The tumor-suppressive promyelocytic leukemia (PML) protein of acute promyelocytic leukemia (APL) has served as one of the defining components of a class of distinctive nuclear bodies (NBs). PML is delocalized from NBs in APL cells and is degraded in cells infected by several viruses. In these cells, NBs are disrupted, leading to the aberrant localization of NB proteins. These results have suggested a critical role for the NB in immune response and tumor suppression and raised the question of whether PML is crucial for the formation or stability of NB. In addition, PML is, among other proteins, covalently modified by SUMO-1. However, the functional relevance of this modification is unclear. Here, we show in primary PML−/− cells of various histologic origins, that in the absence of PML, several NB proteins such as Sp100, CBP, ISG20, Daxx, and SUMO-1 fail to accumulate in the NB and acquire aberrant localization patterns. Transfection of PML in PML−/−cells causes the relocalization of NB proteins. By contrast, a PML mutant that can no longer be modified by SUMO-1 fails to do so and displays an aberrant nuclear localization pattern. Therefore, PML is required for the proper formation of the NB. Conjugation to SUMO-1 is a prerequisite for PML to exert this function. These data shed new light on both the mechanisms underlying the formation of the NBs and the pathogenesis of APL.
The cell nucleus is compartmentalized into highly organized domains that are associated with specific nuclear functions. For example, nucleoli are sites of ribosomal RNA synthesis and processing, whereas coiled bodies contain spliceosomal small nuclear ribonucleoproteins plus a subset of other splicing factors.1 One distinctive class of subnuclear domains, which appears as punctate speckles under immunofluorescence microscopy, was originally identified as autoantigens in primary biliary cirrhosis patients2 and variably named Kremer bodies, nuclear domain 10 (ND10), promyelocytic leukemia protein nuclear bodies (PML NBs), or PODs (PML oncogenic domains) because subsequent studies showed that the PML protein of acute promyelocytic leukemia (APL) is tightly associated with these NBs.3-8 Several NB components have been recently identified, such as SUMO-1/PIC1, Sp100, Daxx, ISG20, and CBP.8 9
PML belongs to a family of proteins characterized by the presence of the RBCC (RING-B-Box-Coiled-coil) motif or tripartite motif,10,11 which consists of a C3HC4 zinc finger motif (RING finger) and 1 or 2 additional cysteine-rich regions (B-boxes) followed by a predicted leucine coiled-coil region. PML is specifically up-regulated at the transcriptional level by type 1 and 2 interferons (IFN), which are potent growth and tumor suppressive cytokines.12,13 IFN increases PML expression as well as the size and the number of NBs.2,8,12-14 More recently, the analysis of mice and cells where PML was inactivated has demonstrated that PML functions as a growth and tumor suppressor in vivo, at least in part, through its ability to act as a transcriptional coactivator and that PML is important for multiple apoptotic pathways.15-17
PML is modified by SUMO-1/PIC1 (hereafter referred to for brevity as SUMO-1), a ubiquitin-like protein that localizes to different cellular compartments and is covalently bound to various proteins. For example, SUMO-1 binds to RanGAP1 at the nuclear pore complex18 or to IκBα in the cytoplasm.19 SUMO-1 is also found in the NB, where it covalently modifies NB proteins such as PML20-22 and Sp100.22 It has been proposed that SUMO-1 modification can determine the targeting of modified proteins such as RanGAP1 to a specific cellular compartment.18
The PML gene is fused to the retinoic acid receptor (RAR)α gene in the t(15;17) chromosomal translocation associated with APL.23 As a result of this chromosomal translocation, an oncogenic PML-RARα fusion protein is generated. The PML-RARα hybrid retains most of the functional domains of its parental proteins and can heterodimerize with PML.24 In APL cells harboring the t(15;17) chromosomal translocation, PML, together with other NB components, is delocalized from NBs into PML-RARα microspeckles.5-7 Thus, PML-RARα, through heterodimerization and delocalization of PML, may cause NB proteins to localize to aberrant nuclear sites. Similarly, upon viral infections, PML is delocalized along with other NB components.8
For these reasons, and because PML is invariably associated with NBs in all cells tested, PML has been used as a defining marker for the NB. However, the role of PML in governing the proper formation or the stability of the NB has not been investigated. Furthermore, whether in pathological conditions the targeting of PML by viruses or oncoproteins is a critical event in the subsequent disorganization of the NB remains unknown. If this were to be the case, disruption of normal PML localization would have a pleiotropic effect on the functions of many NB components. In addition, it is still unclear whether the modification of PML by SUMO-1 plays a role in the localization of PML to the NB and, consequently, in NB organization.
In this article, we demonstrate that PML and its modification by SUMO-1 play a critical role in the proper formation of the NB, thus providing an explanation of the dramatic consequences of PML delocalization in APL cells.
Materials and methods
Primary cell preparation, cell culture, and transfection
Mouse primary embryonic fibroblasts, keratinocytes, and splenocytes were prepared as previously described.16,25Cells were split 1 day prior to transfection, whereupon they reached approximately 70% to 80% confluence. Transfections with pSG5-PML (0.2 μg17), pSG5-3M-PML (0.2 μg), pSG5-PML-RARα, pEGFP-Sp100 (0.2 μg), or pSG5-HA-ISG20 (0.2 μg) were carried out in 6-well clusters using the Superfect reagent (Qiagen, Chatsworth, CA) following the manufacturer's instructions. After 24 hours, cells were trypsinized, transferred to chambered slides, and grown for another 24 hours before further analysis.
Indirect immunofluorescence
Primary mouse embryonic fibroblasts (MEFs) and keratinocytes were grown in appropriate medium, as described above. Splenocytes were cytospun for 5 minutes at 1000 rpm directly onto glass slides. Cells were fixed using 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 minutes at room temperature and were permeabilized by incubating in methanol for 7 minutes at −20°C. After washing 3 times in PBS and blocking in phosphate-buffered saline–Tween 20 (PBST) containing 10% heat-inactivated goat serum, the cells were incubated for 1 hour at room temperature with an anti-PML antibody (rabbit polyclonal, cross-reacts with mouse PML15), and/or an anti-SUMO-1 antibody (mouse monoclonal, cross-reacts with mouse SUMO-1; a gift from Dr Michael Matunis, Johns Hopkins University, Baltimore, MD), and/or an anti-Daxx antibody (mouse monoclonal, Santa Cruz Biotechnologies, Santa Cruz, CA), and/or an anti-Sp100 antiserum (human origin, cross-reacts with mouse Sp100; a gift from Dr Gerd G. Maul, The Wistar Institute, Philadelphia, PA), and/or an anti-CBP antibody (rabbit polyclonal, A22, Santa Cruz), and/or an anti-hemagglutinin (HA) antibody (mouse monoclonal, Roche, Indianapolis, IN) diluted in the blocking buffer. For detection, Texas red- or fluorescein-conjugated goat antirabbit, fluorescein-conjugated horse antimouse, or fluorescein-conjugated mouse antihuman IgG antibodies (PharMingen, San Diego, CA) were diluted in blocking buffer containing 1 ng/mL of DAPI. Cells were incubated with the respective secondary antibody mix for 1 hour at room temperature, washed 3 times in PBS, and covered with Permount mounting medium (Fisher Scientific, Springfield, NJ). Slides were viewed on an Olympus fluorescence microscope or analyzed by confocal microscopy in the institute core facility.
Site-directed mutagenesis
The 3M-PML mutant was derived from a human PML complementary DNA26 (cDNA) cloned in the pSG5 expression vector (Stratagene, Madison, WI). Arginine-to-lysine mutations at amino acid positions 65, 160, and 49027 were created using the QuikChange site-directed mutagenesis kit (Promega, Madison, WI) according to manufacturer's instructions. The construct was verified by DNA sequencing.
Results
Sp100, CBP, ISG20, Daxx, and SUMO-1 fail to accumulate in the NB in the absence of PML
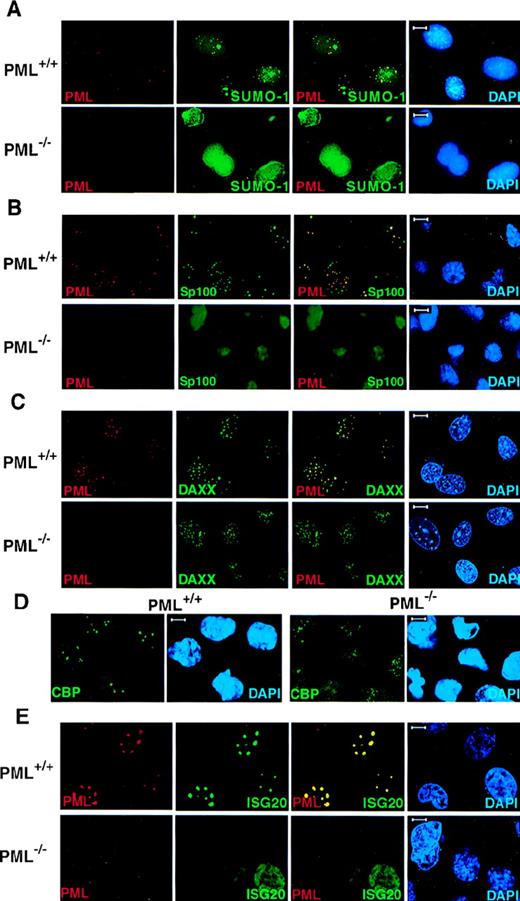
Because PML is covalently modified by SUMO-1, we performed confocal immunofluorescence analysis on primary cells obtained from PML+/+ and PML−/− mice with monoclonal anti-SUMO-1 and polyclonal anti-PML antibodies (see “Materials and methods”). In PML+/+ MEFs, SUMO-1 colocalized with PML in discrete speckled nuclear regions (Figure1A). However, SUMO-1 also displayed a diffuse nuclear and cytoplasmic localization pattern and also accumulated, unlike PML, in larger nuclear regions (Figure 1A). In contrast, in PML−/− MEFs, SUMO-1 did not acquire its distinctive speckled nuclear localization pattern, but it was still readily detected in the nucleus and the cytoplasm in a diffuse distribution or in larger nuclear aggregates (Figure 1A). The pronounced nuclear rim staining of SUMO-1 in PML−/− MEFs may reflect the SUMO-1 modification of RanGAP1, which localizes to the nuclear envelope, whereas the localization of SUMO-1 in the large nuclear domains may be specific to certain cell types, such as MEF, because it was not observed in keratinocytes (Figure 2).
PML is required for the NB localization of SUMO-1, Sp100, Daxx, CBP, and ISG20.
Primary MEFs were grown on chambered slides before immunofluorescence analysis. MEFs were double-labeled for PML and (A) SUMO-1, (B) Sp100, and (C) Daxx. (D) The localization of endogenous CBP in PML+/+ and PML−/− MEFs. (E) The localization of transfected HA-ISG20 in PML+/+ and PML−/− MEFs. Primary cells were transfected with HA-ISG20. After 24 hours, cells were harvested and stained for the HA epitope. Representative confocal micrographs are shown, with the respective immunofluorescent colors (PML, red; SUMO-1/Sp100/Daxx/CBP/HA-ISG20, green; DAPI, blue) labeled in the lower corners of each image. Colocalization is reflected by the yellow color. Bar: 5 μm.
PML is required for the NB localization of SUMO-1, Sp100, Daxx, CBP, and ISG20.
Primary MEFs were grown on chambered slides before immunofluorescence analysis. MEFs were double-labeled for PML and (A) SUMO-1, (B) Sp100, and (C) Daxx. (D) The localization of endogenous CBP in PML+/+ and PML−/− MEFs. (E) The localization of transfected HA-ISG20 in PML+/+ and PML−/− MEFs. Primary cells were transfected with HA-ISG20. After 24 hours, cells were harvested and stained for the HA epitope. Representative confocal micrographs are shown, with the respective immunofluorescent colors (PML, red; SUMO-1/Sp100/Daxx/CBP/HA-ISG20, green; DAPI, blue) labeled in the lower corners of each image. Colocalization is reflected by the yellow color. Bar: 5 μm.
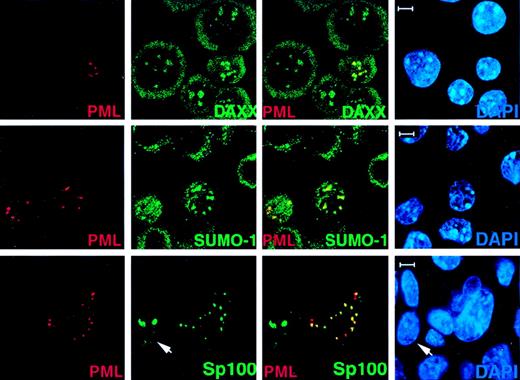
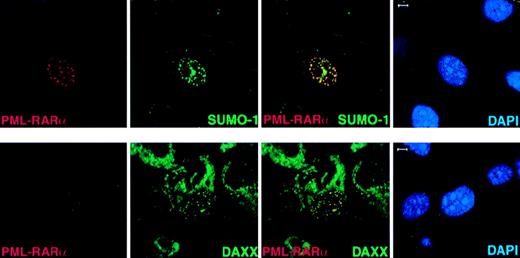
Transfected PML in PML−/− cells recruits Daxx, SUMO-1, and Sp100 to the NBs.
PML−/− keratinocytes were transfected with pSG5-PML alone or cotransfected with pSG5-PML and pEGFP-Sp100. After 24 hours, cells were harvested and cytospun onto glass slides for immunofluorescence staining. Representative confocal micrographs are shown (PML, red; SUMO-1/Daxx/GFP-SP100, green; DAPI, blue). The yellow color reflects colocalization. The arrow points to a cell transfected with GFP-Sp100 only. This represents the localization pattern of transfected Sp100 in PML−/− keratinocytes where this protein aggregates in large patches. Bar: 5 μm.
Transfected PML in PML−/− cells recruits Daxx, SUMO-1, and Sp100 to the NBs.
PML−/− keratinocytes were transfected with pSG5-PML alone or cotransfected with pSG5-PML and pEGFP-Sp100. After 24 hours, cells were harvested and cytospun onto glass slides for immunofluorescence staining. Representative confocal micrographs are shown (PML, red; SUMO-1/Daxx/GFP-SP100, green; DAPI, blue). The yellow color reflects colocalization. The arrow points to a cell transfected with GFP-Sp100 only. This represents the localization pattern of transfected Sp100 in PML−/− keratinocytes where this protein aggregates in large patches. Bar: 5 μm.
To confirm these findings in cells of a different histologic origin, we studied the localization of PML and SUMO-1 in splenocytes obtained from PML+/+ and PML−/− mice untreated or activated with concanavalin A (ConA). ConA activation of splenocytes increases the number and size of the NBs.15 16 In both ConA-treated and -untreated PML+/+ splenocytes, SUMO-1 colocalized with PML in the NB, whereas in PML−/− cells, SUMO-1 displayed an aberrant nuclear diffuse/microspeckled localization pattern and accumulated in larger nuclear regions (not shown).
We next tested whether other NB components, such as Sp100, Daxx, CBP, and ISG20, would be delocalized in the absence of PML.
Sp100 is one of the first NB components identified as an autoantigen that is prevalent in primary cirrhosis.28 Like PML, Sp100 is also up-regulated by IFN,29 and it can play a role in IFN-induced immune responses. However, Sp100 does not appear to physically interact with PML.6 In PML+/+ MEFs, the Sp100 protein colocalized with PML in the NBs, and it acquired an aberrant diffused nuclear distribution pattern in PML−/− MEFs (Figure 1B).
Daxx was cloned as an adaptor molecule that can bind to the death domain of the Fas receptor via its C-terminal end and has been found to play a role in the potentiation of the Fas pro-apoptotic stimulus.30 We have shown that Daxx can physically interact with PML and localize in the NB.9 In PML+/+MEFs, Daxx localized in the NBs together with PML but was detected in a nuclear diffused or patched pattern in PML−/−MEFs and keratinocytes, respectively (Figure 1C and Figure 2). It is noteworthy that in wild-type MEFs Daxx is predominantly nuclear (Figure1C).
The CBP transcriptional coactivator also localizes in the NB.31-33 CBP displayed a speckled as well as a diffuse nuclear localization pattern in PML+/+ MEFs (Figure 1D). However, in PML−/− MEFs, CBP acquired a microspeckled and a diffuse nuclear distribution (Figure 1D).
ISG20 was cloned as an IFN-induced protein, which was found to accumulate in the PML NB.34 Because no antibody against the murine ISG20 is yet available, the nuclear localization of ISG20 was studied in PML+/+ and PML−/− MEFs transiently transfected with an HA-tagged ISG20 expression vector (Figure 1E). HA-ISG20 localized in the NBs in PML+/+ MEFs but was detected in a nuclear diffused pattern in PML−/− MEFs (Figure 1E).
Thus, PML is required for the proper nuclear compartmentalization of NB components.
Transfection of PML in PML−/− cells restores normal localization of NB components
To further characterize the role of PML in the localization of the above proteins in the NB, PML−/− keratinocytes (Figure 2) and MEFs (not shown) were transiently transfected with PML. After 24 hours, cells were fixed and subjected to immunofluorescence analysis with a polyclonal anti-PML antibody (See “Materials and methods”). Using this antibody, transfected or untransfected PML−/− cells could be easily distinguished (Figure 2). In the PML-transfected cells, endogenous NB components such as, for instance, Daxx and SUMO-1, reacquired a speckled pattern, which overlapped with the one of PML (Figure 2). Similarly, whereas transfected GFP-SP100 formed large nuclear aggregates in PML−/− cells, cotransfection of PML along with GFP-Sp100 in PML−/− cells restored the NB localization of GFP-Sp100 (Figure 2).
SUMOylation of PML is a prerequisite for NB formation
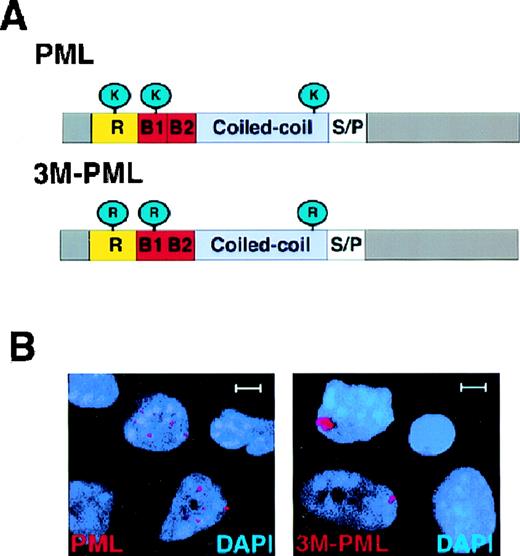
The PML protein forms covalent bonds with SUMO-1 at 3 amino acid positions: 65, 160, and 490 through lysine residues.27Because the SUMOylated PML is tightly associated with the nuclear matrix but the unmodified form of PML is not, it has been proposed that SUMO modification plays an important role for docking PML in the NB.21 To clarify the role of PML SUMOylation in directing PML NB localization, we generated a PML mutant (3M-PML) that is no longer SUMO-modified by mutating amino acid positions 65, 160, and 490 from lysines to arginines (Figure3A). The arginine residue was chosen for its close resemblance to lysine in structure and charge. Western blot analysis of the transfected 3M-PML mutant protein in HeLa and COS-1 cells indicated that this protein was not SUMOylated (not shown). We then transiently transfected wild-type PML or 3M-PML in PML−/− primary cells. After 24 hours, cells were harvested and subjected to immunofluorescence analysis using the anti-PML polyclonal antibody. In PML−/−keratinocytes, transfected 3M-PML accumulated in aberrant nuclear aggregates, and wild-type PML localized in nuclear speckles (Figure3B). Wild-type PML accumulated on average in 14.4 ± 9.5 nuclear speckles per cell (n = 100). The average number of PML NBs in PML+/+ keratinocytes was 15.2 ± 8.3 (n = 100). By contrast, the 3M-PML formed, on average, 1.6 ± 0.9 nuclear aggregates per cell (n = 100). Furthermore, the size of the speckles observed in cells transfected with wild-type PML was similar to the size of PML-NBs in wild-type keratinocytes (0.3-0.5 μm) and much smaller than that of the 3M-PML aggregates (1-2 μm). Thus, SUMOylation of PML is crucial for PML to localize in the NB.
3M-PML forms aberrant nuclear aggregates.
(A) Schematic structure of PML and 3M-PML. Wild-type PML can be modified by SUMO-1 at amino acid positions 65, 160, and 490 through 3 lysines (K). In 3M-PML, the 3 lysines were mutated to arginines (R). R: RING finger domain. B1, B2: B-boxes. Coiled coil: helical coiled-coil region. S/P: serine/proline-rich region. (B) Localization of transfected wild-type PML and 3M-PML. Representative confocal micrographs are shown, with the immunofluorescent colors (PML/3M-PML, red; DAPI, blue) labeled in the lower corners of each image. Bar: 5 μm.
3M-PML forms aberrant nuclear aggregates.
(A) Schematic structure of PML and 3M-PML. Wild-type PML can be modified by SUMO-1 at amino acid positions 65, 160, and 490 through 3 lysines (K). In 3M-PML, the 3 lysines were mutated to arginines (R). R: RING finger domain. B1, B2: B-boxes. Coiled coil: helical coiled-coil region. S/P: serine/proline-rich region. (B) Localization of transfected wild-type PML and 3M-PML. Representative confocal micrographs are shown, with the immunofluorescent colors (PML/3M-PML, red; DAPI, blue) labeled in the lower corners of each image. Bar: 5 μm.
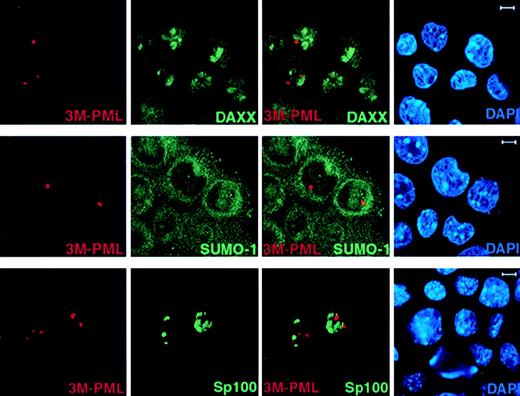
We next studied if unSUMOylated PML can still recruit NB components, such as Daxx, by performing immunofluorescence staining of endogenous Daxx and SUMO-1 in 3M-PML–transfected PML−/−keratinocytes. In contrast to wild-type PML (Figure 2), 3M-PML failed to colocalize with Daxx, SUMO-1, and GFP-Sp100 (Figure4), suggesting that SUMO modification is a prerequisite for PML to recruit other components, such as Daxx and Sp100, to the NB.
3M-PML fails to recruit Daxx, SUMO-1, and Sp100 to the NBs.
PML−/− keratinocytes were transfected with pSG5-3M-PML alone or cotransfected with pSG5-3M-PML and pEGFP-Sp100. After 24 hours, cells were harvested and cytospun onto glass slides for immunofluorescence staining. Representative confocal pictures are shown, with the immunofluorescent colors (PML, red; SUMO-1/Daxx/GFP-Sp100, green; DAPI, blue) labeled in the lower corners of each image. Bar: 5 μm.
3M-PML fails to recruit Daxx, SUMO-1, and Sp100 to the NBs.
PML−/− keratinocytes were transfected with pSG5-3M-PML alone or cotransfected with pSG5-3M-PML and pEGFP-Sp100. After 24 hours, cells were harvested and cytospun onto glass slides for immunofluorescence staining. Representative confocal pictures are shown, with the immunofluorescent colors (PML, red; SUMO-1/Daxx/GFP-Sp100, green; DAPI, blue) labeled in the lower corners of each image. Bar: 5 μm.
PML-RAR can recruit NB components to nuclear microspeckled sites in the absence of PML
In APL cells, PML-RARα delocalizes PML and other NB members, such as SUMO-1 and Daxx, to microspeckled nuclear regions.5-7 It has been suggested that the accumulation of NB components in microspeckles might be due to the ability of PML-RARα to heterodimerize with PML.5-7 To further understand the role of PML in determining the subnuclear localization of PML-RARα and in the ability of the fusion protein to determine aberrant localization of NB components, we transfected PML-RARα inPml+/+ andPml−/− MEFs. In the absence of PML, PML-RARα still formed nuclear microspeckles that were indistinguishable from the ones in Pml+/+cells (Figure 5 and not shown). Endogenous SUMO-1 and Daxx colocalized with overexpressed PML-RARα in nuclear microspeckles in Pml−/− cells (Figure5B). Because PML-RARα contains a large portion of PML and 2 or 3 PML SUMOylation sites depending on the breakpoint within the PML locus23,24 (3 in the PML-RARα isoform that we have used; see “Materials and methods”) and it can be SUMOylated,21 the recruitment of SUMO-1 and Daxx is presumably due to the PML moiety of PML-RARα. It must be noted, however, that in NB4 APL cells expressing PML-RARα, Daxx was found delocalized from the NBs but did not completely colocalize with PML-RARα.9
In Pml−/− cells, PML-RAR accumulates in nuclear microspeckles and recruits endogenous SUMO-1 and Daxx to these aberrant sites.
PML−/− MEFs were transfected with pSG5-PML-RARα. After 24 hours, cells were harvested and cytospun onto glass slides for immunofluorescence analysis. Representative confocal pictures are shown, with the immunofluorescent colors (PML-RARα, red; SUMO-1/Daxx, green; DAPI, blue) labeled in the lower corners of each image. Bar: 5 μm.
In Pml−/− cells, PML-RAR accumulates in nuclear microspeckles and recruits endogenous SUMO-1 and Daxx to these aberrant sites.
PML−/− MEFs were transfected with pSG5-PML-RARα. After 24 hours, cells were harvested and cytospun onto glass slides for immunofluorescence analysis. Representative confocal pictures are shown, with the immunofluorescent colors (PML-RARα, red; SUMO-1/Daxx, green; DAPI, blue) labeled in the lower corners of each image. Bar: 5 μm.
Discussion
The PML NBs have been intensively studied during the past 10 years, mostly because of the dynamics of these subnuclear structures during viral infections and in human cancer. However, the function of the NB remains unclear. The variety of the NB components suggests a wide range of possible biological roles, including tumor and growth suppression, transcription regulation, and cellular immune response via IFN.8 Also, it has been suggested that these bodies can accumulate inactive proteins and release them upon certain signals.35 One fundamental question that needs to be addressed, however, is whether certain proteins act as constitutive components of the NB and are required for the integrity and stability of this nuclear structure. Here, we demonstrate that PML is critical for several proteins to localize in the NB, including Sp100, Daxx, CBP, and ISG20. Other proteins that have been recently found to colocalize with PML in the NB, such as the BLM DNA helicase implicated in the pathogenesis of the Bloom syndrome, are also delocalized in the absence of PML.36 Both BLM and Sp100 do not appear to directly interact with PML.6 36 However, it is possible that BLM and Sp100 interact with other NB components. It remains to be seen if other NB components might aggregate in NB-like structures, even in the absence of PML, and whether these molecules are required for PML accumulation in the NB. The identification of such proteins will further clarify the chain of events required for NB nucleation.
It has been suggested that only the SUMO-1–modified PML is tightly associated with the nuclear matrix and that, therefore, PML is localized to the NB as a consequence of its modification.21This mechanism of recruitment could apply to other NB components. SUMO-1 would therefore serve to direct the proper compartmentalization of proteins into the NB.21 While this article was in revision, it was reported that, in HEp-2 cells, transfected 3M-PML mutant protein accumulated in NBs.37 However, because HEp-2 cells express endogenous PML, it is likely that the 3M-PML mutant protein formed heterodimeric complexes with the endogenous wild-type PML and was thus recruited to the NBs. In contrast, by using PML−/− cells, we conclusively demonstrate that PML needs to be modified by SUMO-1 to acquire an NB-speckled localization pattern. Furthermore, our data also demonstrate that the SUMO-1 modification of Sp100 is not sufficient for this protein to acquire an NB localization in the absence of PML. This is also supported by the finding that SUMOylation of Sp100 is not necessary for its NB localization because an Sp100 mutant that can no longer be SUMOylated still localizes in the NB.38 Thus, although our data support a critical role for SUMOylation in directing the NB localization of modified proteins, they suggest that some of the modified NB components still depend on PML to accumulate into the NB. The notion that SUMO-1 is necessary but not sufficient to determine the localization of its targets is in agreement with the fact that modified proteins have been found in various locations, such as the NB, the nuclear pore (RanGAP118), and the cytosol (IκBα19)b and that, as aforementioned, at least in the case of Sp100, lack of modification does not impair proper nuclear localization.38 It is also noteworthy that not every SUMO-1–modified NB component is essential for the localization of other NB proteins. For example, when NT2 cells differentiate into nerve cells in vitro, Sp100 is not expressed while PML is still localized to the NBs.35 Hence, it is not just the relative amount of SUMO-1–interacting proteins in the NBs but, rather, the specific function of PML that determines the localization of the SUMO-1–modified NB proteins. We therefore propose a hierarchy of events by which PML needs to be first SUMOylated in order to accumulate in the NBs, where it subsequently recruits additional NB components.
Our findings also provide an explanation for why the delocalization of PML from the NB by PML-RARα in APL blasts, or the degradation of PML upon viral infections, results in the delocalization of multiple proteins from the NB.2,5-7 In some instances, PML-RARα could induce the delocalization of NB proteins directly, possibly through the PML moiety. We have recently reported that the PML−/− mice are viable but are more susceptible to tumorigenesis and bacterial infections.15PML−/− cells exhibit a marked growth advantage.15 In addition, PML−/−mice and cells are protected from multiple apoptotic stimuli.16 Thus, PML appears to regulate multiple cellular functions. Some of these tumor-suppressive functions could be attributed to the ability of PML to act as a transcriptional cofactor.17 One implication of the findings herein is that the pleiotropic effects of PML inactivation might be due to the disruption of several pathways regulated by proteins that depend on PML to be properly compartmentalized in the nucleus in order to exert their normal functions. The fact that Daxx and Sp100 are delocalized from the NBs in the absence of PML may be particularly relevant because Daxx may have a role regulating programmed cell death,30-39 whereas Sp100 is up-regulated by interferons29 and thus possibly mediates the growth-suppressive and immunologic functions of these cytokines. Delocalization of CBP in the PML−/−cells may contribute to their impaired responses to retinoic acid–induced transcription.17Therefore, PML might exert some of its biological functions through the regulation of the formation of the NB, and PML-RARα may contribute to leukemogenesis through its ability to interfere with these processes by interacting with and sequestering PML.
Acknowledgments
We thank Michael Antoniou, Maria Barna, Letizia Longo, Katia Manova, Gerd G. Maul, Michael Matunis, and Zhu Gang Wang for advice, materials, and help in some experiments.
Supported by the National Cancer Institute (CA-08748, CA-71692, and CA74031 awarded to P.P.P.). S.M. was supported by a fellowship from the Association for International Cancer Research. S.R. was partially supported by Centro Nazionale per la Ricerca. P.P.P. is a Scholar of the Leukemia Society of America.
Reprints:Pier Paolo Pandolfi, Department of Human Genetics and Molecular Biology Program, Memorial Sloan-Kettering Cancer Center, Sloan-Kettering Division, Graduate School of Medical Sciences, Cornell University, 1275 York Ave, New York, NY 10021; e-mail:p-pandolfi@ski.mskcc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal