The HLA-DR-associated peptides from peripheral blood mononuclear cells of 2 patients with plasmacytoma and 1 with chronic myeloid leukemia were isolated, identified, and compared. Several were identified as derivatives of the defensin family. Defensins (or human neutrophil peptides [HNP]) are antimicrobial, cationic peptides of 29 to 35 amino acids in length and are the major constituents of the azurophilic granules of human neutrophils. Using peripheral blood cells from leukapheresis, containing about 90% of polymorphonuclear cells, we could identify HNP-1, -2, and -4 and propeptides of up to 49 amino acids in length, eluted from HLA class II molecules. Binding of isolated and synthetic defensin peptides to various HLA-DR alleles using an in vitro binding/competition assay based on size exclusion chromatography revealed that defensin may bind into the peptide-binding groove. In a T-cell competition assay, defensins were able to reduce the proliferation of an HLA-DR-restricted T-cell line after preincubation of stimulating cells (CHO-DRB1*0401 transfectants) with defensin. Therefore, binding of defensins might prevent T-cell recognition of HLA class II molecules expressed on different blood precursor cells (all of which are “nonprofessional” antigen-presenting cells) by blocking the HLA peptide-binding groove or, alternatively, might protect defensin-expressing cells from self-destruction.
Only antigen-presenting cells (APC), which express major histocompatibility complex (MHC) class II molecules, are able to present a great variety of peptide antigens derived from proteins entering the endocytic pathway to CD4+ T cells. Constitutive MHC class II gene expression is not only tightly restricted to APC but is also under developmental control. Cells of the B-cell lineage acquire the capacity to express MHC class II genes early during ontogeny but normally lose this property during terminal differentiation into plasma cells.1 This phenomenon is due to a silencing of the transactivator gene CIITA.2Little is known about the function of these temporarily expressed MHC class II molecules. Recently, Harris et al3 reported on MHC class II-associated self-peptides from the hematopoietic progenitor cell line KG-1. This cell line, originating from a patient with acute myelogenous leukemia, is believed to be derived from an oligopotent myeloid progenitor cell and is morphologically heterogeneous, containing myeloblasts and, to a lesser extent, promyelocytes, myelocytes, granulocytes, macrophages, and eosinophils. A number of HLA-DR-associated peptides, mainly derived from intracellular rather than from exogenous or transmembrane protein sources, was identified.3 Furthermore, KG-1 showed a much lower frequency of class II-associated invariant chain peptides (CLIP) on the plasma membrane, compared to professional APCs. Therefore, the authors suggested that exogenous antigen processing may be a developmentally acquired characteristic in the myeloid lineage. Another paper from the same group4 describes naturally processed peptides bound by HLA-DR1 or 3 (or both) from the CD34+ blast cells of a patient with chronic myeloid leukemia (CML). The authors identified a panel of peptides from different protein sources stemming from membrane-associated, luminal, but also cytoplasmic proteins. The peptide lengths varied between 13 and 18 amino acids and 3 of them seemed to be tissue specific. Two of them, namely MRP14 and leukotriene-B4-omega-hydrolase, were derived from proteins specific for myeloid cells; another one, granzyme H, was specific for T lymphocytes and natural killer cells.
To characterize HLA class II-associated peptides stemming directly from patient material, we studied polymorphonuclear cells (PMNs) from a patient with CML and from 2 patients with plasmacytoma. Surprisingly, we found a very high amount of peptides derived from the defensin family associated with HLA-DR molecules from these cell sources.
Defensins or human neutrophil peptides (HNP) are highly potent antimicrobial peptides of about 30 amino acids in length, effective against a wide variety of bacteria, many fungi, some enveloped viruses, and even a wide range of normal and malignant mammalian target cells (for reviews, see Lehrer et al5, Ganz and Lehrer6, and Hancock7). They function by inserting into various cell membranes due to their ability to aggregate, forming voltage-regulated channels.8,9Defensins, which are released from stimulated neutrophils, show further cytotoxic activity to various autologous cells,10induction of histamine release by mast cells,11 and chemotactic activity for monocytes and T cells.12,13Recently, it has been shown that defensins may influence complement activation by binding to C1q.14,15 Defensin has also been shown to stimulate the binding of lipoprotein (a) to human vascular endothelial and smooth muscle cells.16 Very recently, it was reported that defensins are able to enhance serum IgG antibody responses in mice after intranasal delivery.17 The authors showed further that defensins enhanced both proliferative responses and T-helper cytokine secretion profiles of naive CD4+ T cells. Peptides of the defensin family are highly expressed in promyeloic cells and constitute more than 5% of the total cellular protein in human and rabbit PMN.6 Furthermore, defensins are frequently found in Paneth cells, platelets, and macrophages, as well as in mammalian trachea, intestine, and tongue.7 Therefore, members of the defensin family seem to be important components of the innate immune system of a large number of mammals (and also of some plants), active against microbial infections. These findings support the hypothesis that different classes of antimicrobial peptides play an important role in the natural defense against microbes.
Material and methods
Leukapheresis and cell preparation
Patients with plasmacytoma were pretreated with 10 μg/kg granulocyte colony-stimulating factor (G-CSF) per day for 3 days to release stem cells into the periphery. Then, 1 to 2 × 1010 peripheral blood mononuclear cells (PBMC) were isolated by leukapheresis. CD34+ stem cells were removed immediately after leukapheresis by using magnetic beads coated with α-CD34 (MACRO-MACS, Dynal, Oslo, Norway). The residual cells were used for HLA class II isolation. The cell preparation was performed as described previously.18 Briefly, cells were pelleted at 1000g and lysed with 2% NP-40. The lysate was cleared by ultracentrifugation and submitted to affinity chromatography using the monoclonal antibody (mAb) L243 (α-DR, obtained from the American Type Culture Collection [ATCC], Rockville, MD). For preventing nonspecific adsorbance, a mock precolumn made of sepharose material was used. Elution of the HLA molecules and the bound peptides was performed using trifluoracetic acid (TFA)/water, pH 2.0. The peptide-released HLA class II isolates were analyzed for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were judged to be more than 90% pure. Acid-eluted peptides were separated from the HLA molecules by ultrafiltration (Amicon, Danvers, MA) using a 20-kd cutoff membrane (Sartorius, Göttingen, Germany). The peptide-containing fraction was lyophilized and subsequently resolubilized in acetonitrile/water, 1:1 containing 0.1% (v/v) TFA.
High-performance liquid chromatography (HPLC)—purification and identification of eluted peptides
The peptide pool was separated by capillary reversed phase (RP)-HPLC using an RP-C18 300 μm × 150 mm column (LC-Packings, Amsterdam, The Netherlands). The following gradient conditions were used: solvent A, 0.1% TFA; solvent B, 0.08% TFA, 84% acetonitrile; gradient, 0.5% B/minute, starting at 5% B (plasmacytoma 2), 5% B from 0 to 12 minutes, 1% B/minute (plasmacytoma 1), and 1% B/minute starting at 0% B (CML). The absorbance at 215 nm, corresponding to the peptide bonds, and at 295 nm, detecting tryptophane-containing peptides, was measured (UV detector, LC-Packings). All collected peaks were assayed for their peptide masses and for homogeneity by matrix-assisted laser desorption mass spectrometry (MALDI-MS). Pure fractions were submitted directly for microsequencing by N-terminal Edman degradation using an ABI 476A sequencer (Weiterstadt, Germany), but fractions containing more than 1 mass were further purified by capillary RP-HPLC using a 180-μm column (SGE, Frankfurt/M., Germany), collected automatically with a robot (Probot, BAI, Bensheim, Germany), spotted directly on a polyvinylidine difluoride (PVDF) sequencing membrane and subsequently sequenced. Sequencing results were compared with the Swissprot database at the European Bioinformatics Institute (EBI), Hinxton Hall, Cambridge, UK.
Nanospray electrospray ionization mass spectrometry (ESI-MS) of defensin
For exact mass determination, ESI-MS spectrometry was performed with a TSQ 7000 mass spectrometer (Finnigan, La Jolla, CA) equipped with a nano electrospray (nano ESI) ion source. Nanospray capillaries were obtained from PROTANA (Odense, Denmark). An HPLC fraction (0.5 μL) was used for nano ESI mass spectrometry.
Defensin isolation from the promyelocytic human cell line HL60
Defensin isolation was performed initially as described elsewhere,19 using some modifications. Briefly, HL60 cells were cultured in RPMI 1640 with 5% fetal calf serum (FCS) in suspension to collect a total of 1 × 109cells. After harvesting the cells, 10 mL of extraction medium (1 mol/L HCl, 5% [v/v] formic acid, 1% [w/v] NaCl, 1% TFA) was added. Cells were homogenized and pelleted (2000g, 15 minutes, 4°C), and the pellet re-extracted with 3 mL extraction medium. The collected extraction supernatants were then concentrated using a Sep Pac C18 column (Pharmacia, Uppsala, Sweden). After washing with 0.1% TFA, the elution of the bound material was performed stepwise by increasing acetonitrile concentrations: 20% acetonitrile/0.1% TFA, 40% acetonitrile/0.1% TFA, 60% acetonitrile/0.1% TFA, and 80% acetonitrile/0.1% TFA. Each fraction was collected, lyophilized, and subsequently tested for its defensin content by dot blot using the anti-HNP-1,- 2, -3-specific mouse mAb Def-3 (BMA Biomedicals, Augst, Switzerland). Only the fraction eluted with 80% acetonitrile contained defensins. This fraction was further purified by RP-HPLC (column: Vydac C4, 2 × 150 mm, gradient: 0-15% B in 10 minutes, 15-45% B in 20 minutes, 45-70% B in 50 minutes, and 70-100% B in 60 minutes.). All peaks detected at 214 nm were collected and tested for their defensin content by Western blot after running a 16% SDS-PAGE. The detection was performed using the enhanced chemiluminescence method (ECL, Amersham Buchler, Braunschweig, Germany). Most of the HPLC fractions tested were positive for defensin, but different molecular weights corresponding to different precursor forms of defensins were detected. Each fraction containing defensin was analyzed by mass spectrometry (Finnigan MAT), but none of the fractions proved to be homogeneous for one single defensin peptide. However, the fractions contained no other protein contaminants and were more than 95% pure defensin molecules.
In vitro binding/competition assay
For reasons related to solubility of the material, we were unable to label defensins directly with a fluorescence marker (7-amino-4-methylcoumarin-3-acetic acid [AMCA]). Therefore, to investigate binding/competition of defensins to different solubilized HLA class II alleles, an in vitro binding/competition assay based on gel filtration was used, as described previously.18 20Briefly, we used fluorescent AMCA-labeled allele-specific binding peptides and immunoaffinity-purified HLA-DR isolates from different homozygous Epstein-Barr virus (EBV)-transformed cell lines (B-LCLs). As competitors, the defensin-containing fraction from HL60 or a synthetic HNP-2 (Bachem, Heidelberg, Germany) was used in different concentrations.
Defensin competition assay
To determine the competition capacity of isolated defensins on a preformed MHC class II/peptide complex, HLA-DR1 isolate (0.2 μmol/L) was incubated with AMCA-HA 306-18 (1.5 μmol/L) for 24 hours at 37°C in binding buffer. Then defensins (10 and 20 μg, respectively), or unlabeled self-peptides (HA 306-18, IM 19-31, CLIP 80-104, each at a concentration of 30 μmol/L) were added and the competition kinetics were determined using a gel filtration-based competition assay.
Association kinetics assay
To detect the association kinetics of hemagglutinin 306-18 to HLA-DR1 in the presence of defensin, HLA-DR1 (0.16 μmol/L) was preincubated with 10 μg defensin isolate in binding buffer (0.15 mol/L sodium phosphate, 0.1% [w/v] Zwittergent 3-12, protease inhibitor mix, 15% acetonitrile, pH 5.2) at 37°C for 1 hour or overnight. Then, 1.5 μmol/L AMCA-hemagglutinin 306-318 was added to start the association kinetics. The relative protein-bound AMCA fluorescence intensity was detected at different time points. As a control, the same assay was performed without defensin and in the presence of defensin and HLA-DM.
Cell lines and antibodies
Epstein-Barr virus-transformed, homozygous B-LCLs WT100BIS (HLA-DRB1*0101), LD2B (HLA-DRB1*1501/DRB5*0101), COX (HLA-DRB1*0301/DRB3*0101), BSM (HLA-DRB1*0401/DRB4*0101), and DBB (HLA-DRB1*0701/DRB4*0101) served as sources for the HLA-DR alleles. The expression of the HLA-DRB3 and DRB4 alleles was low compared to that of the DRB1 alleles. Cells were cultured in RPMI (Life Technologies, Eggenstein, Germany) supplemented with 5% FCS (Life Technologies), 20 mmol/L HEPES, 2 mmol/L glutamine, and antibiotics and propagated in roller bottles to 1 to 2 × 109 cells. The mAb L243, which recognizes a nonpolymorphic determinant present on HLA-DR dimers, was obtained from ATCC. The HNP1-3 recognizing monoclonal mouse antibody Def-3 was obtained from BMA Biomedicals.
T-cell proliferation assay
The CHO K1 cells were transfected with the genes for HLA-DRB1*0401 and HLA-DRA and cotransfected with the gene coding for CD80 as described previously (kind gifts of Dr D. Sansom, Bath, UK).21 The expression of the human genes was checked by FACS analysis before the cells were used as APCs. For priming of T cells, 1 × 106 PBMC of healthy donors, carrying the HLA-DRB1*0401 allele, were cocultured with 5 × 105CHO cells (fixed with 0.025% glutaraldehyde for 2 minutes) in 16-mm diameter Costar wells in 2 mL RPMI 1640 supplemented with 10% heat-inactivated human male serum. After 7 days of culture, cells were harvested, washed, and plated at 2 × 104 cells/well into round-bottomed microtiter plates, to which 2.5 × 104 CHO cells as stimulators were added. To be used as stimulators, 5 × 105 CHO cells were preincubated with different concentrations of defensin isolate (none, 10, 20, or 30 μmol/L, respectively) in 500 μL RPMI 1640 for 2 hours at 37°C. Pulsed CHO cells were washed twice, fixed with glutaraldehyde (0.025%, 2 minutes), and washed again. Then they were cocultured with the T cells for 1 or 2 days, respectively, at 37°C. After 24 and 48 hours, 37 kBq of tritiated thymidine (3H-TdR; Amersham-Buchler) was added and cells were harvested after 16 hours on glass fiber filtermats using a semiautomatic cell harvester. T-cell proliferation was determined by3H-TdR incorporation by liquid scintillation counting. The anti DR-specific mAb L243 was used to block the HLA-DR molecules in this stimulation system. For 2 different kinetics, 2 identical plates were set up, containing all cultures performed in triplicate. The results are expressed as mean counts per minute (cpm).
Proliferations of T cells, isolated from a normal donor recognizing CHO-DRB1*0401 transfectants, were tested for their response to the stimulator cells with or without different amounts of defensin in the culture medium.
Results
HLA-DR-bound self-peptides
From 9.6 × 109 PBMC (plasmacytoma patient 1; P1) and 1.4 × 1010 PBMC (plasmacytoma patient 2; P2) 690 and 900μg of HLA-DR was isolated, corresponding to 1.15 and 1.5 nmol of MHC protein. In the case of P2, for example, 12 of the most prominent HPLC fractions were directly submitted to the Edman sequencer. A further 14 fractions were sequenced after re-chromatography. The peptide yield of these sequencing runs differed from about 10 to 100 pmol/run for the directly sequenced fractions and from 5 to 20 pmol for the re-chromatographed fractions. Altogether, these 26 fractions contained about 450 pmol of peptides. From these 26 sequencing runs 9 sequences could be clearly identified. From these 9 sequences, 6 were derived from the defensin family. These 6 defensins yielded 110 pmol, which corresponded to 0.24% of the total yield of protein in all 26 fractions. This means that within the high-copy peptides occupying the binding groove of the HLA-DR molecules of this patient, about one fourth were defensins. Table1 summarizes the peptides identified from the 2 patients with plasmacytoma and the single patient with CML. The most striking feature of the HLA-DR-associated peptides identified from these sources was the predominance of peptides from the defensin family. HNP-1, -2, and -4 and several HNP precursors were identified. HNP-1 and -2 differ only in their N-terminal residue, having an alanine in HNP-1 and no residue in HNP-2. The peptides from the defensin family had lengths from 29 to 49 amino acids with masses in the range of 3400 to 5600 d. This is very unusual for HLA-DR-associated self-peptides, which are usually 13 to 25 amino acids in length.22 Ten of 18 identified peptides were derived from the defensin family; 4 of the remaining sequences stemmed from the CLIP region of the invariant chain. One of the sequences of P2 proved to be the N-terminus of the hemoglobin α-chain. Apart from defensin and CLIP, the usual type of MHC class II-binding peptide of 13 to 25 amino acids was almost completely absent from these isolates. There may possibly be 3 additional peptide sequences in these isolates, but their lengths were not determined and the database alignment with known sequences was not perfect. The potential self-peptide candidates may represent sequences derived from the ribosomal protein S26, from the cell cycle protein CDC27HS, and from an unknown source.
Peptides identified in 3 patients
| Source . | Eluted at (% Actonitril) . | Sequence . | Protein Source . | Residues . | Yield (pmol) . | Mass (Found./Theor.) . |
|---|---|---|---|---|---|---|
| P1 | 30.2, 32.8 | ACYCRIPACIAGERRYGTCIYQGRLWAFCC | HNP-1 | 65-94 | 45, 32 | 3454.4/3439.5 |
| P1 | 30.2, 31.2 | CYCRIPACIAGERRYGTCIYQGRLWAFCC | HNP-2 | 66-94 | 45, 32 | 3383.4/3368.4 |
| P1 | 31.2 | NMACYCRIPACIAGERRYGTCIYQGRLWAFCC | pre-HNP-1 | 63-94 | 20 | 3705.4/3684.6 |
| P1 | 31.2, 32.0 | SLAWDESLAPKHPGSRKNMACYCRIPACIAGERRYGTCIYQGRLWAFCC | pre-HNP-1 | 46-94 | 35, 16 | 5554.4/5544.5 |
| P1 | 23.2 | XPVFYVGPEXQI … | Unknown | ? | 15 | 3188.4/? |
| P1 | 28.4 | LPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTE | CLIP | 80-122 | 20 | 4657.9/4666.7 |
| P1 | 35.3 | LPKPPKPVSKMRMATPLLMQALP | CLIP | 80-102 | 15 | 2557.5/2545.3 |
| P1 | 35.3 | KDVALSVLSKDLTDMD … | CDC27HS | 548-? | 10 | nd |
| P1 | 35.7 | PKPPKPVSKMRMATPLLMQALPM | CLIP | 81-103 | 8 | 2564.9/2563.3 |
| P1 | 36.8 | VCSCRLVFCRRTELRVGNCLIGGVSFTYCCTRVD | HNP-4 | 64-97 | 18 | 3818.6/3821.3 |
| P1 | 37.7 | LPKPPKPVSKMRMATPLLMQALPM | CLIP | 80-103 | 12 | 2679.9/2676.5 |
| P2 | 30.0 | ACYCRIAPACIAGERRYGTCIYQGRLWAFCC | HNP-1 | 65-94 | 20 | 3426.3/3439.5 |
| P2 | 30.0, 30.5 | CYCRIAPACIAGERRYGTCIYQGRLWAFCC | HNP-2 | 66-94 | 20, 17 | 3357.6/3368.4 |
| P2 | 30.5 | VLPKLYVKLHYCVSVSCVIHSKV … | Rib prot S26 | 63-? | 25 | nd |
| P2 | 32.8, 36.0, 36.2 | DIPEVVVSLAWDESLAPKHPGSRKNMACY … | pre-HNP-1 | 39-? | 40, 5, 8 | nd |
| P2 | 30.5, 40.5 | VLSPADKTNVKAAWGKVGAH | hemoglobin α | 1-? | 18, 15 | nd |
| CML | 29.9 | ACYCRIPACIAGERRY … | HNP-1 | 65-? | nd | nd |
| CML | 29.9 | CYCRIPACIAGERRY … | HNP-2 | 66-? | nd | nd |
| Source . | Eluted at (% Actonitril) . | Sequence . | Protein Source . | Residues . | Yield (pmol) . | Mass (Found./Theor.) . |
|---|---|---|---|---|---|---|
| P1 | 30.2, 32.8 | ACYCRIPACIAGERRYGTCIYQGRLWAFCC | HNP-1 | 65-94 | 45, 32 | 3454.4/3439.5 |
| P1 | 30.2, 31.2 | CYCRIPACIAGERRYGTCIYQGRLWAFCC | HNP-2 | 66-94 | 45, 32 | 3383.4/3368.4 |
| P1 | 31.2 | NMACYCRIPACIAGERRYGTCIYQGRLWAFCC | pre-HNP-1 | 63-94 | 20 | 3705.4/3684.6 |
| P1 | 31.2, 32.0 | SLAWDESLAPKHPGSRKNMACYCRIPACIAGERRYGTCIYQGRLWAFCC | pre-HNP-1 | 46-94 | 35, 16 | 5554.4/5544.5 |
| P1 | 23.2 | XPVFYVGPEXQI … | Unknown | ? | 15 | 3188.4/? |
| P1 | 28.4 | LPKPPKPVSKMRMATPLLMQALPMGALPQGPMQNATKYGNMTE | CLIP | 80-122 | 20 | 4657.9/4666.7 |
| P1 | 35.3 | LPKPPKPVSKMRMATPLLMQALP | CLIP | 80-102 | 15 | 2557.5/2545.3 |
| P1 | 35.3 | KDVALSVLSKDLTDMD … | CDC27HS | 548-? | 10 | nd |
| P1 | 35.7 | PKPPKPVSKMRMATPLLMQALPM | CLIP | 81-103 | 8 | 2564.9/2563.3 |
| P1 | 36.8 | VCSCRLVFCRRTELRVGNCLIGGVSFTYCCTRVD | HNP-4 | 64-97 | 18 | 3818.6/3821.3 |
| P1 | 37.7 | LPKPPKPVSKMRMATPLLMQALPM | CLIP | 80-103 | 12 | 2679.9/2676.5 |
| P2 | 30.0 | ACYCRIAPACIAGERRYGTCIYQGRLWAFCC | HNP-1 | 65-94 | 20 | 3426.3/3439.5 |
| P2 | 30.0, 30.5 | CYCRIAPACIAGERRYGTCIYQGRLWAFCC | HNP-2 | 66-94 | 20, 17 | 3357.6/3368.4 |
| P2 | 30.5 | VLPKLYVKLHYCVSVSCVIHSKV … | Rib prot S26 | 63-? | 25 | nd |
| P2 | 32.8, 36.0, 36.2 | DIPEVVVSLAWDESLAPKHPGSRKNMACY … | pre-HNP-1 | 39-? | 40, 5, 8 | nd |
| P2 | 30.5, 40.5 | VLSPADKTNVKAAWGKVGAH | hemoglobin α | 1-? | 18, 15 | nd |
| CML | 29.9 | ACYCRIPACIAGERRY … | HNP-1 | 65-? | nd | nd |
| CML | 29.9 | CYCRIPACIAGERRY … | HNP-2 | 66-? | nd | nd |
Listing of HLA-DR-bound endogenous peptides from different sources. The retention times of the two plasmacytoma patient-derived fractions are comparable. Different acetonitrile gradients were used for the HPLC separation of the different experiments (see “Materials and methods”). To compare the corresponding elution times of each peptide, the calculated percentage of acetonitrile is indicated. Sequences were determined by micro-Edman sequencing, masses were determined by MALDI-MS and ESI-MS and the peptide lengths calculated. Some fractions were not submitted to mass spectrometry and therefore the C-terminus of the peptide was not detected.
CLIP indicates class II-associated invariant chain peptides; CML, chronic myeloid leukemia; ESI-MS, electrospray ionization mass spectrometry; HNP, human neutrophil peptide; HPLC, high-performance liquid chromatography; MALDI-MS, matrix-assisted laser desorption mass spectrometry; nd, not determined; P1, plasmacytoma patient 1; P2, plasmacytoma patient 2.
HLA-DR bound autologous peptides with unusual lengths
The length of most of the sequenced peptides could be determined by mass spectrometry. Figure 1 shows an ESI-MS spectrum of HNP from a representative HPLC fraction. The spectrum shows the nested set of HNP-1 and HNP-2 in 3 different charge states [M+2H]2+, [M+3H]3+, and [M+4H]4+. The mean masses deduced from the charge states were 3440.8 d for HNP-1 and 3370.4 d for HNP-2. The theoretical average mass of HNP-1 and HNP-2 are 3439.1 d and 3368.4 d calculated from the deduced sequence including 3 disulfide bonds. The mass difference of both defensins is 70.4 d corresponding just to the N-terminal alanine residue (theoretical average mass: 71 d).
ESI-MS spectrum of a representative HPLC fraction.
The spectrum shows 3 different charge states [M+2H]2+, [M+3H]3+, and [M+4H]4+ of HNP-1 (1721.3, 1148.1, 861.3) and HNP-2 (1685.8, 1124.6, 843.5), respectively. This indicates that these peptides have full lengths of 30 and 29 amino acids, respectively.
ESI-MS spectrum of a representative HPLC fraction.
The spectrum shows 3 different charge states [M+2H]2+, [M+3H]3+, and [M+4H]4+ of HNP-1 (1721.3, 1148.1, 861.3) and HNP-2 (1685.8, 1124.6, 843.5), respectively. This indicates that these peptides have full lengths of 30 and 29 amino acids, respectively.
Competition of defensins with allele-specific binding peptides
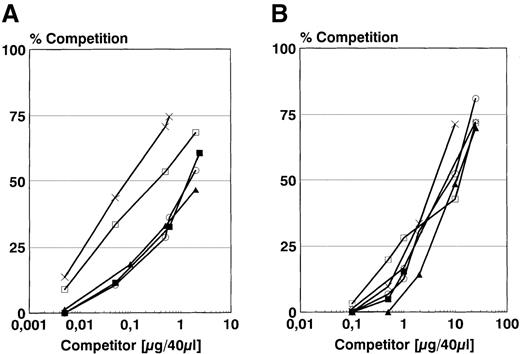
Next, the binding of synthetic HNP-2 and isolated defensins to different solubilized HLA-DR molecules in vitro was investigated. For this purpose, an in vitro binding/competition assay based on gel filtration was used. To be able to use unmodified, native defensin peptides, competition experiments against allele-specific, fluorescently AMCA-labeled binding peptides were performed (Figure2). The competition curves for the synthetic HNP-2 (Figure 2A) revealed allelic differences, showing that HLA-DR3 possessed highest affinity (IC50 5.2 μmol/L), followed by HLA-DR7 (IC50 16.3 μmol/L). HLA-DR1, 2, and 4, on the other hand, showed nearly identical, but markedly decreased affinities (IC50 about 92.5 μmol/L). Isolated defensins, however, proved to be about 10-fold less effective in their ability to compete on the different HLA-DR alleles (Figure 2B). In this case, the allele specificity was less marked, but still detectable. The results were again consistent with best binding to the HLA-DR3 allele. The IC50 values could not be calculated here because a nonhomogeneous mixture of defensins with no defined mass was used.
Competition against allele-specific AMCA-labeled binding peptides.
Competition of synthetic HNP-2 (A) and isolated HNP from HL-60 cells (B) to solubilized HLA-DRB1*0101 (○), HLA-DRB1* HLA-DRB1*1501/DRB5*0101 (▴), HLA-DRB1*0301 (×), HLA-DRB1*0401 (▪) and HLA-DRB1*0701 (□) against fluorescently (AMCA)-labeled binding peptides using a gel filtration assay. As binding peptides, HA 306-18 (PKYVKQNTLKLAT; 1.5 μmol/L) was used for HLA-DR1, -2 and -4. For HLA-DR3, heat shock protein 65, 3-13 (KTIAYDEEARR; 1.5 μmol/L) and for HLA-DR7, heat shock protein 70, 38-52 (TPSYVAFTDTERLIG; 1.5 μmol/L) was used. For (A), the concentration unit μg/40 μL can be transferred to μmol/L by multiplication with the factor 7.4. For (B), however, the μmol/L concentration of the used defensin isolate cannot be determined due to inhomogeneous masses of different defensin molecules.
Competition against allele-specific AMCA-labeled binding peptides.
Competition of synthetic HNP-2 (A) and isolated HNP from HL-60 cells (B) to solubilized HLA-DRB1*0101 (○), HLA-DRB1* HLA-DRB1*1501/DRB5*0101 (▴), HLA-DRB1*0301 (×), HLA-DRB1*0401 (▪) and HLA-DRB1*0701 (□) against fluorescently (AMCA)-labeled binding peptides using a gel filtration assay. As binding peptides, HA 306-18 (PKYVKQNTLKLAT; 1.5 μmol/L) was used for HLA-DR1, -2 and -4. For HLA-DR3, heat shock protein 65, 3-13 (KTIAYDEEARR; 1.5 μmol/L) and for HLA-DR7, heat shock protein 70, 38-52 (TPSYVAFTDTERLIG; 1.5 μmol/L) was used. For (A), the concentration unit μg/40 μL can be transferred to μmol/L by multiplication with the factor 7.4. For (B), however, the μmol/L concentration of the used defensin isolate cannot be determined due to inhomogeneous masses of different defensin molecules.
Defensins have a slow binding/competition kinetics
The competition kinetics of defensins to HLA-DR1-bound AMCA-labeled hemagglutinin (HA) 306-18 peptide were investigated next. After incubating HLA-DR1 with AMCA-HA 306-18 for 24 hours at 37°C, isolated defensins or self-peptides (HA 306-18, influenza matrix protein [IM] 19-31, invariant chain-derived CLIP 80-104) were added and the competition kinetics were determined. As shown in Figure3, the defensin isolate proved to be an effective competitor for the HLA-DR1 binder AMCA-HA 306-18, with a clear dose-dependent effect. After 70 hours, 45% competition using 10 μg defensin isolate versus 64% competition with 20 μg defensin isolate was determined. Using HA 306-18 as the competitor, 86% competition after 70 hours of incubation was observed. Another known HLA-DR-restricted T-cell epitope, IM 19-31,23 however, showed only 40% competition after 70 hours. CLIP 80-104, which has been shown to be a good HLA-DR1 binder,24 gave 73% competition after 70 hours. Interestingly, defensin showed an even slower competition kinetics compared to the other peptides tested and showed no clear saturation even after 70 hours. Half-maximal competition was achieved after about 16 hours for HA 306-18, 12 hours for CLIP 80-104, 13 hours for IM 19-31, but only after 25 or 29 hours for defensin (10 μmol/L; 20 μmol/L). This phenomenon might point to a complicated binding mechanism due to the unusual structure and length of these peptides.
Defensin isolate as competitor for the HLA-DR1 binder AMCA-HA 306-18.
Competition kinetics of defensins (10 μg [○]; 20 μg [×]) and different HLA-DR1-binding peptides (CLIP 80-104: LPKPPKPVSKMRMATPLLMQALPMG (□); IM 19-31: PLKAEIAQRLEDV (▪), and HA 306-18 (PKYVKQNTLKLAT ([▴]), each of them at a concentration of 30 μmol/L) to solubilized HLA-DR using a gel filtration assay; 1.5 μmol/L of AMCA-labeled HA 306-18 was used as a binding peptide.
Defensin isolate as competitor for the HLA-DR1 binder AMCA-HA 306-18.
Competition kinetics of defensins (10 μg [○]; 20 μg [×]) and different HLA-DR1-binding peptides (CLIP 80-104: LPKPPKPVSKMRMATPLLMQALPMG (□); IM 19-31: PLKAEIAQRLEDV (▪), and HA 306-18 (PKYVKQNTLKLAT ([▴]), each of them at a concentration of 30 μmol/L) to solubilized HLA-DR using a gel filtration assay; 1.5 μmol/L of AMCA-labeled HA 306-18 was used as a binding peptide.
Decreased binding capacity of peptide antigens to HLA-DR1 after preincubation with defensins
Reciprocally, the binding kinetics of AMCA-HA 306-18 to HLA-DR1 after preincubation with defensin isolate for 1 hour or overnight and in the presence of 1.5 μmol/L solubilized HLA-DM were determined. Figure 4 shows that although the absolute amount of bound AMCA-HA peptide was clearly influenced by preincubation with defensins, the binding kinetics were not affected (all samples had the same half-maximal association time [t1/2]). Without preincubation with defensins, the relative protein-bound fluorescence (rF), corresponding to the absolute fluorescence divided by the UV signal at 214 nm (protein) concentration was about 8 after 4.5 days. The preincubation with defensin for 1 hour resulted in a 25% decrease of rF to about 6. The overnight preincubation with defensin, however, resulted in a 37% decrease of rF to about 5. After overnight incubation with defensin and HLA-DM, however, an increased rF by 25% of up to nearly 10 after 4.5 days was found. Interestingly, the t1/2 was identical in all cases, being about 18 hours.
Binding kinetics of AMCA-HA 306-18 to HLA-DR1.
Binding kinetics of AMCA-HA 306-18 to HLA-DR1 (□), after preincubation with defensin isolate for 1 hour (▪) or overnight (▴), and in the presence of 1.5μmol/L solubilized HLA-DM (×) using a gel filtration assay. As binding peptide, 1.5 μmol/L AMCA-HA 306-18 was used.
Binding kinetics of AMCA-HA 306-18 to HLA-DR1.
Binding kinetics of AMCA-HA 306-18 to HLA-DR1 (□), after preincubation with defensin isolate for 1 hour (▪) or overnight (▴), and in the presence of 1.5μmol/L solubilized HLA-DM (×) using a gel filtration assay. As binding peptide, 1.5 μmol/L AMCA-HA 306-18 was used.
Defensins are able to decrease the T-cell response of an HLA-DR-restricted T-cell clone
To investigate the functional effects, if any, of defensin binding to class II molecules at the cell surface, HLA-DR-restricted T cells were used. These T cells were generated against CHO cells transfected with HLA-DRA, HLA-DRB1*0401, and CD80. After having shown that defensins alone do not markedly influence T-cell proliferation (data not shown), we used CHO cells pulsed with different defensin concentrations (none, 10, 20, or 30 μmol/L) to stimulate the T cells. After 12 and 24 hours of coculture, 3H-TdR was added and incorporated radioactivity was measured 16 hours later.
As can be seen in Figure 5, the T cells were highly reactive against the CHO-DR/CD80 transfectants, especially after 24 hours of coculture, resulting in about 32,000 cpm. This response could be blocked almost completely (78%) using the HLA-DR-specific antibody L243, showing that the T-cell response was indeed DR restricted. CHO cells pulsed with 10, 20, and 30 μmol/L defensin stimulated T-cell responses that were reduced by 57%, 63%, and 66%, respectively, compared to controls without defensin. Defensin (10, 20, and 30 μmol/L) pulsing of CHO cells combined with the presence of the L243 mAb to the culture medium had an additive effect at the 2 higher defensin concentrations, resulting in 77%, 92%, and 96% blocking of T-cell response. In fact, there was a clearly dose-dependent reduction in T-cell response after pulsing CHO cells with defensin. This finding suggests a blocking of the MHC class II binding groove by defensin molecules to suppress T-cell recognition.
T-cell response of HLA-DR-restricted T-cell clone.
T-cell proliferation assay of an HLA-DR-restricted T-cell clone in the presence of CHO cells as APCs, pulsed with different amounts of defensin isolate (10, 20, 30 μg). White bars show 3H-TdR incorporation at day 1/2, black bars at day 2/3.
T-cell response of HLA-DR-restricted T-cell clone.
T-cell proliferation assay of an HLA-DR-restricted T-cell clone in the presence of CHO cells as APCs, pulsed with different amounts of defensin isolate (10, 20, 30 μg). White bars show 3H-TdR incorporation at day 1/2, black bars at day 2/3.
Discussion
Here we report for the first time that endogenously bound natural peptide antibiotics from the defensin family can be eluted from MHC class II molecules of PBMC of 2 patients with plasmacytoma and 1 with CML. In the 2 plasmacytoma patients defensins were the dominant endogenous HLA-DR-associated peptides and corresponded to about 25% of the high-frequency peptides. Furthermore, in plasmacytoma patient 1, 4 different CLIP peptides with lengths of up to 43 amino acids were found. Interestingly, Harris et al,3 who examined HLA-DR-associated endogenous peptides from the hematopoietic precursor cell line KG-1, found a decreased frequency of CLIP. The finding of the N-terminus of the hemoglobin α-chain from plasmacytoma patient 2 was probably due to contamination of the MHC isolate rather than a specifically binding peptide.
Unlike professional APCs expressing MHC class II molecules constitutively, cells from the sources used in this study need stimulatory factors such as interferon-γ (IFN-γ) or granulocyte macrophage colony-stimulating factor (GM-CSF) for the expression of MHC class II molecules.25-28 The 2 patients with plasmacytoma had been treated with GM-CSF (10 μg/kg body weight) 3 days before harvesting the PBMC. Recent studies have suggested that human PMN express the highest MHC class II levels in the presence of both IFN-γ and GM-CSF.32 However, these cells have also been shown to be unable to present a tetanus toxoid (TT) epitope to TT-specific CD4+ T cells after pulsing with TT protein or a TT peptide. In contrast, HLA-DR-specific T-cell stimulation was supported by PMN incubated with small amounts of bacterial superantigens such as SEA or SEB,29 showing that they can function as accessory cells. We hypothesize that one reason why human PMN are unable to present peptide antigens in the same way as professional APCs may be related to blockade of the peptide-binding groove by very voluminous peptides like the defensins. To this end, using an in vitro binding/competition assay, it is shown here that defensins compete with other peptides for the binding of HLA-DR1 molecules, suggesting that they also associate with the peptide-binding groove defined by x-ray structural analysis.30 The competition kinetics showed even slower defensin binding than measured for other endogenous peptides, suggesting a complicated binding mechanism of these unusual long peptides. This may be related to their intact, 3-dimensional structure having 3 disulfide bonds. Reciprocally, we could show that the in vitro peptide-loading capacity onto preformed HLA-DR1/defensin complexes was markedly reduced compared to binding to HLA-DR1 molecules alone. This also suggests that defensins are able to block the peptide binding groove. However, after preincubation of HLA-DR1 molecules with defensins and HLA-DM, the binding capacity was increased compared to HLA-DR1 molecules alone, suggesting that HLA-DM is able to prevent defensins from binding to the class II molecules. Alternatively, HLA-DM may exchange defensins for other peptide antigens as has been shown for CLIP peptides.31 This is another indication that defensin binding to class II follows essentially the same rules as binding of normal self-peptides.
Regarding the functional consequences of the binding of defensin to class II molecules, the first cellular assays suggest that MHC blockade may be effected by defensins. These experiments used defensin-pulsed CHO cells, transfected with HLA-DRB1*0401 and CD80 as APCs and a DRB1*0401-restricted T-cell line. The results suggest a down-regulation of the T-cell response after pulsing the cells with a defensin isolate. This was demonstrated to be peptide specific and dose dependent, consistent with the hypothesis that defensins block the class II peptide-binding groove and therefore block T-cell responses.
There are different reports concerning the antigen-presenting capacity of a number of human epithelial and endothelial cells, mostly expressing MHC class II only after stimulation (eg, by IFN-γ). Recently, Cunningham et al,32 reported that human lung small airway epithelial cells failed to stimulate proliferation of allogeneic CD4+ T lymphocytes, whereas lung microvascular endothelial cells did stimulate proliferation after IFN-γ stimulation. Further studies demonstrated antigen presentation of peptides in intestinal epithelial cells,33,34 even in the polarized state.35 In some systems, epithelial cells have been shown to induce T-cell hyporesponsiveness, for example, tubular epithelial cells36 and biliary epithelium.37 Thus far, no defensin-derived peptides seem to have been isolated from HLA class II molecules of professional APCs or lymphoblastoid cell lines. On the other hand, a very large number of other endogenous peptides have been isolated and identified (for review, see Rammensee et al38). This consideration raises the possibility that the presence of large amounts of defensins is a feature of HLA class II molecules of nonprofessional APCs after induced expression by stimulation with certain factors. Only very little is known about differences in the peptide-loading compartments or different enzymatic predispositions in different subcellular compartments between professional and nonprofessional APCs. Harris et al3 reported a decreased frequency of invariant chain-derived CLIP peptides on the cell surface of the human myelopoietic progenitor cell line KG-1 and suggested therefore that the antigen-processing pathway of KG-1 is different from that characterized in professional APCs. However, from the cell sources we used, we found class II-associated CLIP peptides from PBMC of both plasmacytoma patients and from PBMC of a CML patient. The plasmacytoma-derived material contained almost exclusively peptides from defensin and CLIP. This was unexpected because we isolated the HLA-DR molecules from a heterogeneous cell population that would normally result in a great variety of different self-peptides. The abundance of peptides derived from the defensin family, however, as well as the finding of incompletely processed peptide variants with unusual lengths of 29 to 49 amino acids, clearly point to a peptide-loading pathway different from professional APCs. Especially the finding of a very long, C-terminally extended CLIP variant 80 to 122 in one of the plasmacytoma patients supports this theory. To determine whether this phenomenon was due to the pathophysiologic characteristics of the patients' diseases, we additionally examined HLA-DR-associated endogenous peptides from PBMC obtained by leukapheresis from a healthy donor who had also been treated with GM-CSF for 3 days. Again, we were able to detect HNP-1 and -2 (data not shown). Therefore, we suggest that the occurrence of defensin peptides in association with MHC class II molecules is a phenomenon found in hematopoietic progenitor cells and probably in other nonprofessional APCs. Because we have no information about the intracellular loading of defensins, we cannot exclude that the MHC-bound defensins bound extracellularly after defensin secretion from the storage vesicles. This extracellular MHC class II binding to defensins could also be a possible protective function of defensin-secreting cells against self-destruction. However, due to the fact that we were able to detect a great deal of preprocessed defensin variants of up to 49 amino acids in length, that are unlikely to be secreted, we believe that this extracellular binding of defensins is unlikely.
Acknowledgments
We thank Drs Robert Busch and Elizabeth Mellins, Stanford University, Palo Alto, CA, for providing us with a HLA-DM isolate. We also thank Dr Martin Deeg for mass spectrometric analyzes, Manuela Braun for defensin isolation, and Tanja Bauer for expert technical assistance. Thanks also to Dr Rupert Handgretinger, University Childrens Hospital, Tübingen for providing us with plasmacytoma patient material and Prof C. A. Müller, Dr Thomas Flad, and Jutta Gamper, Medical Hospital, Tübingen, for providing us with CML cells and helpful discussions.
Supported by grants from the Interdisciplinary Clinical Research Center (IKFZ), University of Tübingen (T.M.H.), DFG grant Pa 361/5-1, E.U. grant BMH4-CT98-3058 (G.P.), and the University of Tübingen Medical Faculty Fortüne grant no. 399 (S.H.).
Reprints:Hubert Kalbacher, Medical and Natural Sciences Research Center (MNF), University of Tübingen, Ob dem Himmelreich 7, D-72074 Tübingen, Germany; e-mail:hubert.kalbacher@uni-tuebingen.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 1. ESI-MS spectrum of a representative HPLC fraction. / The spectrum shows 3 different charge states [M+2H]2+, [M+3H]3+, and [M+4H]4+ of HNP-1 (1721.3, 1148.1, 861.3) and HNP-2 (1685.8, 1124.6, 843.5), respectively. This indicates that these peptides have full lengths of 30 and 29 amino acids, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/9/10.1182_blood.v95.9.2890.009k09_2890_2896/5/m_bloo00909001x.jpeg?Expires=1767715264&Signature=wDC2x3LFFUjjvM6T88lS4tasapQhAulIaQK8dplWU7o~wPRDt8hb2W-VVeBQdg~crrlI6w998sICNyhX143ZCWAWmw5Lgz076IJeq80aGNGdSytlENMSu1DCxlFeOHSW9ismHInUjd-Cpv5wXBR~Mui0PHiy9~9UFfNZqpT~8~ntpMO7xksvLKhWfkm45qiPHj1NHCMKMdwd8LkwyhKW4ETw6aPHh2J7Q-EAUtlUImwLBLFM1ypANwKQwvGim-P6aVYmwvaWiCjHMWObScfTAYDfzdE9-KjoaDAVotU4JJ92VR42nHMXHpr6rydevFeEFqzieS2Q22BQYUjFeVNllw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Defensin isolate as competitor for the HLA-DR1 binder AMCA-HA 306-18. / Competition kinetics of defensins (10 μg [○]; 20 μg [×]) and different HLA-DR1-binding peptides (CLIP 80-104: LPKPPKPVSKMRMATPLLMQALPMG (□); IM 19-31: PLKAEIAQRLEDV (▪), and HA 306-18 (PKYVKQNTLKLAT ([▴]), each of them at a concentration of 30 μmol/L) to solubilized HLA-DR using a gel filtration assay; 1.5 μmol/L of AMCA-labeled HA 306-18 was used as a binding peptide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/9/10.1182_blood.v95.9.2890.009k09_2890_2896/5/m_bloo00909003x.jpeg?Expires=1767715264&Signature=sPa~ApcicsI179E8Or1l9gn8cWgwFkVfaHL-SSQgJGUMpYxuHOf4byqKO-I9WC~GmGzD~ZmqTygEKUTqCfB-RJu21BUIwA046qfE2jgHdoeVT~TKFjn8o9ZRRB3dJy9cFhDe-NA9cS08LkZVcw4BtjRz6jMm78i99dARMDSOLJgiiQt-IitJDg2XqJbKN9powDiWQ2pI5nIRp2hQT~zWnRuie9doSeUTcwnZNQ48MYqMVaRQgaeaubyxT5Tu26GtZVC8ZTUO7p-bWvnu9h7APRyT7Qr02LrJnOmSgsfUqbDlUa9RIRmvG1JGyfGCWO01fvpTa6~n8g2la5YS3mGW3Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal