytoplasmic localization of the fusion protein) suggest that a common mechanism may sustain the oncogenic activity of the rearranged FGFR1 kinase. Further work will aim at the identification of this mechanism and the target cell in which it is abnormally triggered.
Acknowledgments
We thank Drs Mawas and Maraninchi for encouragement and comments. We also thank J. Adélaide and B. Zhang for occasional help in cloning and sequencing the fusion genes, T. Alario for CD34+ cell purification, and V. Ollendorff for helpful advice.
G.G., G.J.M., and C.P. contributed equally to this work.
Supported by Inserm, Institut Paoli-Calmettes, and by grants from the Ligue Nationale contre le Cancer, Comité du Var de la Ligue Nationale contre le Cancer, and FEGEFLUC. G.G. is a recipient of a fellowship from MESR; C.P. is supported by the SociétéFrançaise d'Hématologie; G.J.M. is supported by the Alberta Heritage Foundation for Medical Research; J.B.R. is supported by the National Cancer Institute of Canada, with funds from the Canadian Cancer Society.
Reprints:Marie-Josèphe Pébusque, Laboratoire d'Oncologie Moléculaire, Inserm U119, Institut de Cancérologie et d'Immunologie de Marseille, 27 Boulevard Leı̈ Roure, 13009 Marseille, France; e-mail:pebusque@marseille.inserm.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal