Abstract
The coagulation protease factor Xa induces cellular responses implicated in cardiovascular and inflammatory disease. Effector-cell protease receptor 1 (EPR-1) is a functionally characterized receptor of factor Xa, and the EPR-1complementary DNA (cDNA) was published. Remarkably, the cDNA encoding an inhibitor of apoptosis, survivin, is reportedly identical to that ofEPR-1 except for a few nucleotide differences and its orientation opposite to EPR-1. To isolate the EPR-1cDNA and gene, we surveyed gene databases for expressed sequence tags (ESTs) that could be derived from EPR-1. All ESTs with strong homology to EPR-1/survivin were derived from survivinand could not encode EPR-1. By polymerase chain reaction and Southern blot hybridization, EPR-1 was not detectable in the human or murine genome, but survivin was. Our data suggest that EPR-1 is either highly cell-specific or the published EPR-1 cDNA includes sequences from clones derived from survivin messenger RNA. The means by which factor Xa mediates its cellular effects requires further evaluation.
In addition to their role in maintaining hemostasis, coagulation proteases elicit various cellular responses in vascular, mesenchymal, and inflammatory cell types that have been implicated in cardiovascular disease,1 inflammation,2,3 and tumor metastasis.4 The serine protease factor Xa is one of the major late-stage enzymes in the blood coagulation cascade. Factor Xa acts in concert with the nonenzymatic cofactor, factor Va, to convert prothrombin into thrombin, which induces the formation of the fibrin clot by proteolysis of fibrinogen. In addition, factor Xa has been shown to induce mitogenesis,5vasorelaxation,6 and proinflammatory responses.7 8
A cellular receptor for factor Xa, designated effector-cell protease receptor 1 (EPR-1), was described by D. C. Altieri and colleagues. The human receptor was originally identified and characterized with a monoclonal antibody raised against factor V/Va,9 which also reacted with a second generation of antibodies raised against leukemic T cells.10 The human EPR-1 complementary DNA (cDNA) sequence, published in 1994,10 was obtained by immunoscreening an expression library of a T-cell line (MLT) with one of the second-generation antibodies (2E1). This yielded a partial cDNA clone that was used to screen additional cDNA libraries derived from T-cell lines and human umbilical vein endothelial cell lines. From 28 independent clones, a consensus EPR-1 cDNA sequence of 1165 base pairs (bp) was derived, which was predicted to encode a protein of 337 amino acids with no structural homology to any other known protein class.10 In a subsequent paper, it was reported that there actually exist 2 splice variants of the EPR-1 messenger RNA (mRNA).11 In addition to the cDNA sequence described in the first paper,10 a second variant was described that had retained a 451-bp intervening sequence. The presence of the intervening sequence resulted in preliminary termination of translation of EPR-1, resulting in a truncated protein, designated EPR-1b, that localized in the nucleus.11 More recently, Altieri and coworkers reported the cloning and characterization of the human survivingene, which encodes an inhibitor of apoptosis.12 Most strikingly, the coding strand for survivin was almost identical to the long variant of the EPR-1 mRNA read in the opposite orientation.12
In this report we describe our attempts to isolate the human and murineEPR-1 cDNAs and genes. Our data suggest that EPR-1 is either a unique property of specific cell lines or that the publishedEPR-1 cDNA contains sequence errors or includes sequences from clones that were actually derived from survivin mRNA.
Materials and methods
Northern hybridization
The RNA probes A and B were radiolabeled with α-[32P]UTP using buffers from a RiboQuant Multi-Probe Ribonuclease Protection Assay System (Pharmingen, San Diego, CA). Multiple tissue Northern blots (Clontech, Palo Alto, CA) were prehybridized with 100 μg/mL of denatured salmon sperm DNA (Promega, Madison, WI) and hybridized with riboprobes for 16 hours at 42°C in 50% formamide; 10 × Denhardt's solution; 10% dextran sulfate; 1-mol/L NaCl; 50-mmol/L Tris-HCl, pH 7.5; and 0.1% sodium dodecyl sulfate (SDS). The membranes were washed successively for 5 minutes at 42°C in 2 × SSC, 0.5% SDS; for 15 minutes at 42°C in 2 × sodium chloride sodium citrate (SSC), 0.1% SDS; for 60 minutes at 37°C in 0.1 × SSC, 0.5% SDS; and for 45 minutes at 65°C in 0.1 × SSC, 0.5% SDS; and exposed for autoradiography.
Polymerase chain reaction analysis
Human genomic DNA (Sigma, St Louis, MO) orsurvivin cDNA sequences (from I.M.A.G.E. Clone ID 590560, American Type Culture Collection, Manassas, VA) were amplified with 4 different primer sets. The sequences of the oligonucleotides and their position in the EPR-1cDNA10 and survivin gene sequences12were as follows: primer a: 5′-GTCGTCATCTGGCTCCCAGCC-3′, corresponding to nucleotides 864 to 884 of EPR-1 cDNA and to nucleotides 3258 to 3278 in the survivin gene; primer b: 5′-CACTGGGCCAAGTCTGGCTCGTTCTC-3′, nucleotides 910 to 935 inEPR-1 and 3207 to 3232 in survivin; primer c: 5′-CAAGGACCACCGCATCTCTAC-3′, nucleotides 1017 to 1037 inEPR-1 and 2852 to 2872 in survivin; primer d: 5′-CTGGCCCTTCTTGGAGGGCTGCG-3′, nucleotides 985 to 1007 inEPR-1 and 2882 to 2904 in survivin. Reactions contained 200 ng of each primer and 300 ng of genomic DNA in a total volume of 50 μL. Thirty-five cycles of amplification were carried out with denaturation at 94°C, annealing at 55°C, and extension at 72°C for 30 seconds each step.
Southern hybridization
Probes C and D were separated electrophoretically on 2% agarose gels. Oligonucleotide probe E was end-labeled with γ[32P]ATP. Genomic DNA from mouse embryonic stem cells (15 μg per lane) or P1 artificial chromosome (PAC) genomic DNA (4 μg per lane) was separated on a 0.7% agarose gel following restriction enzyme digestion. After transfer to a nylon filter, the membranes were hybridized in Quik-Hyb (Stratagene, La Jolla, CA) for 1 hour and finally washed with 0.1% SDS, 0.5 × SSC at 55°C. The filters were then exposed for autoradiography.
Results
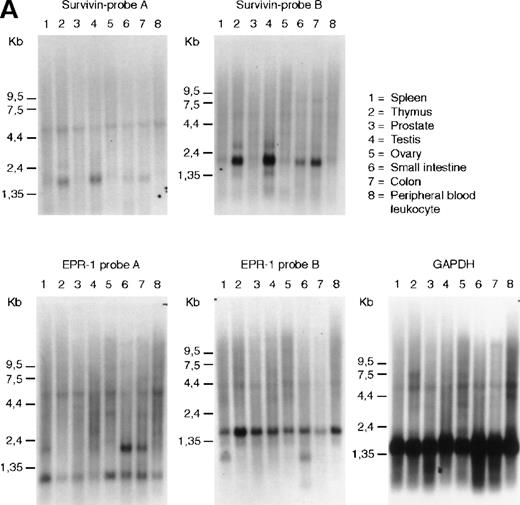
The mRNAs for EPR-1 and survivin can be distinguished by hybidization with single-stranded probes. We have used probes complementary to sequences at the 5′ or 3′ ends of either survivin or EPR-1 to evaluate transcription of the 2 genes from a variety of human tissues by Northern blot analysis. As seen in Figure1A, survivin was detected as a 1.9-kilobase (kb) band in RNA from thymus, in agreement with data from Altieri and coworkers.12 Additionally, the 1.9-kb band was found in RNA from human testis, small intestine, and colon (Figure1A). However, hybridization of the Northern blots with 2EPR-1–specific probes yielded 2 different patterns, indicating that the 2 probes recognized different RNA species. With probe A, directed to the first 226 nucleotides of EPR-1 plus the first 94 nucleotides of the “intervening sequence,” a hybridization pattern similar to that shown by Altieri and coworkers13was obtained. This probe hybridized with a band of 2.0 kb in small intestine and colon and to fainter bands at 0.9 and 6.0 kb in all RNAs on the blot. Probe B, directed to nucleotides 664 to 972 of EPR-1(numbering corresponds to GenBank L26245), hybridized to a band of 1.6 kb in all tissues and 1.0 kb in spleen and small intestine (Figure1A). The identity of the transcripts detected with theEPR-1 probes is unclear; however, if they are derived from a single transcript, they reflect a complex pattern of tissue-specific alternative splicing events.
RNA and genomic analysis of human EPR-1/survivin.
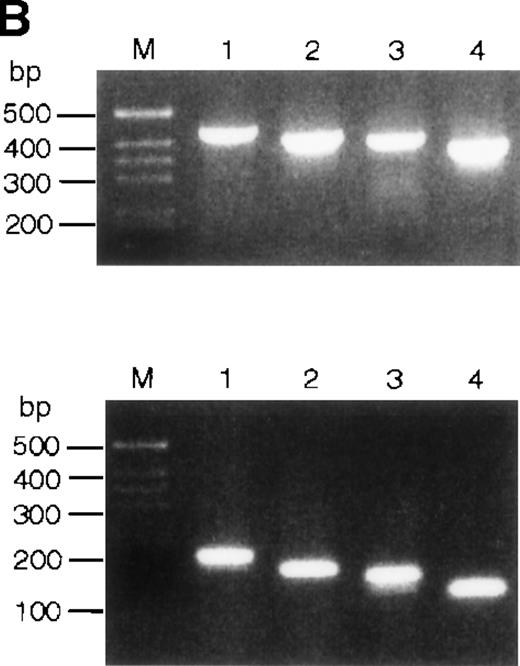
(A) Hybridization of multiple tissue Northern blots (Clontech) withEPR-1 or survivin RNA-specific riboprobes. Probe A is complementary to the first 226 nucleotides of EPR-1 plus the first 94 nucleotides of the intervening sequence. Probe B is complementary to nucleotides 664 to 972 of EPR-1 (GenBank accession number L26245). (B) PCR amplification of survivingenomic DNA sequences from human genomic DNA (upper panel) or of survivin cDNA sequences (lower panel) with 4 different primer sets (see “Materials and methods”). Primer combinations used for the reactions are as follows: lane 1: a-c; lane 2: a-d; lane 3: b-c; lane 4: b-d.
RNA and genomic analysis of human EPR-1/survivin.
(A) Hybridization of multiple tissue Northern blots (Clontech) withEPR-1 or survivin RNA-specific riboprobes. Probe A is complementary to the first 226 nucleotides of EPR-1 plus the first 94 nucleotides of the intervening sequence. Probe B is complementary to nucleotides 664 to 972 of EPR-1 (GenBank accession number L26245). (B) PCR amplification of survivingenomic DNA sequences from human genomic DNA (upper panel) or of survivin cDNA sequences (lower panel) with 4 different primer sets (see “Materials and methods”). Primer combinations used for the reactions are as follows: lane 1: a-c; lane 2: a-d; lane 3: b-c; lane 4: b-d.
Probes A and B were also used in ribonuclease protection experiments to analyze the expression of EPR-1 and survivin in various cell lines, including primary human umbilical vein endothelial cells, human aortic smooth muscle cells (Clonetics), and Jurkat T cells (American Type Culture Collection), which, based on studies with antibodies or functional studies, have previously been reported to express EPR-1 or factor Xa receptors.14-16 Whilesurvivin mRNA could be detected in all cell lines analyzed,EPR-1 mRNA could not be detected (data not shown).
We also surveyed the public and Incyte EST (expressed sequence tag) databases for sequences with strong homology to EPR-1/survivin. Survivin differs from EPR-1 for 5 nucleotide changes and 6 nucleotide insertions and for its directionality of the mRNA.12 In the Incyte database, 210 sequences with strong homology to EPR-1/survivin were identified (LifeSeqGold database, release of October 1999), 149 of which were derived from Incyte clones. Most ESTs in the Incyte database are derived from oligo-dT–primed cDNA libraries that are sequenced in the 5′ to 3′ direction. “Verified reagents” are sequenced in both directions. Based on the position of the polyA tail, most Incyte clones seemed to be derived from survivin coding mRNA. Five verified clones were identified with sequences corresponding to the region encoding the amino terminus of EPR-1. The origin of these cDNAs was checked. All 5 contained an insertion of the same C at position 102 of the published EPR-1 cDNA sequence.10 A sequencing artifact is therefore unlikely. Because of a frame-shift mutation, these clones cannot encode EPR-1. In the public EST database (European Molecular Biology Laboratory, release of September 1999), 30 sequences with strong homology to the amino terminus of EPR-1 were identified. Based on the position of the polyA tail and other sequence features, these clones could not be derived from EPR-1. In the 2 databases, several tens of clones were identified that encompassed the region containing the putative intervening sequence characteristic for EPR-1bmRNA,11 which, in the reverse complementary orientation, is also present in the survivin mRNA.12 All of these clones contained the intervening sequence. Thus, EPR-1 cDNA sequences are absent in both the public and Incyte human gene databases.
It was proposed that EPR-1 and survivin transcripts arise from duplicated genes clustered head to head in opposite orientations.12 However, it was not explained how the positions of intron-exon boundaries could be maintained in the reverse-oriented duplicated genes. Notably and unfortunately, theEPR-1 gene has not been cloned. The simplest explanation for the almost perfect complementarity of the EPR-1 andsurvivin transcripts is that the EPR-1 gene has originated from a chromosomal insertion of a cDNA copy ofsurvivin. If this were true, one would expect that the introns in the survivin gene would be absent in the EPR-1 gene. This is not the case: the first and second exons of thesurvivin gene are separated by a 251-nucleotide intron. The exon-intron boundary lies between the nucleotides corresponding to positions 968 and 969 of the EPR-1 cDNA sequence. This is the part predicted to encode the carboxy terminal end of EPR-1. Using several different primer pairs consisting of a forward primer in exon 1 and a backward primer in exon 2 in polymerase chain reactions (PCRs) on genomic DNA, we tried to identify an intronless survivin gene. As a control, the primer pairs were also used in PCRs with asurvivin cDNA clone. Each primer set yielded a single PCR amplicon on genomic DNA, as determined by agarose gel electrophoresis (Figure 1B). The length and sequences of the fragments corresponded with the products expected from amplification of survivingenomic DNA sequences. In conclusion, we found no evidence for an intronless cDNA copy of the survivin gene in human.
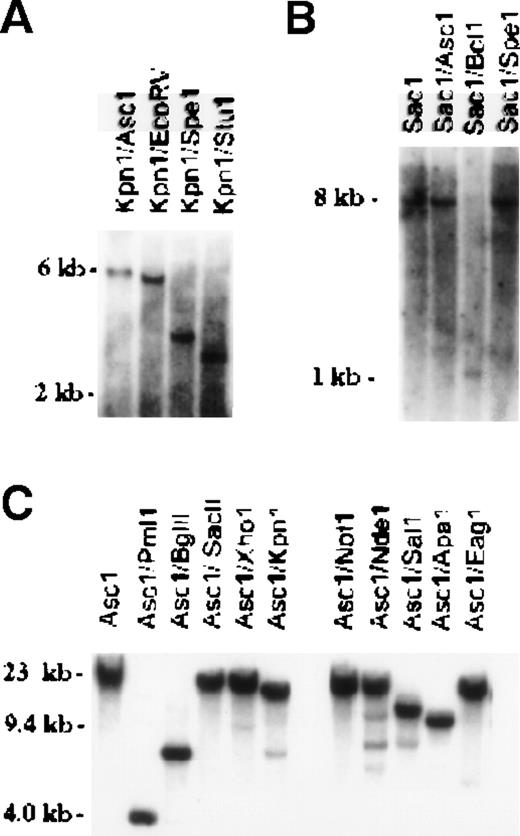
In attempts to identify EPR-1 in the murine genome, we used as a probe a published murine EST (mc61a11.r1) that was originally believed to represent the murine homologue of EPR-1. From murine genomic DNA and several murine cDNA libraries (14.5 dpc embryo, activated T-lymphocyte, adult brain, and adult liver), we could find no evidence of the EPR-1 gene or cDNA. Overlapping genomic clones were obtained from murine λ-phage and PAC genomic DNA libraries and extensively sequenced, and none had sequence consistent with that of EPR-1, in that we were unable to detect the most 5′ coding region sequence (orientation corresponding to that of EPR-1) that is reportedly unique to human EPR-1 and absent in survivin.12 The genomic and cDNA clones, however, were consistent with murine survivin, the genomic structural organization of which is very similar to that of the human gene.12,17 To determine whether murinesurvivin-related genes are present within the mouse genome, embryonic stem cell genomic DNA was analyzed by Southern blot hybridization using different cDNA and genomic DNA probes (Figures2). Probe C, a KpnI-BglII approximately 2.3-kb fragment obtained from the murinesurvivin PAC genomic clone, lies 3.6 kb 5′ of exon 1, and probe D, a SacI-ApaI approximately 1.4-kb fragment, lies 4.5 kb 3′ of exon 4.17 Hybridization of the radiolabeled probes with embryonic stem cell genomic DNA digested with a variety of restriction enzymes identified single, prominent bands, suggestive of the presence of a single gene (Figures2A and 2B). Murine survivin PAC genomic DNA was similarly digested with multiple restriction enzymes and probed with the cDNA oligonucleotide probe E. The sequence of the oligonucleotide (5′-TGGCTGCGCCTTCCTCACTGT), derived from exon 3 of murinesurvivin, is 80% identical to the reverse complement of a portion of the cDNA reportedly encoding human EPR-1. Hybridization signals observed were consistent with previous restriction mapping data and with the known DNA sequence of the murine survivin gene (Figure 2C).17 Overall, we were unable to detect a family of genes related to survivin, nor could an EPR-1 gene be identified in the murine genome.
Genomic analysis of murine EPR-1/survivin.
Southern blotting of murine embryonic stem (ES) cell genomic DNA (A, B) or PAC genomic DNA (C). The membranes were hybridized with probe C in panel A, probe D in panel B, and probe E in panel C, as described in the text. Single hybridization bands, consistent with restriction mapping of the murine survivin gene,17 were detected.
Genomic analysis of murine EPR-1/survivin.
Southern blotting of murine embryonic stem (ES) cell genomic DNA (A, B) or PAC genomic DNA (C). The membranes were hybridized with probe C in panel A, probe D in panel B, and probe E in panel C, as described in the text. Single hybridization bands, consistent with restriction mapping of the murine survivin gene,17 were detected.
Discussion
Taken together, our data suggest that EPR-1 is either a unique property of specific cell lines or that the EPR-1 cDNA contains sequence errors or includes sequences from clones that were actually derived from survivin mRNA. It follows, therefore, that the means by which factor Xa mediates its cellular effects should be further evaluated. There is currently solid evidence supporting the concept that one or more alternative cellular receptors for factor Xa exist and, indeed, cell biological studies show that EPR-1 does not mediate the cellular effects of factor Xa in various cell types.8,18,19 For example, factor Xa has been shown to increase intracellular free calcium levels and phosphoinositide turnover.16,18 The native (zymogen) form of factor X was inactive,18 and a role for PAR2, a member of the protease-activator receptor (PAR) class of G protein–coupled receptors has been suggested.19,20 Another candidate receptor for factor Xa is the recently discovered PAR4,21,22 which was reported to be activated at high (nonphysiologic) concentrations of factor Xa.22 Finally, several cDNAs encoding receptors homologous to the PARs have been identified in EST sequencing projects.23 24 It is anticipated that one or more of these may prove to be a functional receptor for factor Xa, findings that will provide further insights into the biologically wide spectrum of activities of factor Xa and other similar serine proteases.
Acknowledgments
We thank Mr P. Vink, Ms S. Pollefeyt, and Ms M. Steiner-Mosonyi for technical assistance, Dr P. v. d. Spek for bioinformatics, and Incyte Pharmaceuticals for use of the LifeSeqGold database.
Reprints:Guido J. R. Zaman, N.V. Organon, PO Box 20, 5340 BH Oss, The Netherlands; e-mail: g.zaman@organon.oss.akzonobel.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal