Abstract
Previously we reported that the size of the stem cell compartment (measured as LTC-IC) is 11-fold greater in DBA/2 than in C57BL/6 mice, and we identified genes that regulate the size of the stem cell pool. To determine whether stem cell intrinsic or extrinsic events account for these differences, we created chimeras by aggregating morulae from the strains C57BL/6 and DBA/2. In these chimeras stem cells of both genotypes are exposed to a common mixed environment. Thus, an equalization of stem cell frequencies is expected if stem cell extrinsic effects dominate. Conversely, the parental ratio of LTC-IC should be preserved if the regulation is stem cell autonomous. For each chimera, individual LTC-IC were genotyped on the clonal levels by analyzing their progeny. We found that most of the difference that regulates the size of the stem cell compartment was intrinsic.
The mechanisms that control the maintenance of the stem cell pool remain incompletely understood.1 Previously, we showed that the size of the stem cell compartment is regulated by a set of genes, named stem cell frequency regulator (Scfr) genes.2 Several Scfr genes were identified and mapped after the demonstration that the frequency of long-term culture initiating cells (LTC-IC) differs noticeably between inbred strains of mice.2-4
Here we used B6↔D2 aggregation chimeras to examine whether theScfr genes act stem cell intrinsically or extrinsically though the environment. In aggregation chimeras, stem cell behavior can be compared directly without the potential complications inherent in allogeneic transplantation systems. The aggregation chimera system has been used to study stem cell tolerance and repopulation kinetics.5-7 Using a new method in which individual LTC-IC were genotyped in a mixed environment, we show that the control of the size of the stem cell pool is predominantly stem cell intrinsic.
Study design
Mice
B6 and D2 mice (Jackson Laboratory, Bar Harbor, ME) as well as CD-1 female and vasectomized male mice (Charles River Laboratories, Wilmington, MA) were maintained at our facility. Aggregation chimeras were generated using the “darning needle.”7 To avoid complications due to chimeric drift, seen in some aging D2↔B6 chimeras,8 9 all mice were tested before 6 months of age.
Long-term culture initiating cells assay
Genotyping by immunofluorescence
Anti-H-2d and anti-H-2b mAb (Pharmingen, San Diego, CA) were used for FACScan genotyping (Becton Dickinson, Mountain View, CA) of bone marrow and spleen cells and the progeny of LTC-IC. LTC-IC progeny were gated to include only small granulocytic cells as defined by forward and side scatter and expression of the granulocytic marker Gr-1 (mAb 8C5). This excluded macrophages, stromal cells, and other long-lived cells from the analysis. Staining of greater than or equal to 5% above background (isotype-matched controls) was considered positive.
Generation of bone marrow stroma
As described,11 bone marrow stroma was derived by culturing bone marrow cells in Dexter medium at a low density until the adherent cells were confluent. Two treatments with mycophenolic acid removed residual hematopoietic cells.
Semiquantitative polymerase chain reaction genotyping
B6 and D2 contributions to B6↔D2 tissues were quantitated by polymerase chain reaction (PCR) using a microsatellite, D1Mit415. Bands were quantitated using the public domain NIH image program (http://rsb.info.nih.gov/nih-image/).
Optimal model mixing
The chimerism in bone marrow was used to predict the expected ratios of LTC-IC for intrinsic and extrinsic models for each animal. Optimal model mixing (OMM) is based on least-squares fitting. OMM determined optimal weights that quantify the relative contribution of each model to the actual data. Details of the method are available fromhttp://www.skcc.org/skcc-staff/muller/scfrdata.html.
Results and discussion
Genotyping individual LTC-IC
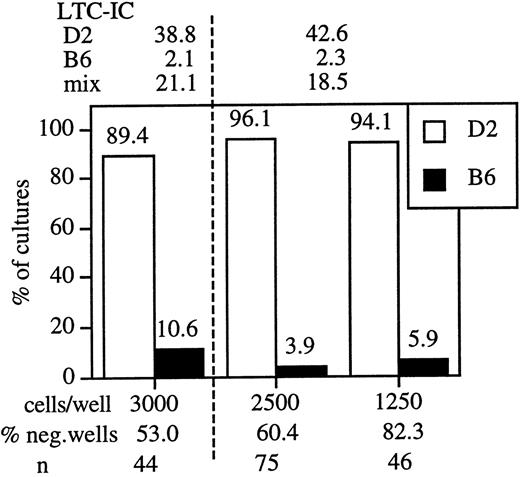
To accurately measure the composition of the LTC-IC compartment in each chimera, we combined a functional stem cell assay with immunofluorescence-based genotyping of individual LTC-IC. The LTC-IC assay that we used to identify the strain differences is linear and measures a single limiting cell, the LTC-IC.2 Thus, in limiting dilution conditions, the cells in each colony are the descendants of a single LTC-IC. Consequently, genotyping the cells in each colony identifies the genotype of the LTC-IC progenitor. As a proof of principle, we mixed equal numbers of B6 and D2 bone marrow cells and plated the cell mixture in limiting dilution cultures. Individual clones (wells) were harvested after 4 weeks of culture, and cells were stained with mAbs specific for H-2b and H-2d. Only cultures that showed at least 37% negative wells were analyzed, and stringent gating assured exclusion of long-lived macrophages and stromal cells from the analysis. Data from 2 independent experiments are depicted in Figure1. In each experiment LTC-IC levels in parental and mixed bone marrow cells were assessed, and these values were compared with the ratio of D2 and B6 LTC-IC found in the mixed bone marrows. The data indicate that B6 and D2 stem cells behaved independently. Thus, this assay is appropriate for the direct examination of LTC-IC frequencies in B6↔D2 chimeras.
Mixtures of D2 and B6 marrow maintain the parental ratio of LTC-IC in culture.
Bone marrow cells from D2, B6, and a 1:1 mixture of the marrows were plated onto S17 stroma. Between 48 and 192 wells were seeded for each cell concentration. The data depicted are from 2 independent experiments, a dashed vertical line separates different experiments. LTC-IC values (per 105 cells seeded) at 4 weeks of culture are indicated above the figure. Positive wells were then harvested and the cells from each well were stained with mAb specific for H2d (D2, white bars) and H2b (B6, black bars). Because the cells in each microculture were derived from an LTC-IC, the genotype of the differentiated progeny establishes the genotype of the LTC-IC. The plating density (cells/well), percentage negative wells, and number of wells tested for each cell dilution (n) is indicated below the figure.
Mixtures of D2 and B6 marrow maintain the parental ratio of LTC-IC in culture.
Bone marrow cells from D2, B6, and a 1:1 mixture of the marrows were plated onto S17 stroma. Between 48 and 192 wells were seeded for each cell concentration. The data depicted are from 2 independent experiments, a dashed vertical line separates different experiments. LTC-IC values (per 105 cells seeded) at 4 weeks of culture are indicated above the figure. Positive wells were then harvested and the cells from each well were stained with mAb specific for H2d (D2, white bars) and H2b (B6, black bars). Because the cells in each microculture were derived from an LTC-IC, the genotype of the differentiated progeny establishes the genotype of the LTC-IC. The plating density (cells/well), percentage negative wells, and number of wells tested for each cell dilution (n) is indicated below the figure.
Long-term culture initiating cell ratios in B6↔D2 chimeras
Six aggregation chimeras that showed between 26% to 74% B6 contribution were analyzed in detail. The extent of chimerism was measured in spleen, bone marrow, bone marrow stroma, coat color, kidney, and heart tissues, and demonstrated that these mice were thorough parental mixtures (Table 1). Heart tissue was chosen because heart muscle and bone marrow stromal cells are developmentally related.12 13 Bone marrow stromal cells from 3 of the chimeras were also PCR typed, and the results confirm a chimeric microenvironment.
Contribution of the parental genotype to individual aggregation chimeras and the genetic composition of LTC-IC in bone marrow
| Mouse . | Coat color . | PCR* . | Staining† . | LTC-IC chimerism . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Measured‡ . | Predicted1-153 . | |||||||||||
| BM . | Kidney . | Heart . | Stroma . | BM . | Spleen . | No. LTC-IC . | Genotype . | Int . | Ext . | OMM . | ||
| 2 | 50 | 30 ± 0.5 | 35 | 50 | nd | 30 | 44 | 70 | 12.0 | 3.8 | 30.0 | 88.8 |
| 3 | 50 | 33 ± 0.5 | 41 | 57 | nd | 40 | 54 | 48 | 11.3 | 5.7 | 40.0 | 87.5 |
| 5 | 70 | 67 ± 4.5 | 76 | 79 | nd | 75 | 76 | 53 | 36.2 | 21.4 | 75.0 | 77.8 |
| 6 | 80 | 63 ± 2.0 | 60 ± 2 | 79 | 30 | 73 | 76 | 130 | 25.1 | 19.6 | 72.8 | 78.8 |
| 8 | 20 | 53 ± 2.0 | 40 ± 0 | 58 | 71 | 68 | 78 | 102 | 21.9 | 16.2 | 68.0 | 80.8 |
| 9 | 20 | 26 | 27 ± 1 | 53 | 42 | 30 | 39 | 120 | 13.7 | 3.8 | 30.0 | 88.8 |
| Controls | ||||||||||||
| B6D2 F1 | 46 ± 3.1 | |||||||||||
| B6 + D21-155 | 44.2 ± 1.9 | |||||||||||
| B6 | 100 | 97 ± 1.6 | 91.1 ± 5.7 | |||||||||
| D2 | 0 | 8 ± 1.6 | 0.2 ± 0.2 | |||||||||
| Mouse . | Coat color . | PCR* . | Staining† . | LTC-IC chimerism . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Measured‡ . | Predicted1-153 . | |||||||||||
| BM . | Kidney . | Heart . | Stroma . | BM . | Spleen . | No. LTC-IC . | Genotype . | Int . | Ext . | OMM . | ||
| 2 | 50 | 30 ± 0.5 | 35 | 50 | nd | 30 | 44 | 70 | 12.0 | 3.8 | 30.0 | 88.8 |
| 3 | 50 | 33 ± 0.5 | 41 | 57 | nd | 40 | 54 | 48 | 11.3 | 5.7 | 40.0 | 87.5 |
| 5 | 70 | 67 ± 4.5 | 76 | 79 | nd | 75 | 76 | 53 | 36.2 | 21.4 | 75.0 | 77.8 |
| 6 | 80 | 63 ± 2.0 | 60 ± 2 | 79 | 30 | 73 | 76 | 130 | 25.1 | 19.6 | 72.8 | 78.8 |
| 8 | 20 | 53 ± 2.0 | 40 ± 0 | 58 | 71 | 68 | 78 | 102 | 21.9 | 16.2 | 68.0 | 80.8 |
| 9 | 20 | 26 | 27 ± 1 | 53 | 42 | 30 | 39 | 120 | 13.7 | 3.8 | 30.0 | 88.8 |
| Controls | ||||||||||||
| B6D2 F1 | 46 ± 3.1 | |||||||||||
| B6 + D21-155 | 44.2 ± 1.9 | |||||||||||
| B6 | 100 | 97 ± 1.6 | 91.1 ± 5.7 | |||||||||
| D2 | 0 | 8 ± 1.6 | 0.2 ± 0.2 | |||||||||
PCR, polymerase chain reaction; BM, bone marrow; LTC-IC, long-term culture initiating cells; int, intrinsic; ext, extrinsic; OMM, optimal model mixing; nd, not determined.
Mouse: Individual aggregation chimeras (identified by number) were genotyped. All data are expressed as percentage of B6 contribution to the indicated tissues.
Coat color was estimated by visual inspection.
Semiquantitative PCR was performed on DNA extracted from the indicated organs and cultured bone marrow stromal cell cultures. Values for chimeras are from 1 to 2 independent PCR assays. Parental control mice were performed 4 times. Level of detection of PCR analysis: 5%.
Immunofluorescence analysis on chimeric bone marrow and spleen was performed once; the corresponding values for the control mice are means from 10 independent experiments. The staining data depict the percent of B6 cells as determined by gating using Lysis software.
Designates the LTC-IC analyzed from each chimeric mouse; the number of microcultures (No. LTC-IC) tested and the proportion of LTC-IC that were of the B6 genotype are given.
The extent of chimerism in bone marrow determined by immunofluorescence before the initiation of the cultures was used to calculate the predicted ratio of LTC-IC if all Scfr genes were to act inside stem cells (intrinsic) or if all activity would be found in the environment (extrinsic). Based on the 11-fold higher frequency of LTC-IC in the parental D2 mice, a factor of 11 was used to multiply the D2 frequency in bone marrow to derive the expected autonomous ratio. The expected extrinsic ratio is identical to the ratio of B6 and D2 cells in the bone marrow of each chimera. OMM was used to calculate for each mouse the percent contribution of the intrinsic model that will give the best fit to the percent B6 actually measured in the LTC-IC. Overall, this yielded 83% ± 4.7% intrinsic regulation.
A mixture of 50% each of D2 and B6 bone marrow was tested; the level of staining for the D2 genotype was 49.9 ± 1.9%. Staining of D2 bone marrow with the H-2d-specific mAb was 94.1 ± 3.1% not significantly different (p = 0.17) from the staining of B6 bone marrow with the H-2b specific mAb.
The chimerism in the LTC-IC compartment was measured by typing the progeny of individual LTC-IC as outlined above. Table 1 depicts the number of clones tested for each animal, the ratio of LTC-IC contributed from each parental strain, and the calculated values expected for the intrinsic and extrinsic models, respectively. In control experiments (n = 9) with parental D2 and B6 mice, we found 37.7 ± 5.8 and 3.4 ± 1.1 LTC-IC per 105 cells seeded, respectively, agreeing well with previously published results.2 Thus, the mean difference of 11-fold in LTC-IC levels was used as a weighting factor to calculate the expected intrinsic values. For example, the chimera M2 showed 30% B6 cells in marrow (Table 1). Thus, if the regulation is extrinsic, 30% of the LTC-IC should be of B6 genotype. For intrinsic regulation, we expect 1 × 30% B6 and 11 × 70% D2 LTC-IC, resulting in 3.8% B6 contribution.
It is apparent that the LTC-IC ratios fit neither model well. Rather, the data suggest a mixed model, although weighted toward the intrinsic model. To quantitate the contributions of the extrinsic and autonomous models to the data, we devised a mathematical method, based on least-square fitting. This analysis showed that the model with the best fit to the data comprises 83.8% ± 4.7% intrinsic and 16% extrinsic contributions. Thus, the model supports the conclusion that the Scfr genes act mostly, but not exclusively, in a stem cell autonomous manner to control the size of the stem cell pool.
We assumed that stem cells from both parental strains have equal access to cellular and humoral influences from both parents in the aggregation chimeras. This is probable for secreted molecules from both the macro- and the microenvironment. Similarly, it is likely that stem cells will seed to and will use stromal niches of both genotypes. During development, establishment of the stroma precedes seeding by stem cells.14,15 Moreover, stromal cells and hematopoietic cells can differ in genotype without affecting their interaction.16
The discovery that the size of the stem cell pool differs amongst humans17 and inbred strains of mice2,3galvanizes a search for genes regulating stem cell self-renewal and differentiation. Our prior work indicated the effect of multiple genes.2 Now we show that the regulation of the size of the stem cell compartment is mostly stem cell intrinsically regulated; this will focus efforts to clone the relevant genes.
Acknowledgment
The excellent technical assistance of America Mauhar is gratefully acknowledged.
Supported through grants DK48015 and DK52177 from the National Institutes of Health.
Reprints:Christa E. Müller-Sieburg, Sidney Kimmel Cancer Center, 10835 Altman Row, San Diego, CA 92121; email:cmuller@skcc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal