Abstract
Investigation of the molecular basis of a severe factor VII (fVII) deficiency revealed compound heterozygosity in the fVII gene. On the paternal allele the patient had 3 structural gene abnormalities frequently associated with fVII deficiency. A new mutation, a C to T transition at position −55 relative to the translational start site, was found on the maternal allele. The study demonstrates that this mutation partially impeded binding of the transcriptional activator, hepatic nuclear factor 4, to the fVII promoter while greatly reducing reporter gene expression in hepatic cells.
Introduction
We have recently characterized 2 independent point mutations within the 5′ regulatory region of the factor VII (fVII) gene in patients with severe fVII deficiency. One mutation, a T to G substitution at position −61, completely eliminates interaction with the orphan nuclear receptor, hepatic nuclear factor 4 (HNF4).1,2 The second mutation, a C to G transversion at position −94, prevents binding and transactivation by Sp1 and other nuclear proteins.3 These regulatory regions are particularly important for expression of the fVII gene.4-6In this report, we describe a new mutation in the fVII promoter that reduced HNF4 binding but nevertheless contributed to severe fVII deficiency.
Study design
Genetic analyses
Informed consent for this study, which was approved by the Human Studies Committee of the Brockton-West Roxbury Department of Veterans Affairs Medical Center, was obtained from both the patient and his mother. Plasma levels of fVII coagulant activity (VII:C) and fVII antigen (VII:Ag) are expressed as a percentage of normal levels.7 Leukocyte genomic DNA8 was used for polymerase chain reaction (PCR) of the fVII 5′ flanking region and exons with surrounding splice sites.1 Sequence analysis was done on a 373A DNA Sequencer (Applied Biosystems, Foster City, CA).9 PCR fragments spanning nucleotides −185 to +1 were subcloned into a promoterless reporter plasmid containing the human growth hormone (hGH) structural gene (Nichols Diagnostics Institute, San Juan Capistrano, CA).1 6
Analysis of promoter mutation
HepG2 cells (ATCC HB 8065) were cultured and transfected with 3 μg of reporter plasmid and 1.5 μg of pRSV-β-galactosidase plasmid (Promega Corp, Madison, WI). A total of 48 hours post-transfection, media were assayed with the hGH assay kit (Nichols Diagnostics Institute) and lysates with a colorimetric β-galactosidase assay kit (Promega).1 For cotransfection, 6 μg of pCDNAI-HNF4, encoding rat α-HNF4, or inert pUC-19 were also present. In vitro transcribed/translated HNF4 was prepared in TNT-wheat germ system (Promega) and used in electrophoretic mobility shift assays with [γ-32P]-labeled oligonucleotides.10 Factor VII oligonucleotides spanned residues −77 to −47 and had either wild-type (WT) sequence, C to T at position −55 (MT55), or T to G at position −61 (MT61). A control oligonucleotide (residues −92 to −67 of human apolipoprotein [Apo] CIIIB)11 was also used. In supershift assays, polyclonal anti-HNF4 antibody was added to binding reactions. Reactions were run on 5% (wt/vol) polyacrylamide gels in Tris-borate buffer and analyzed by autoradiography.
Results and discussion
The patient is a 25-year-old Polish male with a severe bleeding disorder manifested by spontaneous deep muscle hematomas, hemarthroses, ecchymoses, and epistaxis. He has required transfusions of fresh frozen plasma prophylactically and to control severe bleeding episodes. His plasma levels of VII:Ag and VII:C were less than 2% of normal. Sequence analysis of the coding regions and intron/exon boundaries of his fVII gene identified several structural gene mutations on the paternal allele: Ala294Val, Arg353Gln, and a frameshift mutation in codon 404. These mutations are frequently linked in Polish patients with fVII deficiency.7 The fVII protein translated from the paternal allele is expected to be poorly secreted and functionally abnormal.7,12-14 Heterozygosity for a neutral dimorphism in codon His115 15 was also noted. Examination of 404 base pairs of the 5′ flanking region indicated heterozygosity for base substitutions at positions −55 (C to T) and −402 (G to A). Inheritance of these alterations was confirmed by genotyping the patient's mother, who exhibited modest reductions in plasma levels of VII:Ag (38%) and VII:C (52%). The base substitution at position −402, recently reported as a polymorphism associated with slight increases in the plasma levels of VII:Ag and VII:C,16seems unlikely to influence fVII expression, given the severe phenotype of the patient.
Position −55 is in close proximity to the fVII transcriptional start site and lies within a region essential for hepatic expression.4-6 Therefore, fragments of the 5′ flanking region extending from position −185 to position +1 (the translational start site) were prepared by PCR of the patient's genomic DNA and inserted into the promoterless reporter plasmid. The WT and MT55 plasmids were then used to direct expression of hGH in transiently transfected HepG2 cells, a human hepatoma cell line producing fVII and other coagulation proteins.17 Experiments in which 12 dishes were transfected with each reporter plasmid indicated that MT55 plasmid exhibited 9.7% ± 5.4% (1 SD) of the activity observed with WT plasmid.
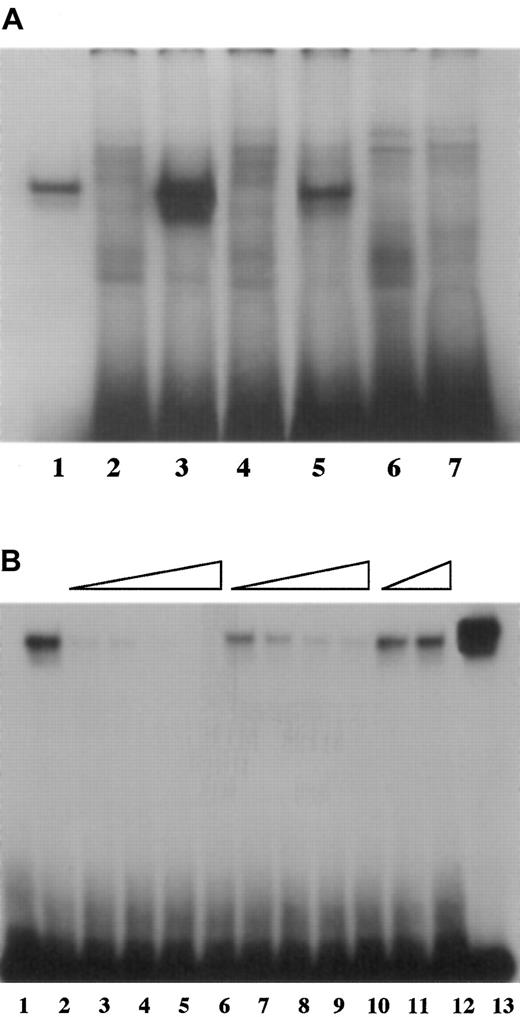
The deleterious effect of the −55 mutation on promoter activity appeared to be due to decreased interaction with HNF4, as demonstrated by electrophoretic mobility shift assays. Direct binding of fVII oligonucleotides to HNF4 protein made by in vitro transcription/translation was strongest with WT oligonucleotide, intermediate with MT55 oligonucleotide and, as expected from prior analysis,1 undetectable with MT61 oligonucleotide (Figure1A). The relative capacities of fVII oligonucleotides to compete with WT oligonucleotide for HNF4 mirrored their abilities to bind HNF4 (Figure 1B). Thus, the WT oligonucleotide served as the strongest competitor, with the MT55 oligonucleotide a weaker competitor, and the MT61 oligonucleotide competed poorly, if at all.
Binding of fVII oligonucleotides.
(A) The −55 C to T mutation diminished binding of HNF4 to fVII promoter sequence. A radiolabeled WT oligonucleotide encompassing the HNF4 binding site (−77 to −47 base pairs prior to the translation start site) in the human fVII promoter region showed binding to HNF4 protein prepared by in vitro transcription/translation (lane 3), and corresponding oligonucleotides having the −55 mutation (MT55, lane 5) or the −61 mutation (MT61, lane 7) showed weaker and undetectable binding, respectively. All 3 oligonucleotides bound nonspecifically to components of the mock transcription/translation reaction mixture (lanes 2, 4, 6), which migrated to various different positions on the gel. The binding reaction with a control oligonucleotide (the HNF4 binding site from the ApoCIIIB promoter) is shown in lane 1. The amounts of WT, MT55, and MT61 oligonucleotides used per lane were identical, but less control oligonucleotide was used to compensate for its comparatively strong binding to HNF4. (B) Competition assays show specificity of binding to HNF4. The radiolabeled WT oligonucleotide bound to HNF4 prepared by in vitro transcription/translation (lane 2), and binding was subject to competition by inclusion of increasing concentrations of unlabeled WT oligonucleotide (5 ×, 10 ×, 50 ×, and 100 × relative to the concentration of the labeled oligonucleotide, lanes 3-6 successively). There was also competition, though less effective, by unlabeled MT55 oligonucleotide (10 ×, 50 ×, 100 ×, and 200 ×, lanes 7-10 successively) but not at all by unlabeled MT61 oligonucleotide (100 × and 200 ×, lanes 11 and 12). Binding of the control ApoCIIIB-labeled oligonucleotide to the HNF4 protein is shown in lane 13, and binding between the WT fVII probe and components of the mock transcription/translation reaction mixture is shown in lane 1.
Binding of fVII oligonucleotides.
(A) The −55 C to T mutation diminished binding of HNF4 to fVII promoter sequence. A radiolabeled WT oligonucleotide encompassing the HNF4 binding site (−77 to −47 base pairs prior to the translation start site) in the human fVII promoter region showed binding to HNF4 protein prepared by in vitro transcription/translation (lane 3), and corresponding oligonucleotides having the −55 mutation (MT55, lane 5) or the −61 mutation (MT61, lane 7) showed weaker and undetectable binding, respectively. All 3 oligonucleotides bound nonspecifically to components of the mock transcription/translation reaction mixture (lanes 2, 4, 6), which migrated to various different positions on the gel. The binding reaction with a control oligonucleotide (the HNF4 binding site from the ApoCIIIB promoter) is shown in lane 1. The amounts of WT, MT55, and MT61 oligonucleotides used per lane were identical, but less control oligonucleotide was used to compensate for its comparatively strong binding to HNF4. (B) Competition assays show specificity of binding to HNF4. The radiolabeled WT oligonucleotide bound to HNF4 prepared by in vitro transcription/translation (lane 2), and binding was subject to competition by inclusion of increasing concentrations of unlabeled WT oligonucleotide (5 ×, 10 ×, 50 ×, and 100 × relative to the concentration of the labeled oligonucleotide, lanes 3-6 successively). There was also competition, though less effective, by unlabeled MT55 oligonucleotide (10 ×, 50 ×, 100 ×, and 200 ×, lanes 7-10 successively) but not at all by unlabeled MT61 oligonucleotide (100 × and 200 ×, lanes 11 and 12). Binding of the control ApoCIIIB-labeled oligonucleotide to the HNF4 protein is shown in lane 13, and binding between the WT fVII probe and components of the mock transcription/translation reaction mixture is shown in lane 1.
Binding and supershifted complexes of identical electrophoretic mobilities were formed between HepG2 nuclear extract, or HNF4 protein made by in vitro transcription/translation, and the ApoCIIIB, WT, and MT55 oligonucleotides (data not shown). The MT55 oligonucleotide interacted with HNF4 present in the nuclear extracts more weakly than did the WT oligonucleotide.
The HNF4 binding ability of these oligonucleotides reflected their relative capacity to be transactivated by this transcription factor. Compared with the WT promoter fragment, reporter plasmids containing MT55 and MT61 caused similar reductions in promoter activity as determined by transient transfection assays (9.7% and 6.7% 1 of expression observed with WT reporter plasmid, respectively). Without cotransfected HNF4, MT55 plasmid exhibited negligible expression relative to WT plasmid. However, inclusion of an HNF4 expression vector elevated hGH expression from both reporter plasmids, although to a significantly lesser extent with the MT55 plasmid than with the WT plasmid (Figure2). This is in contrast to the MT61 plasmid, for which we previously observed no transactivation by an HNF4 expression vector.1
Cotransfection of an HNF4 expression vector alters expression of reporter gene from vectors with WT and MT55 fVII promoter sequence.
A total of 3 μg of WT or MT55 reporter vectors were transiently transfected into HepG2 cells, with or without 6 μg of the pCDNAI-HNF4 expression vector or pUC-19 plasmid DNA, as noted. The growth hormone values were corrected for expression from the promoterless pOGH vector under parallel conditions, and for transfection efficiency, then normalized to expression from the WT vector in the absence of coexpressed HNF4. The results shown (3.97% ± 2.87% [1 SD] for MT55 plasmid versus 100% ± 13.1% [1 SD] for WT plasmid, in the absence of pCDNAI-HNF4; 187% ± 99% [1 SD] for MT55 plasmid versus 610% ± 185% [1 SD] for WT plasmid, in the presence of pCDNAI-HNF4) were the averages from 3 experiments. The total number of replicates in each group is shown.
Cotransfection of an HNF4 expression vector alters expression of reporter gene from vectors with WT and MT55 fVII promoter sequence.
A total of 3 μg of WT or MT55 reporter vectors were transiently transfected into HepG2 cells, with or without 6 μg of the pCDNAI-HNF4 expression vector or pUC-19 plasmid DNA, as noted. The growth hormone values were corrected for expression from the promoterless pOGH vector under parallel conditions, and for transfection efficiency, then normalized to expression from the WT vector in the absence of coexpressed HNF4. The results shown (3.97% ± 2.87% [1 SD] for MT55 plasmid versus 100% ± 13.1% [1 SD] for WT plasmid, in the absence of pCDNAI-HNF4; 187% ± 99% [1 SD] for MT55 plasmid versus 610% ± 185% [1 SD] for WT plasmid, in the presence of pCDNAI-HNF4) were the averages from 3 experiments. The total number of replicates in each group is shown.
Even though cotransfected HNF4 interacted functionally with the mutant promoter fragment in vitro, native HNF4 appears unable to affect expression from the mutant allele in vivo. This may be due to a limited amount of HNF4 or more avid binding between available HNF4 and other promoter elements. In human liver, the fVII promoter is rather weak compared with other promoters whose expression is HNF4-dependent, and a poor interaction of this transcription factor with the WT fVII promoter was suggested as the cause.5 The C to T mutation at position −55 would reduce ability of HNF4 to bind the fVII promoter still further, thereby resulting in a negligible in vivo interaction.
Acknowledgments
We thank Dr Margarita Hadzopoulou-Cladaras of Boston University School of Medicine, Boston, MA, both for generously providing pCDNAI-HNF4 and the human ApoCIII HNF4-binding oligonucleotide and for helpful advice during this study. We also appreciate the gift of polyclonal antibody directed against an epitope in the C-terminus of rodent and human HNF4, given by Dr Frances Sladek of the University of California, Riverside, CA.
Supported by the Medical Research Service of the Department of Veterans Affairs (K.A.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kenneth A. Bauer, VA Boston Healthcare System, 1400 VFW Parkway, West Roxbury, MA 02132.

![Fig. 2. Cotransfection of an HNF4 expression vector alters expression of reporter gene from vectors with WT and MT55 fVII promoter sequence. / A total of 3 μg of WT or MT55 reporter vectors were transiently transfected into HepG2 cells, with or without 6 μg of the pCDNAI-HNF4 expression vector or pUC-19 plasmid DNA, as noted. The growth hormone values were corrected for expression from the promoterless pOGH vector under parallel conditions, and for transfection efficiency, then normalized to expression from the WT vector in the absence of coexpressed HNF4. The results shown (3.97% ± 2.87% [1 SD] for MT55 plasmid versus 100% ± 13.1% [1 SD] for WT plasmid, in the absence of pCDNAI-HNF4; 187% ± 99% [1 SD] for MT55 plasmid versus 610% ± 185% [1 SD] for WT plasmid, in the presence of pCDNAI-HNF4) were the averages from 3 experiments. The total number of replicates in each group is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/13/10.1182_blood.v96.13.4370/5/m_h82400460002.jpeg?Expires=1769336043&Signature=BTBioSnvLTgOwvsyrOZ~7hK~duAxvc7umhTCaF4mSOeGfK53K2IP8tlCGauk~eCirHcIgV22R39oU7KCmYouWw6bQlKsp3r~PGjDjpgGOlJaYZMbBdRffLpprzOvLPpq4Lxg90GcFjSb~~2SfWlejp7hCwXRPLquCoRw1TeYGoCVFJO7tT2wl3a9XZNjM6mcoIcuUQM13Bqi1oMCzi1~w6H2b8WwU0~ZwrYyvrWFEwhxfs2YGX9NhCJoibTQDZVQ~45rrwEb5JXHNk3d~8ihxl-JFKBkWeEarGZoiQ0~17IbjctO9gRKpzkoPAvAAh6oXLad-Ajs0sVlbXGnsh3NwQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal