Abstract
Folate receptor (FR) type β is expressed in the myelomonocytic lineage, predominantly during neutrophil maturation and in myeloid leukemias. FR-β expression was elevated up to 20-fold by all-trans retinoic acid (ATRA) in KG-1 myeloid leukemia cells in a dose-dependent and reversible manner in the absence of terminal differentiation or cell growth inhibition. ATRA also increased FR-β expression in vitro in myeloid leukemia cells from patient marrow. FR-β was not up-regulated in KG-1 cells treated with phorbol ester, dexamethasone, 1,25-dihydroxy vitamin D3, or transforming growth factor β. ATRA did not induce FR-β expression in receptor negative cells of diverse origin. The ATRA-induced increase in FR-β expression in KG-1 cells occurred at the level of messenger RNA synthesis, and in 293 cells containing a stably integrated FR-β promoter–luciferase reporter construct, ATRA induced expression of the reporter. From experiments using retinoid agonists and antagonists and from cotransfection studies using the FR-β promoter and expression plasmids for the nuclear receptors retinoic acid receptor (RAR)α, RARβ, or RARγ, it appears that the retinoid effect on FR-β expression could be mediated by ligand binding to RARs α, β, or γ, but not to retinoid X receptors. Furthermore, there was apparent cross-talk between RARα and RARγ selective agonists or antagonists, suggesting a common downstream target for RAR isoforms in inducing FR-β expression. Thus, blocks in the RARα-specific pathway of retinoid-induced differentiation may be bypassed during retinoid induction of FR-β expression. The results suggest that to facilitate FR-targeted therapies, retinoids may be used to modulate FR-β expression in myeloid leukemia cells refractory to retinoid differentiation therapy.
Introduction
The human folate receptor (FR) is a single polypeptide glycoprotein encoded by a family of 3 genes.1-4 FR-α and FR-β are attached to the cell surface by a glycosyl-phosphatidylinositol (GPI) membrane anchor,5 whereas FR-γ is constitutively secreted.3 The membrane-anchored proteins mediate internalization of receptor-bound folate compounds and folate conjugates.6,7 The FR isoforms exhibit narrow tissue and tumor specificities. FR-α is expressed in certain normal epithelial cells and is vastly elevated in various carcinomas, particularly those of gynecological tissues.8-10 In recent preclinical and clinical studies, primarily of ovarian cancer, FR-α has shown considerable promise as a potential means of delivering a wide variety of novel folate-mediated and non–folate-mediated therapeutic agents to tumor cells and as a tumor and serum marker.11 FR-β is highly expressed in placentas,2 mature neutrophils,12 and activated monocytes and macrophages,13 and in more than half of all myeloid leukemias12,14,15 (also unpublished data, 2000) and is virtually absent in other tissues. In normal hematopoiesis, FR-β is expressed in the myelomonocytic lineage in a subpopulation of CD34+ cells, and its expression is further increased about 5-fold during neutrophil maturation.11,16 In normal hematopoietic cells, FR-β does not bind folic acid with a high affinity even though its messenger RNA (mRNA) sequence is unaltered16 (also unpublished data, 2000). The function of FR-β in normal hematopoietic cells is unclear since deletion of a gene presumably corresponding to this receptor isoform did not produce an altered phenotype in mice.17Nevertheless, the restricted expression of FR-β in later stages of normal myelopoiesis renders this FR isoform a potential target for novel FR-mediated therapies in myeloid leukemia.
At present, the most effective treatment for myeloid leukemia is retinoid differentiation therapy, for which acute promyelocytic leukemia (APL) is the paradigm, since in various studies 72% to 95% of APL patients have shown complete remission owing to treatment with all-trans retinoic acid (ATRA).18 Other types of acute myeloid leukemia (AML), which constitute about 90% of AML cases, do not respond well to ATRA therapy. Furthermore, in APL, the initial favorable response to ATRA treatment is followed by a significant incidence of relapse, at which time the patients are refractory to ATRA differentiation therapy.18 In those cases, ATRA resistance has frequently been related to mutations that cause blocks in the pathway of retinoid-induced differentiation.19
We report here that in KG-1 myeloid leukemia cells and also in leukemic blasts from patients with non-APL AMLs, ATRA induces FR-β expression and that this effect occurs in the absence of terminal differentiation or inhibition of cell proliferation. The cell type, inducing reagent, and nuclear receptor specificities of this effect support the feasibility of developing more effective therapies for myeloid leukemia by combining FR-β targeting and retinoid treatment.
Materials and methods
Cells and reagents
NB4 and KCL22 cells were kindly provided by Dr H. Philip Koeffler (University of California, Los Angeles). All of the other cell lines were purchased from American Type Culture Collection (Rockville, MD). Cells were cultured at 37°C in 5% CO2. The cell-culture media were supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 292 μg/mL L-glutamine (Gibco-BRL, Grand Island, NY). Caki-1, CCRF-CEM, Hela, KCL22, and NB4 cells were grown in folate-free RPMI-1640 medium (FFRPMI-1640) (JRH Bioscience, Lenexa, KS) with 10% fetal bovine serum (FBS) (Gibco-BRL). K562, L1210, JAR, SKOV3, and 293 cells were grown in RPMI-1640 medium with 10% FBS. For transient transfection experiments, 293 cells were grown in Eagle's Minimum Essential Medium (MEM) (Gibco-BRL). HL60, KG-1, and KG-1a cells were grown in FFRPMI-1640 medium with 20% FBS. MG-63 cells were grown in Dulbecco's Modified Eagle's Medium (D-MEM) (Gibco-BRL) with 10% FBS. ATRA, 9-cis retinoic acid (9-cis RA), dexamethasone, phorbol 12-myristate 13-acetate (PMA), and 1,25(OH)2vitamin D3 (vit D3) were purchased from Sigma-Aldrich (St. Louis, MO). Transforming growth factor β (TGF-β) was purchased from R&D Systems, Inc (Minneapolis, MN). We dissolved 9-cis RA, dexamethasone, and vit D3 in ethanol to achieve stock solutions of 5 mmol/L. Compounds LG101093, TTNPB, LG100364, and LG100629 were a kind gift from Dr Elizabeth Allegreto (Ligand Pharmaceuticals Inc, San Diego, CA). ATRA and all of the compounds in the LG series were dissolved in a mixture of 50% ethanol and 50% dimethyl sulfoxide (DMSO). Compounds CD2314, CD2503, CD2665, CD2781, CD336, and CD417 were kindly provided by Dr Uwe Reichert (Galderma Research and Development, Valbonne, France). These compounds were dissolved in DMSO to achieve stock solutions of 5 × 10−2 mol/L. All the stock solutions were stored in aliquots at −80°C.
Treatment of cells with retinoid compounds or other differentiation reagents
For suspension cultures, cells growing in log phase were seeded into 60-mm or 100-mm tissue-culture plates at a density of 2.5 × 105 cells per milliliter in 4 mL or 8 mL of media, respectively. For monolayer cultures, the cells were seeded at 10% to 20% confluence into 6-well tissue-culture plates. After a 6-hour preculture, various retinoids or other differentiation reagents were added to the medium, either directly or after appropriate dilution, to obtain the final concentration of the reagent. After the desired period of incubation, KG-1 cells and other suspension cells were harvested by sedimenting at 400g for 10 minutes and suspending in phosphate-buffered saline (PBS) (10 mmol/L sodium phosphate, pH 7.5, per 150 mmol/L NaCl) at 4°C. This washing procedure was repeated once before the cells were used for immunostaining, cell lysate preparation, or other procedures described below. The monolayer cells were washed in the culture plates twice with PBS and subjected to either [3H] folic acid binding assay or cell lysate preparation.
Treatment of leukemic blasts with ATRA
Leukemic blasts from patient bone marrow aspirates, separated by Ficoll density gradient centrifugation, were received frozen from the Pediatric Oncology Group tissue bank (St. Jude's Children's Hospital, Memphis, TN). The cells were rapidly thawed under hot running water and immediately resuspended at 37°C in RPMI-1640 containing 30% FBS. The cells were sedimented at 400g for 10 minutes and seeded at a density of 2.5 × 105 cell/mL in RPMI-1640 medium containing 20% FBS with the inclusion of interleukin 3 (IL-3) (20 ng/mL), human stem cell factor (cKit ligand) (20 ng/mL), and granulocyte-macrophage colony–stimulating factor (GM-CSF) (10 ng/mL) (R&D Systems, Inc). The cells were treated with ATRA as described above.
Folate binding assay
Cells were washed with ice-cold acid buffer (10 mmol/L sodium acetate, pH 3.5, containing 150 mmol/L NaCl) and cold PBS sequentially. The cells were then incubated with 1 mL PBS containing a mixture of 5 pmol of [3H] folic acid (20.2 Ci/mmol) (Moravek Biochemicals, Brea, CA) and 25 pmol of unlabeled folic acid at 4°C for 30 minutes. After the incubation, the cells were washed twice with cold PBS. The acid buffer (1 mL) was used to extract bound [3H] folic acid, and the radioactivity was measured by liquid scintillation counting.
Preparation of cell lysate and Western blot analysis
Cells (1 × 106) were lysed at 37°C for 45 minutes in 30 μL of PBS containing 1% Triton X-100 and 1 mmol/L phenylmethanesulfonyl fluoride.20 After sedimenting the insoluble material by centrifugation at 10 000g for 10 minutes, we mixed 25 μL of the supernatant (cell lysate) with an equal volume of 2 × sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (62.5 mmol/L Tris-HCl, pH 6.8, containing 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.00125% bromophenol blue). The samples were electrophoresed on 12% SDS-PAGE gels and transferred to nitrocellulose filters. The blots were probed with affinity-purified rabbit antihuman FR-β antibody followed by alkaline phosphatase–conjugated goat antirabbit immunoglobulin G (IgG) (Promega, Madison, WI) as previously described.2 20
Immunostaining for flow cytometry analysis
Cells were washed and resuspended in PBS at a density of 1 × 107 cells per milliliter. Aliquots (100 μL) of the cell suspension were incubated with either rabbit anti–FR-β antibody or normal rabbit serum (1:20 dilution in PBS) on ice for 60 minutes with intermittent gentle agitation. The cells were washed twice with PBS and incubated with fluorescein isothiocyonate (FITC)–conjugated goat antirabbit IgG (1:500 dilution) in 500 μL PBS on ice for 30 minutes with intermittent gentle agitation (Cappel, Durham, NC). After 2 more washes with PBS, the cells were fixed in 300 μL of 1% paraformaldehyde and stored at 4°C. The samples were examined in a flow cytometer within 24 hours.
MTT assay
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) was purchased from Sigma (St. Louis, MO), and MTT cell proliferation assay was carried out following the supplier's protocol. Briefly, 90 μL aliquots of KG-1 or HL60 cells were added to each well of a 96-well plate followed by the addition of 10 μL of MTT stock solution (5 mg/mL) and incubation at 37°C for 3 to 4 hours. The reaction was stopped by adding 100 μL of 0.1 N HCl in isopropanol. The optical density was measured at 570 nm with background subtraction at 630 to 690 nm.
Nitroblue tetrazolium reduction assay and α-naphthyl acetate esterase staining
The cells were harvested and suspended at a density of 1 × 106/mL. The cell suspensions (100 μL) were treated for the measurement of nitroblue tetrazolium (NBT) reduction activity as described.21 We spread 20 μL of the cell suspensions evenly onto microscope slides (Labcraft, distributed by Curtin Matheson Scientific, Inc, Houston, TX). The slides were stained by means of the α-naphthyl acetate esterase (NAE) kit (Sigma) following the manufacturer's protocol.
Treatment with phosphatidylinositol-specific phospholipase C
Cells (1 × 106) were incubated in 500 μL of phosphatidylinositol-specific phospholipase C (PI-PLC) buffer (25 mmol/L Tris-HCl, pH 7.5, containing 250 mmol/L sucrose, 10 mmol/L glucose, and 1% bovine serum albumin) at 37°C for 1 hour in either the presence or the absence of 0.15 U of PI-PLC (Boehringer Mannheim Corp, Indianapolis, IN) with mild agitation. Cells were then washed twice with PBS, and cell lysates were prepared for Western blot analysis as described above. Alternatively, crude membranes were prepared from the PI-PLC–treated cells as described previously.20
Isolation of total RNA and Northern blot analysis
Total RNA was isolated with the use of Trizol reagent (Gibco-BRL) following the protocol provided by the vendor. Total RNA (30 μg) from each sample was analyzed by Northern blot as described previously.9 The blot was hybridized separately with [α-32P] deoxyadenosine triphosphate (3000Ci/mmol, DuPont–New England Nuclear, Boston, MA) labeled complementary DNA (cDNA) fragments of FR-β (nucleotides 591 to 933), FR-α (nucleotides 558 to 900), or γ-actin.
Nuclear run-on assay
This procedure is a modification of the one described by Garber et al.22 Briefly, cells (5 × 107) were lysed in 5 mL of lysis buffer A (15 mmol/L Hepes, 60 mmol/L KCl, 15 mmol/L NaCl, 0.15 mmol/L spermin, 0.5 mmol/L spermidine, 0.5 mmol/L egtazic acid, 2 mmol/L EDTA, 0.3 mol/L sucrose, and 14 mmol/L β-mercaptoethanol) containing 0.1% nonidet P-40. The nuclei were separated by centrifugation and finally resuspended gently in nuclear storage buffer (50 mmol/L Tris, pH 8.3, containing 40% glycerol, 5 mmol/L MgCl2, and 0.1 mmol/L EDTA), snap-frozen on dry ice and stored at −80°C up to 4 weeks. For each reaction, 107 nuclei were thawed on ice and resuspended with 300 μL reaction buffer (50 mmol/L Tris, pH 8.0, containing 150 mmol/L KCl, 5 mmol/L MgCl2, 0.5 mmol/L MnCl2, 2 mmol/L dithiothreitol, 0.1 mmol/L EDTA, 10% glycerol), and 60 U RNase inhibitor from Roche Diagnostic Corp (Indianapolis, IN). The reaction was initiated by the addition of ribonucleotides to a final concentration of 0.5 mmol/L adenosine triphosphate, 0.5 mmol/L guanosine 5′-triphosphate, 0.5 mmol/L cytidine 5′-triphosphate, and 100 μCi α-32P] uridine triphosphate (800 Ci/mmol, DuPont–New England Nuclear). The reaction was first incubated in a water bath at 30°C followed by digestion with 100 U DNase I (Roche Diagnostics Corp) and 33 μg of proteinase K in 33 μL of 10 × second incubation buffer (100 mmol/L Tris-HCl, pH 7.5, containing 5% SDS, 50 mmol/L EDTA) at 37°C or 42°C, respectively. Each incubation step lasted for 30 minutes. The newly transcribed RNA was extracted with phenol-chloroform, precipitated with sodium acetate and ethanol, and purified on micro bio-spin columns P-30 Tris (Bio-Rad Laboratories, Hercules, CA). The purified RNA probes (1 × 106 to 3 × 106cpm) were hybridized to FR-β or γ-actin cDNA, which was immobilized on a nitrocellulose membrane, in 500 μL hybridization buffer at 45°C for 48 hours. Then the membrane was washed with 2 × SSPE at 45°C and sequentially washed with the same buffer containing either RNase A (Roche Diagnostics Corp) or proteinase K at 37°C. The membrane was then exposed to x-ray film.
DNA constructs
For FR-β promoter–luciferase constructs, the FR-β genomic DNA fragment −478nt to +64nt containing the transcription start-site at nucleotide +1 was inserted into the pGL3 basic vector (Promega) in the polylinker immediately upstream of the luciferase reporter. The retinoid acid receptor (RAR)α, RARβ, and RARγ cDNA constructs were kindly provided by Dr Pierre Chambon (Institute de Genetique et de Biologie Moleculaire et Cellulaire, Cude Strasbourg, France).
Transient transfection and luciferase assay
We transfected 293 cells, at 50% to 70% confluence in 6-well tissue-culture plates, with 1 μg each of the FR-β promoter–luciferase construct, pSV-β-gal (Promega), and RARα, RARβ, or RARγ cDNA constructs using lipofectamine (Life Technologies, Inc, Gaithersburg, MD) according to the vendor's protocol. At 8 hours after transfection was initiated, the medium containing DNA complexes was removed and 2 mL of regular MEM medium containing ATRA (1 μmol/L) or the vehicle alone was added to each well. At 48 hours after transfection, the cells were harvested in the reporter lysis buffer provided in the luciferase assay system (Promega) and centrifuged at 14 000g for 2 minutes at room temperature. The supernatant was assayed for luciferase, β-galactosidase activity, and total protein concentration, as previously described.23
Stable transfection and isolation of transfectant clones
We cotransfected 293 cells, at 60% confluence in a 100-mm plate, with 9 μg of the FR-β promoter–luciferase construct and 3 μg of pcDNA1/Neo plasmid (Invitrogen, Carlsbad, CA) using lipofectamine. At 40 hours after transfection, the cells were transferred into MEM medium containing G418 (0.5 mg/mL) (Gibco-BRL). At 72 hours after transfection, the cells were split at different ratios (1:3 to 1:24) and cultured in 100-mm plates. After about 2 more weeks, individual colonies were picked and cultured.
Results
Specific and reversible up-regulation of FR-β by ATRA in KG-1 cells
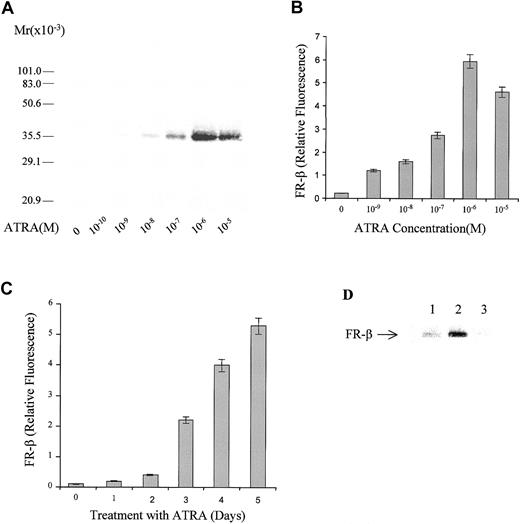
KG-1 leukemia cells express FR-β, which may be detected on the cell membrane by FR-β–specific antibodies (Figure1). When the cells were grown in the presence of ATRA, there was a dramatic dose-dependent increase in FR-β expression up to a concentration of 10−6 mol/L ATRA. The elevation in FR-β could be demonstrated by an increase in the intensity of a protein of apparent molecular weight of approximately 36 000 on a Western blot (Figure 1A) probed with a rabbit antibody that is specific for FR-β but that is not cross-reactive with FR-α.12 Flow cytometric quantitation of the cell-surface–associated FR-β showed an elevation of the receptor by approximately 20-fold in the presence of 10−6mol/L ATRA (Figure 1B). The ATRA-induced increase in FR-β occurred progressively over a 5-day period (Figure 1C). Withdrawal of ATRA at the end of a 5-day treatment resulted in an eventual decline in FR-β (Figure 1D), indicating that the effect of ATRA on FR-β expression is reversible. Treatment with 1 μmol/L ATRA for 5 days did not induce FR-β expression or appreciably alter FR-α expression in a variety of established cell lines, including myeloid leukemia cells, which either did not express FR or expressed only FR-α. The cell lines tested and their FR expression pattern are indicated in Table1.
Dose response and time course of FR-β induction by ATRA in KG-1 cells.
(A) KG-1 cells were grown in the presence of the indicated concentrations of ATRA for 5 days. Crude membranes from the treated cells were probed by Western blot with the use of rabbit antihuman FR-β antibody and alkaline phosphate (AP)–conjugated goat-antirabbit secondary antibody. (B) KG-1 cells were grown in the presence of ATRA as in panel A. On day 5, cells were washed with PBS and stained with anti–FR-β antibody and FITC-conjugated secondary antibody and then examined by flow cytometry. (C) KG-1 cells were treated with 1 μmol/L ATRA for different periods, and cell surface FR-β expression was quantitated by flow cytometry as in panel B. (D) KG-1 cells were grown in the absence (lane 1) or presence (lane 2) of 1 μmol/L ATRA for 5 days, and the cell lysates analyzed by Western blot. Alternatively, the ATRA-treated cells were washed and resuspended in medium without ATRA and grown for a further 7 days before Western blot analysis of the cell lysates (lane 3). The Western blot was probed with rabbit anti–FR-β antibody and AP-conjugated secondary antibody.
Dose response and time course of FR-β induction by ATRA in KG-1 cells.
(A) KG-1 cells were grown in the presence of the indicated concentrations of ATRA for 5 days. Crude membranes from the treated cells were probed by Western blot with the use of rabbit antihuman FR-β antibody and alkaline phosphate (AP)–conjugated goat-antirabbit secondary antibody. (B) KG-1 cells were grown in the presence of ATRA as in panel A. On day 5, cells were washed with PBS and stained with anti–FR-β antibody and FITC-conjugated secondary antibody and then examined by flow cytometry. (C) KG-1 cells were treated with 1 μmol/L ATRA for different periods, and cell surface FR-β expression was quantitated by flow cytometry as in panel B. (D) KG-1 cells were grown in the absence (lane 1) or presence (lane 2) of 1 μmol/L ATRA for 5 days, and the cell lysates analyzed by Western blot. Alternatively, the ATRA-treated cells were washed and resuspended in medium without ATRA and grown for a further 7 days before Western blot analysis of the cell lysates (lane 3). The Western blot was probed with rabbit anti–FR-β antibody and AP-conjugated secondary antibody.
Effect of all-trans retinoic acid on folate receptor expression in various cell lines
| Cell line . | Cell type* . | FR expression† . | ||
|---|---|---|---|---|
| Before treatment . | After treatment . | |||
| FR-α . | FR-β . | |||
| KG-1 | Acute myelogenous leukemia | — | 0.02 ± 0.01 | 0.5 ± 0.1 |
| KG-1a | Acute myelogenous leukemia | — | — | — |
| NB4 | Acute promyelocytic leukemia | — | — | — |
| HL60 | Acute promyelocytic leukemia | — | — | — |
| KCL22 | Myeloblastic leukemia | — | — | — |
| K562 | Erythroleukemia | — | — | — |
| CCFR-CEM | Acute lymphoblastic leukemia | — | — | — |
| L1210 | Mouse lymphoblastic leukemia | — | — | — |
| SVG | Transformed fibroblast | — | — | — |
| MG-63 | Osteosarcoma | — | — | — |
| 293 | Transformed primary embryonal kidney epithelial cell | — | — | — |
| SKOV3 | Ovarian adenocarcinoma | 0.25 ± 0.02 | — | 0.26 ± 0.03 |
| JAR | Placental choriocarcinoma | 1.50 ± 0.20 | — | 1.10 ± 0.10 |
| HeLa | Cervical carcinoma | 1.70 ± 0.20 | — | 1.20 ± 0.20 |
| Caki-1 | Kidney carcinoma | — | — | — |
| Cell line . | Cell type* . | FR expression† . | ||
|---|---|---|---|---|
| Before treatment . | After treatment . | |||
| FR-α . | FR-β . | |||
| KG-1 | Acute myelogenous leukemia | — | 0.02 ± 0.01 | 0.5 ± 0.1 |
| KG-1a | Acute myelogenous leukemia | — | — | — |
| NB4 | Acute promyelocytic leukemia | — | — | — |
| HL60 | Acute promyelocytic leukemia | — | — | — |
| KCL22 | Myeloblastic leukemia | — | — | — |
| K562 | Erythroleukemia | — | — | — |
| CCFR-CEM | Acute lymphoblastic leukemia | — | — | — |
| L1210 | Mouse lymphoblastic leukemia | — | — | — |
| SVG | Transformed fibroblast | — | — | — |
| MG-63 | Osteosarcoma | — | — | — |
| 293 | Transformed primary embryonal kidney epithelial cell | — | — | — |
| SKOV3 | Ovarian adenocarcinoma | 0.25 ± 0.02 | — | 0.26 ± 0.03 |
| JAR | Placental choriocarcinoma | 1.50 ± 0.20 | — | 1.10 ± 0.10 |
| HeLa | Cervical carcinoma | 1.70 ± 0.20 | — | 1.20 ± 0.20 |
| Caki-1 | Kidney carcinoma | — | — | — |
FR indicates folate receptor. FR expression was detected by [3H] folic acid binding, immunoblotting with the use of antibody to FR-α or FR-β, and Northern blot by means of complementary DNA probes specific for FR-α or FR-β.
The cell lines are of human origin unless indicated otherwise.
Expressed as picomoles per 106 cells; a dash indicates that FR expression was undetectable.
PI-PLC sensitivity of ATRA-induced FR-β in KG-1 cells
When KG-1 cells were grown in the presence of ATRA to induce FR-β expression and then treated with PI-PLC, there was a quantitative decrease in the FR-β detected by Western blot in the cell lysate (results not shown). The ability of PI-PLC to release a protein from the cell surface is a diagnostic test for the presence of a GPI membrane anchor in proteins including FR-β. This result further confirms the cell surface localization and identity of the ATRA-induced band on Western blots of cell lysates from KG-1 cells.
Effect of ATRA and other differentiating agents on growth, differentiation, and FR-β expression in KG-1 cells
In contrast to the growth-inhibitory effect of ATRA on HL60 cells (approximately 50% growth inhibition in 5 days), the compound did not produce a significant inhibition in the growth of KG-1 cells (results not shown). Furthermore, in contrast to HL60 cells, ATRA did not cause differentiation of KG-1 cells along the granulocytic lineage as monitored by the NBT reduction assay (Table2). Other differentiating agents, ie, phorbol ester and dexamethasone, also did not induce granulocytic differentiation in KG-1 cells (Table 2). On the other hand, phorbol ester, but not ATRA or dexamethasone, could induce KG-1 cells to differentiate along the monocytic lineage as determined by NAE staining (Table 2). In contrast to ATRA, however, a variety of differentiating agents (ie, PMA, dexamethasone, vit D3, and TGF-β) had no apparent effect on FR-β expression in KG-1 cells (Table 2).
Effects of various differentiation reagents on KG-1 cells
| Treatment . | NBT reduction (times control) . | NAE staining (% positive cells) . | FR-β (relative fluorescence) . |
|---|---|---|---|
| None | 1.00 ± 0 | 6.8 ± 0.34 | 1.7 ± 0.03 |
| ATRA* | 0.87 ± 0.04 | 6.2 ± 0.31 | 21.6 ± 0.05 |
| PMA | 0.93 ± 0.05 | 28.7 ± 1.43 | 0.90 ± 0.05 |
| DEX | 1.10 ± 0.05 | 6.2 ± 0.31 | 1.7 ± 0.08 |
| Vit D3 | — | — | 1.8 ± 0.01 |
| TGF-β | — | — | 1.4 ± 0.01 |
| Treatment . | NBT reduction (times control) . | NAE staining (% positive cells) . | FR-β (relative fluorescence) . |
|---|---|---|---|
| None | 1.00 ± 0 | 6.8 ± 0.34 | 1.7 ± 0.03 |
| ATRA* | 0.87 ± 0.04 | 6.2 ± 0.31 | 21.6 ± 0.05 |
| PMA | 0.93 ± 0.05 | 28.7 ± 1.43 | 0.90 ± 0.05 |
| DEX | 1.10 ± 0.05 | 6.2 ± 0.31 | 1.7 ± 0.08 |
| Vit D3 | — | — | 1.8 ± 0.01 |
| TGF-β | — | — | 1.4 ± 0.01 |
NBT indicates nitroblue tetrazolium; NAE, α-naphthyl acetate esterase; ATRA, all-trans retinoic acid; PMA, phorbol 12-myristate 13-acetate; DEX, dexamethasone; Vit D3, 1,25(OH)2 vitamin D3; and TGF, transforming growth factor. Dash indicates not determined.
HL60 cells were used as a positive control for ATRA-induced differentiation, giving an NBT reduction value of 4.3 ± 0.22 times control.
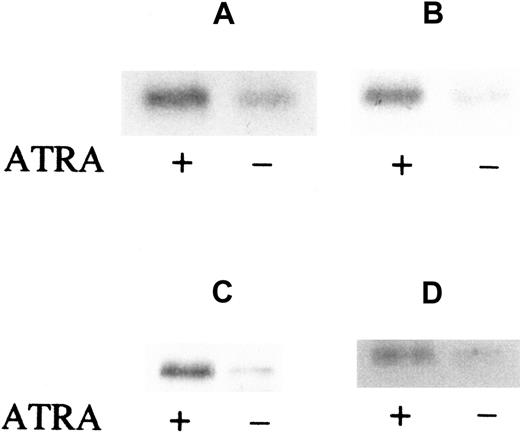
Induction of FR-β by ATRA in myeloid leukemia cells from patient marrow
The effect of ATRA on FR-β expression in leukemic cells in the bone marrow of AML patients was tested under cell-culture conditions in the presence of GM-CSF, IL-3, and cKit ligand to ensure cell viability. Leukemic blasts and mononuclear cells were obtained from the patient bone marrow aspirates by Ficoll density gradient centrifugation and cultured in suspension in either the presence or absence of 10−6 mol/L ATRA. The leukemic cells were derived from 2 patients with French-American-British (FAB)–M2 type (AML with maturation) leukemia and 2 with FAB-M4 type (acute myelomonocytic) leukemia. Subsequent Western blot analysis of the cell lysates showed that ATRA induction of FR-β expression occurred in all of those samples (Figure2). Furthermore, the increase in FR-β expression could be observed as early as 6 hours after the introduction of ATRA (Figure 2A) and could persist up to 5 days of treatment (Figure2D). These results extend the observed ATRA induction of FR-β in the KG-1 cell line to primary cultures of leukemic cells derived from AML patients.
ATRA induction of FR-β in AML blasts from patient marrow.
Cells (1 × 106) were cultured and treated with or without ATRA (1 μmol/L) for 6 hours (panels A and B), 24 hours (panel C), or 5 days (panel D). The samples were obtained from separate patients with FAB-M2 (panels A and C) or FAB-M4 (panels B and D) type AML. Cell lysates were then examined by Western blot with the use of rabbit anti–FR-β and AP-conjugated secondary antibody.
ATRA induction of FR-β in AML blasts from patient marrow.
Cells (1 × 106) were cultured and treated with or without ATRA (1 μmol/L) for 6 hours (panels A and B), 24 hours (panel C), or 5 days (panel D). The samples were obtained from separate patients with FAB-M2 (panels A and C) or FAB-M4 (panels B and D) type AML. Cell lysates were then examined by Western blot with the use of rabbit anti–FR-β and AP-conjugated secondary antibody.
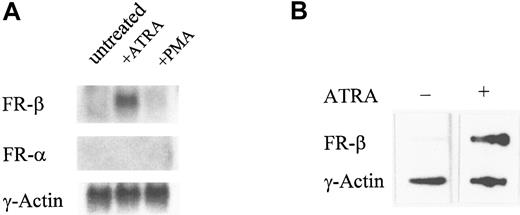
Effect of ATRA on FR-β gene transcription
The induction of FR-β expression by ATRA in KG-1 cells was accompanied by a specific increase in the FR-β, but not FR-α, transcript observed in Northern blots (Figure3A). From nuclear run-on experiments, ATRA increased the transcription rate of the FR-β gene in KG-1 cells (Figure 3B). Under similar conditions, neither the mRNA level nor the transcription rate was altered for a control housekeeping gene, the γ-actin gene (Figure 3). It appears, therefore, that the induction of FR-β by ATRA in KG-1 cells may be accounted for, at least in part, by direct or indirect modulation of the FR-β gene by ATRA.
Effect of ATRA on the expression of the FR-β transcript.
(A) KG-1 cells were treated with ATRA (1 μmol/L), PMA (0.05 μmol/L), or the vehicle alone for 5 days. The total RNA was purified and probed on a Northern blot with cDNA for FR-β, FR-α, or γ-actin. (B) KG-1 cells were treated with ATRA (1 μmol/L) for 6 hours. Nuclei were isolated and nuclear run-on reactions carried out as described in “Materials and methods.” Labeled RNA isolated from each reaction was used to probe immobilized cDNA for FR-β or γ-actin as described in “Materials and methods.”
Effect of ATRA on the expression of the FR-β transcript.
(A) KG-1 cells were treated with ATRA (1 μmol/L), PMA (0.05 μmol/L), or the vehicle alone for 5 days. The total RNA was purified and probed on a Northern blot with cDNA for FR-β, FR-α, or γ-actin. (B) KG-1 cells were treated with ATRA (1 μmol/L) for 6 hours. Nuclei were isolated and nuclear run-on reactions carried out as described in “Materials and methods.” Labeled RNA isolated from each reaction was used to probe immobilized cDNA for FR-β or γ-actin as described in “Materials and methods.”
Modulation of FR-β expression in KG-1 cells by retinoid analogues
Retinoid agonists and antagonists with different specificities for nuclear receptors were tested in KG-1 cells to determine the type(s) of nuclear receptors for retinoids that may mediate ATRA induction of FR-β. The retinoid agonists 9-cis RA (pan-RAR, pan–retinoid X receptors [pan-RXR]), TTNPB (pan-RAR), CD336 (RARα), LG101093 (RARβ and RARγ), and CD2781 (RARγ) induced FR-β expression, albeit to a lesser extent than ATRA (pan-RAR), whereas CD417 (RARβ), CD2314 (RARβ), and LG100364 (pan-RXR) had no detectable effect on the level of FR-β (Figure4; Table3). Among the retinoid antagonists tested, LG100629 (RARα), CD2503 (RARα), and CD2665 (RARβ/RARγ) counteracted retinoid-agonist–induced FR-β expression (Figure4; Table 3). These results indicate that in KG-1 cells, up-regulation of FR-β by retinoids may be mediated by the nuclear receptors, RARα, and/or RARγ but not by RARβ or by RXRs.
Effects of various retinoid compounds on FR-β expression in KG-1 cells.
(A) KG-1 cells were treated with ATRA, 9-cis-RA, LG101093, LG100364, or TTNPB at a concentration of 1 μmol/L or with the vehicle alone for 5 days. Lysates from 1 × 106 cells from each sample were probed on Western blots with rabbit anti–FR-β antibody and AP-conjugated secondary antibody. (B) KG-1 cells were treated with different concentrations (10−7 or 10−6 mol/L) of CD2665 and CD2503, either alone or in combination with ATRA (1 × 108 mol/L), for 5 days. The cell lysates from each sample were subjected to Western blot analysis as in panel A. (C) KG-1 cells were treated with CD336 or LG101093 at a concentration of 10−7 mol/L, either alone or in combination with CD2665 or CD2503 (10−6 mol/L). The cell lysates were analyzed as in panel A. Panels A-C show representative Western blots from at least 3 experiments.
Effects of various retinoid compounds on FR-β expression in KG-1 cells.
(A) KG-1 cells were treated with ATRA, 9-cis-RA, LG101093, LG100364, or TTNPB at a concentration of 1 μmol/L or with the vehicle alone for 5 days. Lysates from 1 × 106 cells from each sample were probed on Western blots with rabbit anti–FR-β antibody and AP-conjugated secondary antibody. (B) KG-1 cells were treated with different concentrations (10−7 or 10−6 mol/L) of CD2665 and CD2503, either alone or in combination with ATRA (1 × 108 mol/L), for 5 days. The cell lysates from each sample were subjected to Western blot analysis as in panel A. (C) KG-1 cells were treated with CD336 or LG101093 at a concentration of 10−7 mol/L, either alone or in combination with CD2665 or CD2503 (10−6 mol/L). The cell lysates were analyzed as in panel A. Panels A-C show representative Western blots from at least 3 experiments.
Nuclear receptor isoform specificities of retinoid analogues and their effects on FR-β expression
| Agonist (specificity) . | Antagonist (specificity) . | FR-β expression . |
|---|---|---|
| ATRA (pan-RAR) | — | Increase |
| TTNPB (pan-RAR) | — | Increase |
| 9-cis RA (pan-RXR, pan-RAR) | — | Increase |
| CD336 (RARα) | — | Increase |
| CD417 (RARβ) | — | No change |
| CD2314 (RARβ) | — | No change |
| LG101093 (RAR β and RARγ) | — | Increase |
| CD2781 (RARγ) | — | Increase |
| LG100364 (pan-RXR) | — | No change |
| ATRA (pan-RAR) | LG100629 (RARα) | Decrease |
| ATRA (pan-RAR) | CD2503 (RARα) | Decrease |
| ATRA (pan-RAR) | CD2665 (RARβ, RARγ) | Decrease |
| CD336 (RARα) | LG100629 (RARα) | Decrease |
| CD336 (RARα) | CD2503 (RARα) | Decrease |
| CD336 (RARα) | CD2665 (RARβ, RARγ) | Decrease |
| LG101093 (RARβ and RARγ) | LG100629 (RARα) | Decrease |
| LG101093 (RARβ and RARγ) | CD2503 (RARα) | Decrease |
| LG101093 (RARβ and RARγ) | CD2665 (RARβ, RARγ) | Decrease |
| Agonist (specificity) . | Antagonist (specificity) . | FR-β expression . |
|---|---|---|
| ATRA (pan-RAR) | — | Increase |
| TTNPB (pan-RAR) | — | Increase |
| 9-cis RA (pan-RXR, pan-RAR) | — | Increase |
| CD336 (RARα) | — | Increase |
| CD417 (RARβ) | — | No change |
| CD2314 (RARβ) | — | No change |
| LG101093 (RAR β and RARγ) | — | Increase |
| CD2781 (RARγ) | — | Increase |
| LG100364 (pan-RXR) | — | No change |
| ATRA (pan-RAR) | LG100629 (RARα) | Decrease |
| ATRA (pan-RAR) | CD2503 (RARα) | Decrease |
| ATRA (pan-RAR) | CD2665 (RARβ, RARγ) | Decrease |
| CD336 (RARα) | LG100629 (RARα) | Decrease |
| CD336 (RARα) | CD2503 (RARα) | Decrease |
| CD336 (RARα) | CD2665 (RARβ, RARγ) | Decrease |
| LG101093 (RARβ and RARγ) | LG100629 (RARα) | Decrease |
| LG101093 (RARβ and RARγ) | CD2503 (RARα) | Decrease |
| LG101093 (RARβ and RARγ) | CD2665 (RARβ, RARγ) | Decrease |
Interestingly, the induction of FR-β by the RARα agonist, CD336, was inhibited by the RARβ/γ antagonist, CD2665, and conversely, the RARα antagonists, CD2503 and LG100629; both counteracted the up-regulation of FR-β by the RARγ agonist, LG101093 (Figure 4; Table 3). These results show that even though RARα and RARγ may independently mediate retinoid induction of FR-β, there is an apparent cross-talk between them.
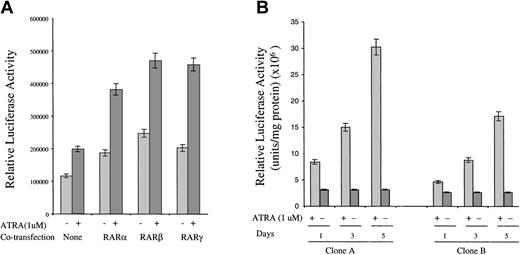
Ability of RAR isoforms to mediate ATRA activation of the FR-β promoter
The inability of RARβ agonists to induce FR-β expression in KG-1 cells noted above could be attributed to the known lack of RARβ in KG-1 cells.24 To test whether RARβ can mediate the effect of ATRA on FR-β expression, as well as to complement the studies with retinoid compounds described in the previous section, it was of interest to test the effect of ATRA on the activity of the FR-β gene promoter and the effect of transiently expressing individual RAR isoforms on the action of ATRA. Owing to the technical difficulty of transfecting KG-1 cells, 293 cells were chosen for these studies. For testing the FR-β promoter activity, a luciferase reporter was attached downstream of a 542–base pair genomic fragment of FR-β (−478nt to +64nt), encompassing the transcription start site at +1nt and the known upstream cis elements required for promoter activity.25 ATRA treatment resulted in an increase in the reporter activity in 293 cells transiently transfected with the promoter construct (Figure 5A). Co-expression of the nuclear receptors, RARα, RARβ, and RARγ, all resulted in an enhancement of the ATRA effect (Figure 5A). Overexpression of the nuclear receptors also resulted in relatively small but significant increases in the FR-β promoter activity, even in the absence of ATRA treatment (Figure 5A); this effect is presumably due to enhancement of the activity of endogenous retinoids in the cells caused by overexpression of RARs. The results suggest that ATRA induction of FR-β may be mediated at least in part by activation of the proximal promoter for FR-β and that the RAR types α, β, and γ are all capable of mediating this effect. Here, it may be noted that in keeping with the narrow tissue specificity of FR-β discussed earlier, the endogenous FR-β gene in 293 cells was not expressed under any of the conditions in Figure 5A.
Effect of ATRA on FR-β promoter–luciferase reporter activity in transfected 293 cells.
(A) The 293 cells were cotransfected with the FR-β promoter linked to a luciferase reporter gene and an expression plasmid for RARα, RARβ, or RARγ, together with a β-galactosidase expression plasmid as described in “Materials and methods.” At 8 hours after transfection, ATRA (1 μmol/L) was added, and the cells were cultured for a further 40 hours. Cells were then harvested and lysed with luciferase reporter buffer, and the luciferase activity was determined as described in “Materials and methods.” The relative luciferase activity was normalized to β-galactosidase activity. (B) Representative clones A and B of recombinant 293 cells stably transfected with the FR-β promoter–luciferase reporter construct (described in “Materials and methods”) were treated with ATRA (1 μmol/L) for up to 5 days in 6-well plates. The cells were harvested and lysed on days 1, 3, and 5 of the treatment, and the luciferase activity in the lysate was determined as described in “Materials and methods.” The relative luciferase activity was normalized to 1 mg of total protein.
Effect of ATRA on FR-β promoter–luciferase reporter activity in transfected 293 cells.
(A) The 293 cells were cotransfected with the FR-β promoter linked to a luciferase reporter gene and an expression plasmid for RARα, RARβ, or RARγ, together with a β-galactosidase expression plasmid as described in “Materials and methods.” At 8 hours after transfection, ATRA (1 μmol/L) was added, and the cells were cultured for a further 40 hours. Cells were then harvested and lysed with luciferase reporter buffer, and the luciferase activity was determined as described in “Materials and methods.” The relative luciferase activity was normalized to β-galactosidase activity. (B) Representative clones A and B of recombinant 293 cells stably transfected with the FR-β promoter–luciferase reporter construct (described in “Materials and methods”) were treated with ATRA (1 μmol/L) for up to 5 days in 6-well plates. The cells were harvested and lysed on days 1, 3, and 5 of the treatment, and the luciferase activity in the lysate was determined as described in “Materials and methods.” The relative luciferase activity was normalized to 1 mg of total protein.
Response of the FR-β promoter to ATRA when stably integrated in the 293-cell genome
Following stable integration of the FR-β promoter–luciferase reporter fragment into the 293-cell genome, the majority of randomly selected recombinant 293 sublines (21 of 25) showed an induction of the reporter luciferase activity upon ATRA treatment. In representative clones (Figure 5B), ATRA treatment produced up to a 10-fold increase in luciferase activity progressively over a 5-day period, mimicking ATRA induction of FR-β in KG-1 cells. This result provides evidence that modulation of the FR-β promoter in the chromosome is a major mechanism of FR-β induction by ATRA.
Discussion
Even though FR-β is expressed in myeloid leukemia and in several sarcomas, the in vitro expression patterns of the receptor do not generally appear to reflect its in vivo tissue specificity, perhaps owing to events associated with the immortalization of the cell lines in culture9,12 (also, unpublished data, 2000). Among the available myeloid leukemia cell lines tested, KG-1 cells alone expressed FR-β. The KG-1 cell line is an AML cell line in which myeloblasts and promyelocytes constitute the predominant population.26
KG-1 cells cannot be induced to produce terminal differentiation by ATRA,27,28 but do differentiate into macrophages in response to phorbol ester.29 Treatment of KG-1 cells with ATRA for a 5-day period resulted in a progressive and dramatic increase in FR-β expression; optimal induction of FR-β was observed at about 1 μmol/L ATRA, and the effect was reversed upon withdrawal of the reagent. The optimal conditions for FR-β induction by ATRA thus occurred under conditions in which myeloid leukemia cells that are therapeutically responsive to ATRA would show growth inhibition and terminal differentiation. The identity of the ATRA-induced protein as cell-surface–associated FR-β was established from its reactivity with an antibody specific for this FR isoform,12 its apparent molecular weight of approximately 36 000, and its release by treatment of intact cells with PI-PLC. In contrast to the control HL60 cells, the induction of FR-β in KG-1 cells was not associated with significant inhibition of the cell growth in suspension culture and terminal differentiation into either granulocytes or monocytes; the KG-1 cells, however, retained their ability to differentiate into monocytes in response to treatment with phorbol ester. The induction of FR-β in KG-1 cells was specific for ATRA since other known differentiating agents (ie, PMA, dexamethasone, vit D3 and TGF-β) did not significantly alter FR-β expression. The observation of ATRA induction of FR-β expression in KG-1 cells was extended to leukemic blasts obtained from AML patient bone marrow aspirates. Of the 4 AML samples tested, 2 were characterized as M2 type AML and 2 as M4 type AML, according to the FAB classification; it may be noted that neither subclass of AML is known to respond favorably to ATRA differentiation therapy. Since FR-β is a terminal differentiation marker for neutrophils in normal hematopoiesis, its expression in response to ATRA treatment of leukemic cells in the absence of terminal differentiation may be viewed as a partial differentiation response. Indeed, in the majority of AML cells from different FAB subclasses, ATRA treatment in short-term primary cultures has been reported to alter the expression patterns of myeloid differentiation antigens such as CD33, CD14, and p124 in a manner consistent with changes during normal myelopoiesis.30
The increase in the expression of FR-β upon ATRA treatment in KG-1 cells was accompanied by a parallel increase in the FR-β mRNA. Furthermore, nuclear run-on data showed that the elevated mRNA level was at least in part caused by an increased rate of transcription of the FR-β gene. Retinoid responsive genes that are transcriptionally regulated are known to contain consensus retinoic acid response elements (RAREs) within their promoter sequences.31,32Inspection of the FR-β gene sequence did not reveal consensus RARE sequences, suggesting that activation of the FR-β promoter by ATRA may occur by a trans effect. The effects of retinoid agonists and antagonists with different nuclear receptor specificities on FR-β expression in KG-1 cells showed that RARα and RARγ, but not RXRs, could mediate FR-β induction. The inability of RARβ-specific retinoids to influence FR-β expression may be due to the apparent lack of RARβ in KG-1 cells.24 This conclusion is supported by the results in transiently transfected 293 cells in which RARs α, β, and γ were all able to mediate activation of the FR-β promoter in response to ATRA. Even though the endogenous FR-β gene in 293 cells is inactive, in stable recombinant 293-cell sublines expressing a reporter luciferase under the influence of the FR-β promoter, ATRA strikingly stimulated the reporter expression, demonstrating transcriptional control of the FR-β gene by ATRA in the chromosomal context. Thus far, our results using established cell lines of diverse cell type and leukemic blasts indicate that up-regulation of the endogenous FR-β gene by ATRA does not occur in cells in which the gene is not expressed, presumably because ATRA cannot overcome the repressive mechanisms that ensure tissue specificity of FR-β. FR-α was down-regulated by ATRA in a squamous cell carcinoma cell line33 and up-regulated in mouse embryonic stem cells,34 but FR-β was not expressed in those cells.
The effect of ATRA in promoting differentiation is mediated primarily by the nuclear receptor isoform RARα, which is also linked to the molecular mechanism of leukemogenesis in APL.19,35 36 In KG-1 cells, there was apparent cross-talk between retinoid agonists and antagonists with different RAR isoform specificities with respect to FR-β regulation, suggesting that a common downstream target for the receptors mediates trans activation of the FR-β gene. The pathway for FR-β induction by ATRA is therefore clearly distinct from that leading to terminal differentiation, which is mediated principally by RARα.
The refractoriness of ATRA-resistant APL cells, as well as other malignant cell types, to retinoid differentiation therapy has been attributed to various resistance mechanisms. Reduced intracellular availability of retinoids due to either accelerated in vivo clearance37 or increased levels of cellular retinoic acid binding protein38-40 may occur in the resistant cells. More recent studies have implicated alterations in the level of RARα or PML/RARα41-43 and mutations in PML/RARα44-48 as a major cause of in vitro as well as clinical resistance to ATRA. As discussed above, retinoids may use any of the 3 RAR isoforms to induce FR-β expression, and therefore retinoid regulation of FR-β would be unaffected by differences in RARα levels or by mutations that decrease the affinity of ATRA for RARα.
Recent studies with variant APL-derived cell lines, as well as primary cultures of non-APL myeloid leukemia cells, have characterized the cell growth inhibition, terminal differentiation, and apoptosis resulting from ATRA treatment of APL as independent events.49-51 The therapeutic action of ATRA in APL is, therefore, the net result of its pleiotropic actions in the cell. It follows that a block in any one critical pathway of ATRA action leading to ATRA resistance should not affect its ability to modulate a variety of genes. The studies described in this article would predict that ATRA could up-regulate FR-β in various retinoid-resistant myeloid leukemias as well as in APL cells harboring minimal residual disease following ATRA therapy. The elevated FR-β may serve as an effective target for therapy following retinoid treatment.
Acknowledgments
We thank Dr Elizabeth Allegreto (Ligand Pharmaceuticals Inc, San Diego, CA) and Dr Uwe Reichert (Galderma Research and Development, Valbonne, France) for sharing various retinoid compounds; Dr Pierre Chambon (Institute de Genetique et de Biologie Moleculaire et Cellulaire, Cude Strasbourg, France) for providing the RAR cDNAs; Jenny Zak and Anna Chlebowski for typing the manuscript; and Thomas Sawyer for his invaluable help in the use of the flow cytometer.
Supported by National Institutes of Health R01 grants CA80183 and CA70873 to M.R.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Manohar Ratnam, Medical College of Ohio, Department of Biochemistry and Molecular Biology, 3035 Arlington Ave, Toledo, OH 43614-5804; e-mail: mratnam@mco.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal